Growth inhibition induced by short hairpin RNA to silence survivin gene in human pancreatic cancer cells
2010-12-14YongMeiShenXiaoChunYangMiaoLiSongChenHaoQinChenYangandYiHuiSun
Yong-Mei Shen, Xiao-Chun Yang, Miao-Li Song, Chen-Hao Qin, Chen Yang and Yi-Hui Sun
Suzhou, China
Growth inhibition induced by short hairpin RNA to silence survivin gene in human pancreatic cancer cells
Yong-Mei Shen, Xiao-Chun Yang, Miao-Li Song, Chen-Hao Qin, Chen Yang and Yi-Hui Sun
Suzhou, China
(Hepatobiliary Pancreat Dis Int 2010; 9: 69-77)
pancreatic neoplasms;short hairpin RNA;survivin;pGenesil-1 vector
Introduction
Pancreatic cancer is a leading cause of cancerrelated death, and is characterized by rapid disease progression, a highly invasive tumor phenotype,and resistance to chemotherapy. Despite signi fi cant advances in the diagnosis and surgical management of the disease, the 5-year survival rate remains below 5%and the median survival time after diagnosis is only 4-6 months.[1-3]Thus, the search for novel treatment strategies to improve the prognosis is an urgent task.
Survivin is a member of the inhibitors of the apoptosis protein family, which inhibits apoptosis and regulates cell division. At present, the overexpressionof survivin has been reported in various human malignancies including pancreatic cancer, but not in most normal adult tissues, which has made survivin a promising gene target for anticancer therapies.[4,5]Studies have demonstrated that the inhibition of survivin expression by antisense oligonucleotides,[6,7]dominant negative mutation,[8]or ribozymes[9-11]reduces the growth of pancreatic cancer cells. However, their ef fi ciency is far from satisfactory because of nuclease degradation.
Small interfering RNA (siRNA) involves posttranscriptional gene silencing via a process in which double-stranded RNA (dsRNA) inhibits gene expression in a sequence-dependent manner through degradation of the corresponding mRNA. It has been veri fi ed as a powerful tool to knock down the expression of a target gene in mammalian cells.[12-14]Recent reports have indicated that siRNA has advantages over other posttranscriptional gene silencing techniques, such as antisense oligonucleotides and gene knockout technology in vitro and in vivo.[15-17]It is easier to deliver, requires only small doses of siRNA to produce its silencing effect,resists nuclease degradation, and can inactivate a gene at almost any stage in development.[18]In this study,siRNA targeted to the survivin gene was introduced into pancreatic cancer cells and its effect on cancer cell growth was investigated.
MethodsCell culture
The human pancreatic cancer cell line Patu8988 used in this study was maintained in our laboratory. The cells were grown in a suspension of RPMI 1640 medium(Gibco, USA) supplemented with 10% heat-inactivated fetal bovine serum (Sijiqing, Hangzhou, China), 5 mmol/L HEPES, 100 U/ml penicillin, and 100 U/ml streptomycin in 5% atmospheric CO2at 37 ℃. The cells were passaged every three days, checked routinely, and found to be free of contamination. When the cells grew to 75% con fl uence, they were digested and used for in vitro and in vivo studies.
shRNA design and construction of survivin-shRNA expression vector
According to the short hairpin RNA (shRNA)design principle,[19]two 19bp sequences in survivin cDNA from GenBank accession number NM-001168 as our target sites were designed by using siRNA design software downloaded from the internet (http://www.ambion.com). One was 5'-GGA CCA CCG CAT CTC TAC A-3' which corresponds to nucleotides 166-184,and the other was 5'-GCA TTC GTC CGG TTG CGC T-3', which corresponds to the nucleotides 358-376. The secreted sequences were submitted to BLAST search to assure that only the secreted gene was targeted. The negative control scrambled sequence 5'-GAC TTC ATA AGG CGC ATG C-3', which has no signi fi cant homology to mouse or human gene sequences, was used to eliminate nonspeci fi c effects. The oligonucleotides contained a sense strand of 19 nucleotides followed by the loop sequence TTC AAG AGA, the antisense strand,transcription terminator TTT TT, the identi fi cation restriction enzyme EcoRⅠ/SacⅠ/SalⅠ site, as well as terminal BamHⅠ and Hind Ⅲ restriction enzyme sites(Table). Double complementary shRNA DNA segments were gained through annealing, named survivin-1-shRNA, survivin-2-shRNA, and CN-shRNA, and then inserted into the BamHⅠ and Hind Ⅲ sites of plasmid pGenesil-1 vector. The recombinant plasmids were evaluated by restriction enzyme cutting and sequencing,and then were designated as pGenesil-1-survivin-1,pGenesil-1-survivin-2, and pGenesil-1-NC. In order to construct a double interfering RNA vector, pGenesil-1-survivin-2 plasmid was digested with restriction enzymes SacⅠ and SalⅠ. Small bands about 400bp containing U6 Promotor+survivin-2-shRNA were purifi ed by agarose gel extraction, and ligated overnight with SacⅠ/SalⅠ-linearized pGenesil-1-survivin-1 vector(puri fi ed by agarose gel extraction) in a total volume of 10 μl with 3 units of T4 DNA ligase (Promega, USA) at 22 ℃. Following the ligation, 5 μl of recombinant DNA was transfected into competent DH5a cells which were then seeded on solid LB medium containing kanamycinand cultured overnight at 37 ℃. Monoclonal colonies were picked out and seeded in 3 ml LB culture fl uid containing 0.05 g/L kanamycin and cultured overnight at 37 ℃ in a rocking bed. The recombinant plasmids were con fi rmed by digestion with restriction enzymes and DNA sequence, and then were designated as pGenesil-1-survivin-1+2.

Table. DNA sequences of insertion fragments for shRNA
Transfection with the shRNA expression vector
For transfection, 2×105cells were seeded into wells of a six-well plate and divided into fi ve groups: group 1,normal cultured Patu8988 cells; group 2, Patu8988 cells transfected with pGenesil-1-survivin-1; group 3, Patu8988 cells transfected with pGenesil-1-survivin-2; group 4,Patu8988 cells transfected with pGenesil-1-survivin-1+2;and group 5, Patu8988 cells transfected with negative control plasmid vector pGenesil-1-NC. When the cells were 60%-70% con fl uent, the common complete medium was replaced by antibody-free medium containing serum and transfection was carried out according to the manufacturer's protocol. Survivin plasmid (5 μg)was diluted with 100 μlOPTiMEM (Invitrogen, USA)or 10 μl lipofectamineTM2000 (Invitrogen, USA) with 100 μlOPTiMEM. After 5 minutes, the dilutions were mixed together and incubated at room temperature for 20 minutes, then dispensed into each well. Four hours after the transfection, the medium was replaced by the common complete medium again. Transiently transfected cells were harvested at 48 hours after transfection and analyzed by inverse fl uorescence microscopy, RT-PCR,Western blotting, and fl ow cytometry. All transfection experiments were conducted in triplicate. Cells stably expressing shRNA were established by selection with medium fi rst containing 600 μg/ml G418. The medium was renewed every 3 days. After 15-20 days selection, the resistant colonies were further selected by a huge dose of G418 (1800 μg/ml) for one week in order to exclude the possibility of non-transfected but G418-resistant colonies.And then the resistant colonies were ampli fi ed for cell proliferation assay and subcutaneous tumorigenesis assay in nude mice.
Survivin mRNA expression detected by semi-quantitative RT-PCR
Transfected cells, as well as untreated cells, were collected and washed twice with PBS. Total cellular RNA was extracted using Trizol reagent (Invitrogen, USA)according to the manufacturer's instructions. The purity and concentration were determined by measuring the absorbance (A) at 260 nm and 280 nm (A260/A280). To generate fi rst-strand cDNA, an oligo(dT)18 was used as primer, and 2 μg RNA was reverse-transcribed using the MMLV First Strand cDNA Synthesis Kit (Fermentas,USA) protocols. And then aliquots of 5 μl cDNA were ampli fi ed in a total volume of 25 μl using the polymerase chain reaction (PCR) kit (Fermentas, USA).The sense and antisense primers for β-actin, which served as an internal control, were 5'-GTG GAC ATC CGC AAA GAC-3' and 5'-AAA GGG TGT AAC GCA ACT AA-3' (302bp). The sense and antisense primers for survivin were 5'-CAG ATT TGA ATC GCG GGA CCC-3'and 5'-CCA AGT CTG GCT CGT TCT CAG-3' (208bp).The cycling conditions were 94 ℃ for 5 minutes,followed by 30 cycles at 94 ℃ for 45 seconds, at 51 ℃ for 45 seconds and at 72 ℃ for 1 minute, and then a fi nal extension at 72 ℃ for 10 minutes. PCR products were separated on 1.5% agarose gels, and viewed by ethidium bromide staining. Densitometric scanning of the bands was performed and the relative amount of survivin gene mRNA expression was estimated by normalization to the β-actin mRNA detected in the same sample.
Survivin protein expression detected by fl ow cytometry
Transfected cells, as well as untreated cells, were collected and washed twice with PBS, and then fi xed in 1% paraformaldehyde for 5 minutes and permeabilized with 0.1% Triton X-100 for 5 minutes. After two additional rinses with PBS, the cells were incubated with primary survivin mAb for 30 minutes, followed by PE-conjugated goat anti-mouse IgG for 30 minutes at room temperature in the dark. The cells were then rinsed twice with PBS containing 2% FBS and analyzed by fl ow cytometry. Controls consisted of incubation with no primary antibody or incubation but with only the secondary antibody. The experiments were performed in triplicate and the results were expressed as mean±SD.
Western blotting analysis
Cytoplasmic proteins of the groups above were extracted using cytoplasmic extraction reagents(Beyotime Biotechnology, China). Cell lysates were centrifuged at 15 000 rpm for 5 minutes at 4 ℃. Protein content in the supernatants was determined by a BCA protein assay kit. Equal amounts of lysate protein were separated by 12% SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked in 5% skimmed milk in TBST buffer at room temperature for 1-2 hours with gentle shaking and then incubated overnight at 4 ℃ with mouse anti-human survivin IgG mAb (1∶2000, Santa Cruz, USA) or mouse anti-human β-actin IgG mAb (1∶1000, Santa Cruz,USA). After washing with TBST, the membranes were incubated with HRP-conjugated goat anti-mouse IgG antibody (1∶5000). Bands were developed using ECL substrate and exposed to X-ray fi lm. The expression levels of survivin and β-actin protein were quanti fi ed by densitometry. The signal strength of each survivin signal was normalized against the corresponding β-actin control.
Cell proliferation assay
Patu8988 cells stably transfected with pGenesil-1-survivin-1, pGenesil-1-survivin-2, pGenesil-1-survivin-1+2,pGenesil-1-NC, and untreated cells were plated in 96-well plates at a density of 2×103cells/well and incubated for 1-7 days. Each group contained three wells.At the end of incubation, 20 μl of 5 g/L MTT (Sigma,USA) in PBS was added into each well, followed by incubation for 4 hours at 37 ℃. Formazan crystals were dissolved in DMSO for 15 minutes at 37 ℃. Absorbance was determined with an enzyme-linked immunosorbent assay reader at 570 nm. The cell proliferation curves were drawn according to the absorbance.
Treatment in vivo
To investigate whether survivin shRNA would alter the tumorigenicity, an equal number (3×107cells in 0.2 ml PBS) of Patu8988 cells stably transfected with pGenesil-1-survivin-1, pGenesil-1-survivin-2, pGenesil-1-survivin-1+2, pGenesil-1-NC, and untreated cells were harvested, washed with PBS and injected subcutaneously into 4-week-old male BALB/c nude mice (six mice for each group). The mice were kept in a pathogen-free environment. Tumor sizes were measured every four days and calculated by the formula: volume (mm3)=1/2(width)2×length. Thirty-two days after injection, the mice were sacri fi ced and the weights of the tumors were recorded.
Statistical analysis
All experiments were performed in triplicate and data were expressed as mean±SD. Statistical analyses were conducted with Student's t test and performed with SPSS 10.0 software. P<0.05 was considered statistically signi fi cant.
ResultsIdenti fi cation of survivin-shRNA expression vector
Because the restriction site of enzyme PstⅠ in plasmid pGenesil-1 (Fig. 1) was replaced by the inserted DNA sequence, the reconstructed plasmids could not be digested by PstⅠ. In the inserted target gene template DNA of survivin-1-ShRNA, survivin-2-ShRNA, and NC-shRNA, an EcoRⅠ/SacⅠ/SalⅠ enzyme digest site was designed between Bam HⅠ and Hind Ⅲ. Meanwhile,the pGenesil-1 plasmid itself carried EcoRⅠ and SalⅠdigest sites. If the insertion was correct, a band about 400bp should be cut off by EcoRⅠ or SalⅠ, and a linear band about 5000bp should appear with SacⅠ digestion.The pGenesil-1-survivin-1, pGenesil-1-survivin-2, and pGenesil-1-NC plasmids were digested by the restriction enzymes PstⅠ and EcolⅠ/SacⅠ/SalⅠ. Agarose gel (1.0%)electrophoresis showed that EcoⅠ and SalⅠ digestion produced a fragment of 400bp, and SacⅠ digestion produced a linear band of 5000bp, while PstⅠ did not.pGenesil-1-survivin-1+2 was constructed by ligation of SacⅠ/SalⅠ-linearized pGenesil-1-survivin-1 vector and U6 Promotor+Survivin-2-shRNA; both parts had a BamHⅠdigest site. When pGenesil-1-survivin-1+2 plasmids were digested by BamHⅠ, 1.0% agarose gel electrophoresis showed that a fragment about 400bp was cut off, but it could not be digested by PstⅠ (Fig. 2). DNA sequencing con fi rmed that the plasmids were reconstructed successfully (Fig. 3).

Fig. 1. Structure of plasmid pGenesil-1 vector. The shRNA encoding template was inserted between the Hind Ⅲ and BamHⅠ restriction sites, and the PstⅠ restriction site of plasmid pGenesil-1 was replaced.
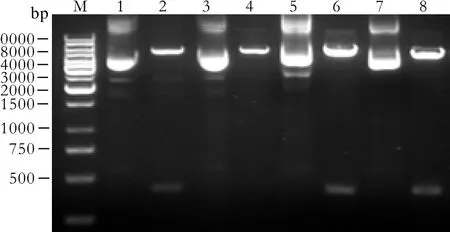
Fig. 2. Identification of recombinant plasmids by restriction enzyme digestion. M: 1 kb DNA marker; 1, 3, 5, 7: pGenesil-1-survivin-1, pGenesil-1-survivin-2, pGenesil-1-survivin-1+2, and pGenesil-1-NC digested by PstⅠ, respectively. Because the restriction site of enzyme PstⅠ in plasmid pGenesil-1 was replaced by the inserted DNA segment, the reconstructed plasmids could not be digested by PstⅠ. 2: An EcoRⅠ site was designed in the inserted segment for pGenesil-1-survivin-1 and a band about 400bp was cut off by EcoRⅠ. 4: A SacⅠ site was designed in the inserted segment for pGenesil-1-survivin-2 and a linear band about 5000bp was cut off by SacⅠ. 6: SacⅠ/SalⅠ-linearized pGenesil-1-survivin-1 vector and U6 Promotor+shRNA-survivin-2 both had a BamHⅠ digest site for pGenesil-1-survivin-1+2 and a band about 400bp was cut off by BamHⅠ. 8: A SalⅠ site was designed in the inserted segment for pGenesil-1-NC and a band about 400bp was cut off by SalⅠ.
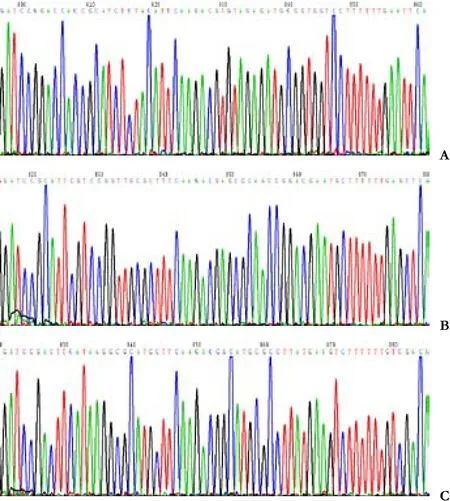
Fig. 3. Recombinant plasmids identi fi ed by DNA sequence analysis(the insertion segment). A: survivin-1-shRNA; B: survivin-2-shRNA; C: NC-shRNA.
Transfection
After the recombinantplasmid vectors with enhanced green fl orescence protein (EGFP) were transfected to the Patu8988 cells, they were seen as a green fl uorescent wave. When the cells were examined under a fl uorescence microscope after 48 hours transfection, more than 60% cells showed green fl uorescence (Fig. 4). This was con fi rmed by fl ow cytometry. The results showed ef fi cient shRNA transfection.
Inhibition of survivin mRNA expression by survivin shRNA
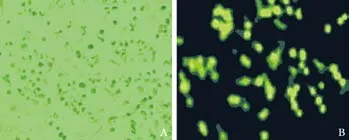
Fig. 4. Light and fluorescent images of Patu8988 cells transfected with recombinant plasmid at 48 hours (original magni fi cation ×400).A: Light image; B: Fluorescent image.
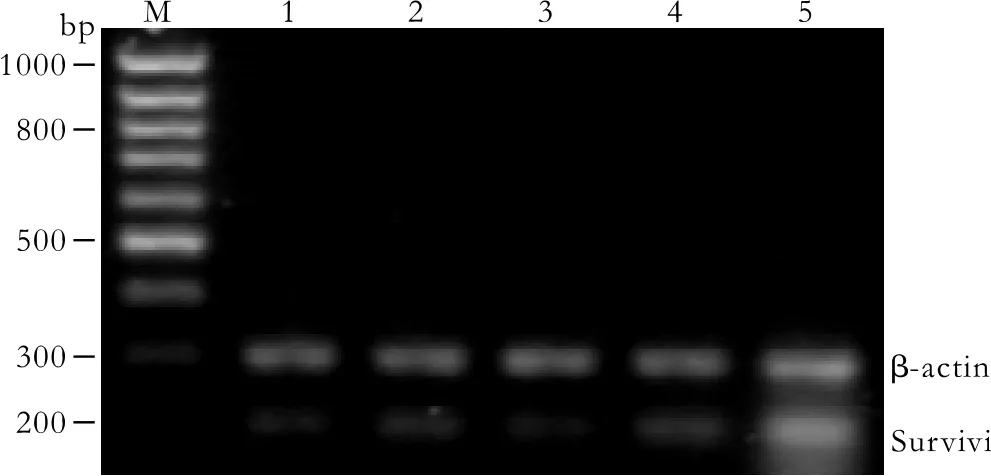
Fig. 5. Survivin mRNA expression in Patu8988 cells detected by RT-PCR at 48 hours post-transfection. M: 100bp DNA marker;1: pGenesil-1-survivin-1 group; 2: pGenesil-1-survivin-2 group;3: pGenesil-1-survivin-1+2 group; 4: pGenesil-1-NC group; 5:untreated control group. Survivin products were quanlified relative to the internal control β-actin.
The mRNA expression intensities of survivin genes,inhibited by speci fi c survivin shRNAs in the Patu8988 cells, were analyzed by semiquantitive RT-PCR. The mRNA levels were normalized to the internal control,β-actin (Fig. 5). The rate of survivin/β-actin mRNA was 59.35±3.46%, 20.40±1.80%, 26.77±5.16%, 20.79±3.12%and 55.14±5.50%, respectively in the control, pGenesil-1-survivin-1, pGenesil-1-survivin-2, pGenesil-1-survivin-1+2, and pGenesil-1-NC groups. The statistical analysis showed that survivin mRNAs of Patu8988 cells in the pGenesil-1-survivin-1, pGenesil-1-survivin-2, and pGenesil-1-1+2 groups were reduced compared with the control group (P<0.05). The inhibition rate reached 65.62%,54.89%, and 64.97%, respectively in the pGenesil-1-survivin-1, pGenesil-1-survivin-2, and pGenesil-1-survivin-1+2 groups. However, the pGenesil-1-NC group showed no inhibition of survivin mRNA expression (P>0.05, vs.control).
Knockdown of survivin protein expression by survivin shRNA
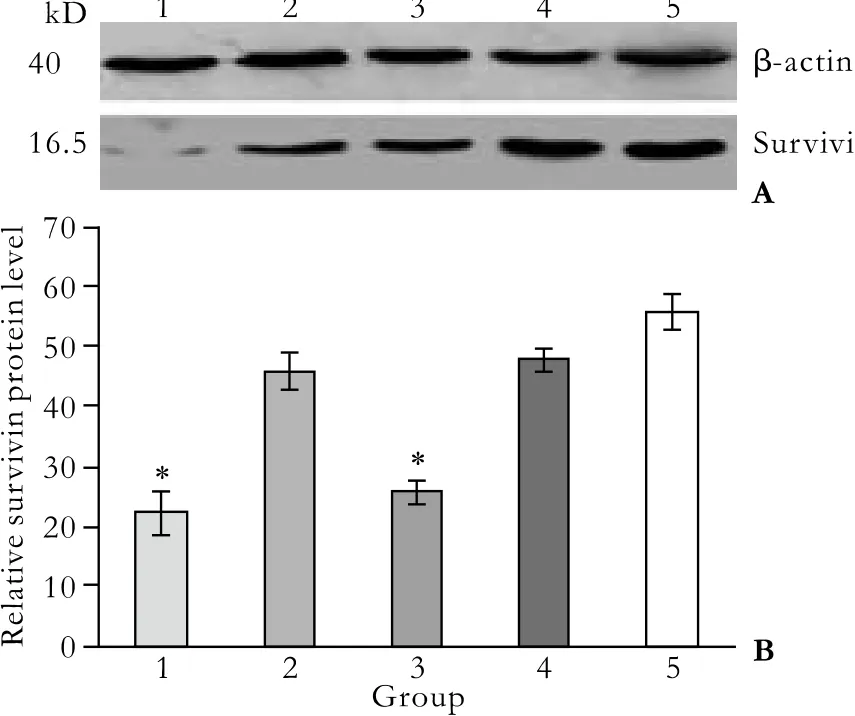
Fig. 6. Survivin protein expression detected by Western blotting.1: pGenesil-1-survivin-1 group; 2: pGenesil-1-survivin-2 group;3: pGenesil-1-survivin-1+2 group; 4: pGenesil-1-NC group; 5:untreated control group. A: Western blots of survivin protein levels.B: Quantitative expression of survivin levels. Bands corresponding to survivin and β-actin protein were scanned and the intensity was determined by optical density measurement. Each value represents the mean±SD from triplicate determinations. *: P<0.05, vs.untreated control group.
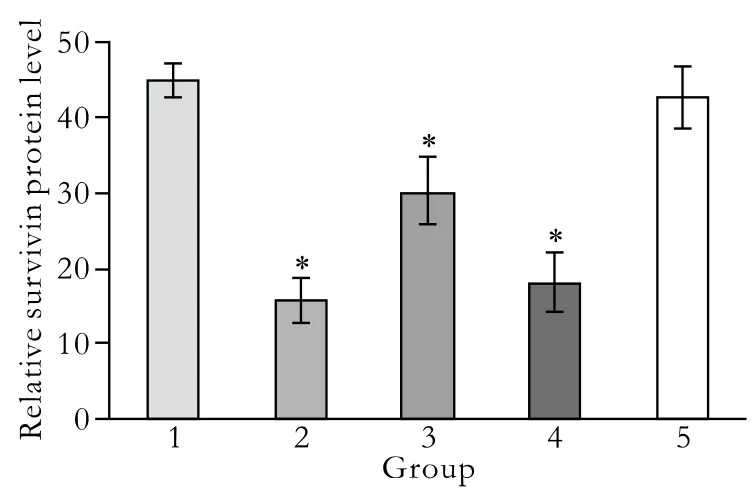
Fig. 7. Quantitative expression of survivin protein levels detected by flow cytometry. 1: untreated control group; 2: pGenesil-1-survivin-1 group; 3: pGenesil-1-survivin-2 group; 4: pGenesil-1-Survivin-1+2 group; 5: pGenesil-1-NC group. Data were the mean±SD. *: P<0.05, vs. untreated Patu8988 cells.
The effect of survivin-shRNA on survivin protein expression at 48 hours post-transfection was evaluated by Western blotting. Protein levels of survivin in the control and pGenesil-NC groups were similar, but reduced by 60.04% and 54.22% in the pGenesil-1-survivin-1 and pGenesil-survivin-1+2 groups, respectively (P<0.05, vs.control; Fig. 6). As expected, there was no difference in the reduction of survivin protein expression between the control and pGenesil-NC groups (P>0.05).However, no difference was seen between the pGenesil-1-survivin-2 and control groups (P>0.05). Next, we performed fl ow cytometry using human survivinspeci fi c antibody to quantify the amount of survivin protein expression in each group. The results showed that survivin levels were 44.70±2.24%, 15.82±2.96%,30.09±4.46%, 18.11±3.96% and 42.74±4.08%, in the control, pGenesil-1-survivin-1, pGenesil-1-survivin-2,pGenesil-1-survivin-1+2, and pGenesil-NC groups, respectively (Fig. 7). Consistent with the Western blotting results, the statistical analysis showed that both the pGenesil-1-survivin-1 and pGenesil-1-survivin-1+2 groups down-regulated the survivin protein expression in Patu8988 cells (P<0.05, vs. control), and the inhibition rates were 64.61% and 59.49%, respectively (P<0.05, vs control). However, unlike the Western blotting results,down-regulation of survivin protein was also seen in the pGenesil-1-survivin-2 group (P<0.05, vs. control).In contrast, the pGenesil-NC had no inhibitory effect on survivin expression (P>0.05, vs. control). The above results showed that transient transfection with survivinshRNAs signi fi cantly decreased the survivin protein expression levels in Patu8988 cells.
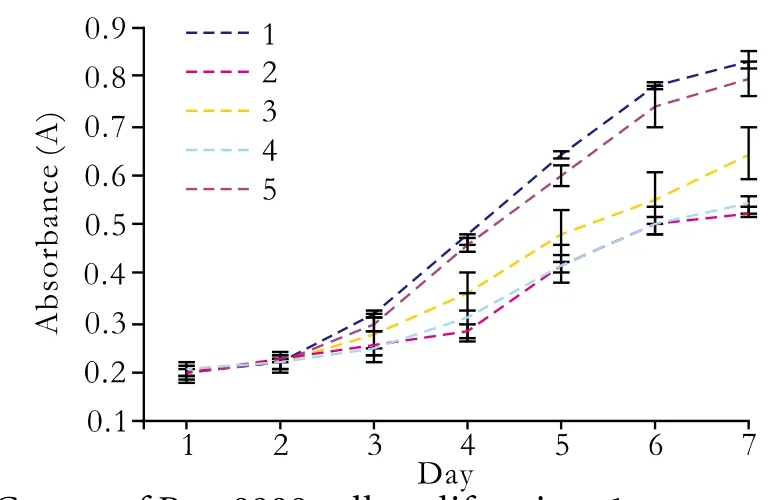
Fig. 8. Curves of Patu8988 cell proliferation. 1: untreated control group; 2: pGenesil-1-suvivin-1 group; 3: pGenesil-1-survivin-2 group; 4: pGenesil-1-survivin-1+2 group; 5: pGenesil-1-NC group.
Inhibition of Patu8988 cell proliferation by survivin shRNA
Because the silencing effect of survivin expression by transient transfection may not last over 72 hours,we evaluated the proliferation of Patu8988 cells stably transfected with pGenesil-1-survivin-1, pGenesil-1-survivin-2, pGenesil-1-survivin-1+2, and pGenesil-1-NC.
When Patu8988 cells were exposed to pGenesil-1-survivin-1, pGenesil-1-survivin-2, or pGenesil-1-survivin-1+2, the growth of the cells was suppressed compared with untreated cells (P<0.01; Fig. 8). However, no statistical difference was found between the control and pGenesil-1-NC groups. The Patu8988 cell growth was inhibited at a higher rate in the pGenesil-1-survivin-1 and pGenesil-survivin-1+2 treatment than that in pGenesil-survivin-2 at different transfection times (P<0.05).The inhibition peak was reached on day 4. Subsequently,the inhibition wore off and the tumor cells returned to proliferation.
Inhibition of in vivo tumor growth by survivin shRNA
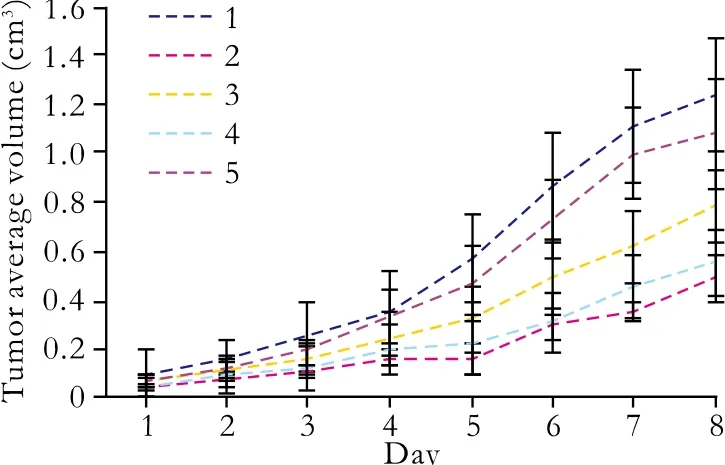
Fig. 9. Tumor growth curves of transplanted pancreatic cancer cells. 1: untreated control group; 2: pGenesil-1-survivin-1 group; 3:pGenesil-1-survivin-2 group; 4: pGenesil-1-survivin-1+2 group; 5:pGenesil-1-NC group; Data were expressed as mean±SD.
With the above fi ndings of the effects of survivinshRNAs in vitro, we next explored whether survivinshRNAs play a critical role in tumor formation in vivo,and whether it can be used in clinical gene therapy.Equal numbers (3×107cells/ml) of Patu8988 cells transfected with pGenesil-1-survivin-1, pGenesil-1-survivin-2, pGenesil-1- survivin-1+2, pGenesil-NC, and untreated cells were injected subcutaneously into nude mice. Untreated cells and those with pGenesil-1-NC grew rapidly, resulting in palpable tumors 4-5 days after injection. In contrast, tumor formation was signi fi cantly slower after inoculation of pGenesil-1-survivin-1,pGenesil-survivin-2, or pGenesil-1-survivin-1+2 clones(P<0.01). The tumors were smaller than those in either the untreated control or pGenesil-1-NC groups (P<0.05;Fig. 9). These results indicated that survivin-shRNAs mediating survivin downregulation exerted a strong growth-suppressive effect on pancreatic cancer in vivo.
Discussion
Previous studies have demonstrated that survivin is overexpressed in pancreatic cancer, and its overexpression is considered to participate in the development and progression of pancreatic cancer by suppressing apoptosis and regulating cell division.[20]In addition, survivin overexpression is associated with the insensitivity of pancreatic cancer to chemotherapy and radiotherapy as a marker of poor prognosis.[4]All these studies suggest that survivin remains an attractive target for pancreatic cancer treatment.
Reported strategies for targeting survivin include antisense oligonucleotides,[6,7]dominant-negative mutation,[8]ribozymes,[9-11]an anticancer vaccine,[21]and RNA interference.[22,23]Among these methods, RNA interference is becoming a powerful technique for its high ef fi ciency, high speci fi city, low toxicity, and great resistance to nuclease degradation. When small interfering RNA is introduced into mammalian cells,sequence-speci fi c destruction of endogenous target mRNA occurs and gene expression is effectively suppressed.[12-14]Nowadays, two kinds of RNA interference techniques, chemically synthesized siRNA and vector-mediated expression of shRNA, are mainly used in silencing gene expression in mammalian cells with their respective characteristics.[13,24]Compared with shRNA vectors, chemically synthesized siRNA is more easily transfected into cancer cell lines and is apparently more effective in silencing targeted genes.However, chemically synthesized siRNA is costly and has a relatively short half-life with only transient inhibition (less than one week) of the target gene.[25]shRNA expressing vectors can be used to transcribe and generate shRNA in vivo and their advantage is that the expression of target genes is reduced for weeks or even months, which allows analysis of the consequences of stably silencing a gene.[13]
In this study, we used the vector-based shRNA technique and constructed recombinant plasmids expressing survivin-shRNA to transfect pancreatic cancer Patu8988 cells. The results of RT-PCR and Western blotting demonstrated that speci fi c survivinshRNA successfully suppressed survivin mRNA and its corresponding protein expression in transiently transfected Patu8988 cells when compared with pGenesil-1-NC and untreated Patu8988 cells (P<0.05).When the Patu8988 cells transfected with pGenesil-1-survivin-1, pGenesil-1-survivin-2, and pGenesil-1-survivin-1+2 were injected into nude mice, tumor growth was slower than that in pGenesil-1-NC transfected cells and untreated cells (P<0.01), and the tumor weights were less than that those in either the pGenesil-1-NC group or untreated Patu8988 cell group (P<0.05). All these data indicated that survivin shRNA-mediated survivin downregulation exerted a strong growth inhibition on pancreatic cancer in vitro and in vivo. However, unlike the report by Wu et al[26]of an enhanced inhibition of colon cancer cells after combined knockdown of two targets of the EGFR gene,our study demonstrated that the inhibitory effects of pGenesil-1-survivin-1 on the survivin gene was the best among the three survivin-shRNAs, and pGenesil-1-survivin-1+2 was better than pGenesil-1-survivin-2.However, the inhibition ef fi ciency of double interfering sites on survivin did not reach the sum of two single interfering sites. Further studies on this are required.
It has been identi fi ed in several reports that survivin shRNA can silence survivin mRNA and protein expression, increase cell apoptosis, and inhibit transfected cell proliferation in different tumor models, such as hepatocellular carcinoma,[27]colon tumor,[28]lymphoma[29]and gastric carcinoma.[30]In our study, the inhibition of survivin mRNA and protein expression was consistent with the above reports. However, the results in our study showed that less than 10% transfected Patu8988 cells underwent apoptosis (data not shown). The reason for these differences remains unclear. The blockade of apoptotic signaling by survivin involves inhibition of the activation of caspase-9, which subsequently activates effector caspases, including caspase-3 and -7.[31]Whitacre et al[32]reported that procaspase-3 was absent in MCF-7 human breast cancer cells because of partial deletion of the CASP-3 gene. Therefore, one possible explanation might be that, like MCF-7 human breast cancer cells, human pancreatic cancer Patu8988 cells also had low caspase expression undergoing cell cycle arrest after treatment with survivin speci fi c siRNA.Further studies are needed to con fi rm this.
In conclusion, using vector-based survivin-shRNAs signi fi cantly inhibits the levels of survivin mRNA and its protein in human pancreatic cancer Patu8988 cells,with dramatic inhibition of cell proliferation in vitro and in vivo. Thus, knockdown of the expression of survivin based on RNAi may be a potential alternative method for the treatment of human pancreatic cancer.
Funding: This study was supported by grants from the Social Bureau Foundation of Suzhou (SZD0614) and the Foundation of Health Bureau of Jiangsu Province (Z200622).
Ethical approval: Not needed.
Contributors: YXC and YC proposed the study. SYM wrote the fi rst draft. SML performed the research. SML, QCH, and SYH analyzed the data. All authors contributed to the design and interpretation of the study and to further drafts. SYM is the guarantor.
Competing interest: No bene fi ts in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Coppola D. Molecular prognostic markers in pancreatic cancer. Cancer Control 2000;7:421-427.
2 Welsch T, Kleeff J, Friess H. Molecular pathogenesis of pancreatic cancer: advances and challenges. Curr Mol Med 2007;7:504-521.
3 Fryer RA, Galustian C, Dalgleish AG. Recent advances and developments in treatment strategies against pancreatic cancer. Curr Clin Pharmacol 2009;4:102-112.
4 Lopes RB, Gangeswaran R, McNeish IA, Wang Y, Lemoine NR. Expression of the IAP protein family is dysregulated in pancreatic cancer cells and is important for resistance to chemotherapy. Int J Cancer 2007;120:2344-2352.
5 Capalbo G, Rodel C, Stauber RH, Knauer SK, Bache M,Kappler M, et al. The role of survivin for radiation therapy.Prognostic and predictive factor and therapeutic target.Strahlenther Onkol 2007;183:593-599.
6 Dai DJ, Lu CD, Lai RY, Guo JM, Meng H. Effect of survivin targeting on cell proliferation and apoptosis in pancreatic cancer cells. Zhonghua Yi Xue Za Zhi 2004;84:1894-1898.
7 Rödel F, Frey B, Leitmann W, Capalbo G, Weiss C, Rödel C.Survivin antisense oligonucleotides effectively radiosensitize colorectal cancer cells in both tissue culture and murine xenograft models. Int J Radiat Oncol Biol Phys 2008;71:247-255.
8 Zhang R, Wang T, Li KN, Qin WW, Chen R, Wang K, et al. A survivin double point mutant has potent inhibitory effect on the growth of hepatocellular cancer cells. Cancer Biol Ther 2008;7:547-554.
9 Pennati M, Colella G, Folini M, Citti L, Daidone MG,Zaffaroni N. Ribozyme-mediated attenuation of survivin expression sensitizes human melanoma cells to cisplatininduced apoptosis. J Clin Invest 2002;109:285-286.
10 Choi KS, Lee TH, Jung MH. Ribozyme-mediated cleavage of the human survivin mRNA and inhibition of antiapoptotic function of survivin in MCF-7 cells. Cancer Gene Ther 2003;10:87-95.
11 Pennati M, Binda M, De Cesare M, Pratesi G, Folini M, Citti L, et al. Ribozyme-mediated down-regulation of survivin expression sensitizes human melanoma cells to topotecan in vitro and in vivo. Carcinogenesis 2004;25:1129-1136.
12 Chen Y, Huang L. Tumor-targeted delivery of siRNA by nonviral vector: safe and effective cancer therapy. Expert Opin Drug Deliv 2008;5:1301-1311.
13 Walchli S, Sioud M. Vector-based delivery of siRNAs: in vitro and in vivo challenges. Front Biosci 2008;13:3488-3493.
14 Gondi CS, Rao JS. Concepts in in vivo siRNA delivery for cancer therapy. J Cell Physiol 2009;220:285-291.
15 Bertrand JR, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvy C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem Biophys Res Commun 2002;296:1000-1004.
16 Miyagishi M, Hayashi M, Taira K. Comparison of the suppressive effects of antisense oligonucleotides and siRNAs directed against the same targets in mammalian cells.Antisense Nucleic Acid Drug Dev 2003;13:1-7.
17 Aoki Y, Cioca DP, Oidaira H, Kamiya J, Kiyosawa K. RNA interference may be more potent than antisense RNA in human cancer cell lines. Clin Exp Pharmacol Physiol 2003;30:96-102.
18 Kubo T, Zhelev Z, Bakalova R, Ohba H. Enhancement of gene silencing potency and nuclease stability by chemically modi fi ed duplex RNA. Nucleic Acids Symp Ser (Oxf) 2007;51:407-408.
19 Tuschl T. Expanding small RNA interference. Nat Biotechnol 2002;20:446-448.
20 Lens SM, Vader G, Medema RH. The case for Survivin as mitotic regulator. Curr Opin Cell Biol 2006;18:616-622.
21 Tsuruma T, Hata F, Torigoe T, Furuhata T, Idenoue S,Kurotaki T, et al. Phase I clinical study of anti-apoptosis protein, survivin-derived peptide vaccine therapy for patients with advanced or recurrent colorectal cancer. J Transl Med 2004;2:19.
22 Chen XM, Luan XY, Lei DP, Ma XJ, Liu XX, Liu J, et al.Suppression of survivin expression by short hairpin RNA induces apoptosis in human laryngeal carcinoma cells. ORL J Otorhinolaryngol Relat Spec 2008;70:168-175.
23 Shen J, Liu J, Long Y, Miao Y, Su M, Zhang Q, et al.Knockdown of survivin expression by siRNAs enhances chemosensitivity of prostate cancer cells and attenuates its tumorigenicity. Acta Biochim Biophys Sin (Shanghai) 2009;41:223-230.
24 Shiota M, Ikeda Y, Wadhwa R. The factors that contribute to the long-term expression of siRNA. Nucleic Acids Symp Ser(Oxf) 2006;:243-244.
25 Scherr M, Eder M. Gene silencing by small regulatory RNAs in mammalian cells. Cell Cycle 2007;6:444-449.
26 Wu X, Deng Y, Wang G, Tao K. Combining siRNAs at two different sites in the EGFR to suppress its expression, induce apoptosis, and enhance 5- fl uorouracil sensitivity of colon cancer cells. J Surg Res 2007;138:56-63.
27 Cheng SQ, Wang WL, Yan W, Li QL, Wang L, Wang WY.Knockdown of survivin gene expression by RNAi induces apoptosis in human hepatocellular carcinoma cell line SMMC-7721. World J Gastroenterol 2005;11:756-759.
28 Cai M, Wang GB, Tao KX, Cai CX. Enhanced chemotherapy sensitivity of human colon cancer cells to 5- fl uorouracil by siRNA recombinant expression vector targeting survivin gene. Chin Med Sci J 2009;24:97-101.
29 Congmin G, Mu Z, Yihui M, Hanliang L. Survivin--an attractive target for RNAi in non-Hodgkin's lymphoma,Daudi cell line as a model. Leuk Lymphoma 2006;47:1941-1948.
30 Miao GY, Lu QM, Zhang XL. Downregulation of survivin by RNAi inhibits growth of human gastric carcinoma cells.World J Gastroenterol 2007;13:1170-1174.
31 Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res 2008;14:5000-5005.
32 Whitacre CM, Satoh TH, Xue L, Gordon NH, Oleinick NL.Photodynamic therapy of human breast cancer xenografts lacking caspase-3. Cancer Lett 2002;179:43-49.
BACKGROUND: Survivin is known to be overexpressed in various human malignancies, including pancreatic cancer,and mediates cancer cell proliferation and tumor growth, so the regulation of this molecule could be a new strategy for treating pancreatic cancer. In this study, short hairpin RNAs(shRNAs) speci fi c to survivin were introduced into human pancreatic cancer Patu8988 cells to investigate the inhibitory effects on survivin expression and cell proliferation in vitro and in vivo.
METHODS: Three kinds of shRNA speci fi c to the survivin gene were designed and cloned into eukaryotic expression plasmid pGenesil-1 vector. Subsequently the recombinant plasmids were transfected into human pancreatic cancer Patu8988 cells with lipfectamineTM2000 reagent. The mRNA and protein expressions of survivin in the transiently transfected Patu8988 cells were determined by RT-PCR, fl ow cytometry, and Western blotting analysis. The proliferation inhibition rates of stably transfected Patu8988 cells were determined by MTT assay. The antitumor activities of the three kinds of survivin-shRNA plasmids were evaluated in BALB/c nude mice inoculated with Patu8988 cells and bearing human pancreatic cancer.
RESULTS: The three survivin-shRNA plasmids named pGenesil-1-survivin-1, pGenesil-1-survivin-2 and pGenesil-1-survivin-1+2(with double interfering RNA sites) were successfully constructed, and were con fi rmed by restriction enzyme cutting and sequencing. At 48 hours after transfection, the expression of survivin mRNA and protein was inhibited in Patu8988 cells transfected with pGenesil-1-survivin-1,pGenesil-1-survivin-2, and pGenesil-1-survivin-1+2 when compared with that of either pGenesil-1-NC (with scrambled small interfering RNA) transfected cells or control cells(P<0.05). The MTT results showed that the proliferation rates of Patu8988 cells stably transfected with survivin-shRNA plasmids were reduced when compared with that of either pGenesil-1-NC transfected cells or control cells (P<0.01).Furthermore, when Patu8988 cells stably transfected with survivin-shRNA were injected into BALB/c nude mice,tumor growth was dramatically lower and the tumor was smaller than that of either pGenesil-1-NC transfected cells or control cells (P<0.01). The inhibitory effect of pGenesil-1-survivin-1 was the best among the three kinds of survivinshRNA plasmids, but no combination of inhibitory effects was found in pGenesil-1-survivin-1+2.
CONCLUSIONS: shRNAs speci fi c to survivin have gene silencing effects and inhibit pancreatic cancer cell proliferation. shRNA activity against survivin could be of potential value in gene therapy for pancreatic cancer. However, shRNAs with double combining sites did not signi fi cantly enhance the interference compared with single site shRNAs, therefore further studies on this are needed.
Author Af fi liations: Radioimmunoassay Center & Clinical Laboratory(Shen YM, Song ML and Qin CH), Department of MRI (Yang XC), and Department of General Surgery (Sun YH), Second Af fi liated Hospital,Soochow University, Suzhou 215004, China; Radioimmunoassay Center, the Municipal Hospital of Suzhou, Suzhou 215002, China (Yang C)
Xiao-Chun Yang, PhD, Department of MRI,Second Af fi liated Hospital, Soochow University, Suzhou 215004, China(Tel: 86-512-67783462; Fax: 86-512-68284303; Email: szsxy@yahoo.com.cn)© 2010, Hepatobiliary Pancreat Dis Int. All rights reserved.
September 9, 2009
Accepted after revision December 30, 2009
Painting is silent poetry, and poetry is a apeaking picture.
— Simonides
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Critical fl icker frequency for diagnosis and assessment of recovery from minimal hepatic encephalopathy in patients with cirrhosis
- Risk factors for early recurrence of smallhepatocellular carcinoma after curative resection
- Pathological changes at early stage of multiple organ injury in a rat model of severe acute pancreatitis
- Potential etiopathogenesis of seventh day syndrome following living donor liver transplantation: ischemia of the graft?
- Comparatively lower postoperative hepatolithiasis risk with hepaticocholedochostomy versus hepaticojejunostomy
- Effect of sodium salicylate on oxidative stress and insulin resistance induced by free fatty acids
