Fatty acid binding protein 5 is a novel therapeutic target for hepatocellular carcinoma
2024-03-08YanLiWilliamLeeZhenGangZhaoYiLiuHaoCuiHaoYuWang
Yan Li,William Lee,Zhen-Gang Zhao,Yi Liu,Hao Cui,Hao-Yu Wang
Abstract BACKGROUND Hepatocellular carcinoma (HCC) is an aggressive subtype of liver cancer and is one of the most common cancers with high mortality worldwide.Reprogrammed lipid metabolism plays crucial roles in HCC cancer cell survival,growth,and evolution.Emerging evidence suggests the importance of fatty acid binding proteins (FABPs) in contribution to cancer progression and metastasis;however,how these FABPs are dysregulated in cancer cells,especially in HCC,and the roles of FABPs in cancer progression have not been well defined.AIM To understand the genetic alterations and expression of FABPs and their associated cancer hallmarks and oncogenes in contributing to cancer malignancies.METHODS We used The Cancer Genome Atlas datasets of pan cancer and liver hepatocellular carcinoma (LIHC) as well as patient cohorts with other cancer types in this study.We investigated genetic alterations of FABPs in various cancer types.mRNA expression was used to determine if FABPs are abnormally expressed in tumor tissues compared to non-tumor controls and to investigate whether their expression correlates with patient clinical outcome,enriched cancer hallmarks and oncogenes previously reported for patients with HCC.We determined the protein levels of FABP5 and its correlated genes in two HCC cell lines and assessed the potential of FABP5 inhibition in treating HCC cells.RESULTS We discovered that a gene cluster including five FABP family members (FABP4,FABP5,FABP8,FABP9 and FABP12) is frequently co-amplified in cancer.Amplification,in fact,is the most common genetic alteration for FABPs,leading to overexpression of FABPs.FABP5 showed the greatest differential mRNA expression comparing tumor with non-tumor tissues.High FABP5 expression correlates well with worse patient outcomes (P < 0.05).FABP5 expression highly correlates with enrichment of G2M checkpoint (r=0.33,P=1.1e-10),TP53 signaling pathway (r=0.22,P=1.7e-5) and many genes in the gene sets such as CDK1 (r=0.56,P=0),CDK4 (r=0.49,P=0),and TP53 (r=0.22,P=1.6e-5).Furthermore,FABP5 also correlates well with two co-expressed oncogenes PLK1 and BIRC5 in pan cancer especially in LIHC patients (r=0.58,P=0;r=0.58,P=0;respectively).FABP5high Huh7 cells also expressed higher protein levels of p53,BIRC5,CDK1,CDK2,and CDK4 than FABP5low HepG2 cells.FABP5 inhibition more potently inhibited the tumor cell growth in Huh7 cells than in HepG2 cells.CONCLUSION We discovered that FABP5 gene is frequently amplified in cancer,especially in HCC,leading to its significant elevated expression in HCC.Its high expression correlates well with worse patient outcome,enriched cancer hallmarks and oncogenes in HCC.FABP5 inhibition impaired the cell viability of FABP5high Huh7 cells.All these support that FABP5 is a novel therapeutic target for treating FABP5high HCC.
Key Words: Hepatocellular carcinoma;Fatty acid binding protein;Novel target;Amplification;Correlated expression
lNTRODUCTlON
Cancer cells are heavily dependent on cellular metabolism pathways for their disease malignancy and progression.Lipid metabolism has been increasingly recognized to be reprogrammed and plays crucial roles in cancer cell survival,growth,and evolution[1].
There is emerging evidence suggesting the critical roles of fatty acid binding proteins (FABPs) in contribution to cancer progression and metastasis[2,3].FABPs are a family of chaperone proteins that bind to long-chain fatty acids,retinoids,and other hydrophobic molecules[3].There are ten FABP genes identified in the human genome,each with restricted tissue distribution in healthy individuals.Some of the family members includingFABP4,FABP5,andFABP7are abnormally expressed in cancer cells beyond tissue expression restriction and play important roles in cancer malignancy and progression[2,3].FABP4is normally expressed in adipocytes at high levels and its expression is controlled by peroxisome proliferator-activated receptor gamma (PPARγ)[4].Accumulating evidence shows that FABP4 plays important roles in cancer progression in multiple cancer types,including breast cancer[5],ovarian cancer[6],and colon cancer[7].In contrast,loss ofFABP1expression in colorectal cancer is associated with poor patient outcome[8].FABP5is normally expressed in cells of epidermal origin and emerging evidence shows that it functions to regulate fatty acid trafficking,lipid metabolism and cell growth[9].FABP5was found to be upregulated in many cancer types[10].However,the mechanism leading to abnormal FABP expression in cancer is not clear and the roles of these FABPs in contributing to cancer progression have not been well defined.In this study,we aimed to investigate the genetic alterations leading to abnormal expression of FABPs in cancer and the missing links of abnormal FABP expression to cancer gene signatures and patient survival.
MATERlALS AND METHODS
Cells and reagents
Hepatocellular carcinoma (HCC) cell lines HepG2 and Huh7 were obtained from our laboratory for long-term storage and cultured in high glucose Dulbecco’s modified Eagle medium containing 10% fetal bovine serum and 1% penicillin/streptomycin.SBFI-26 (S9957) was purchased from Selleck Chemicals (Houston,TX,United States).The antibodies against FABP5 (39926),p53 (9282),BIRC5 (2808),CDK1 (77055),CDK2 (2546),CDK4 (12790),MCL-1 (94296) and GAPDH (2118) were purchased from Cell Signaling Technology (Danvers,MA,United States).A CellTiter-Glo 2.0 cell viability assay kit (G9241) was purchased from Promega (Madison,WI,United States).
Collection of datasets and bioinformation analysis platforms
We performed data analysis for The Cancer Genome Atlas (TCGA) pan-cancer and liver hepatocellular carcinoma (LIHC) patient cohort through cBioPortal (https://www.cbioportal.org/) and a well-established web bioinformatic platform Gene Expression Profiling Interactive Analysis 2 (GEPIA2,http://gepia2.cancer-pku.cn/#index),developed by Zhang Lab,Peking University[11].GEPIA2 collected RNA sequencing data of 9,736 tumors and 8,587 non-tumor samples from the TCGA and the Genotype-Tissue Expression projects.
Identification of genetic alterations of FABP family members
To find out the genetic alterations of FABPs (FABP1,FABP2,FABP3,FABP4,FABP5,FABP6,FABP7,PMP2,FABP9andFABP12) in cancer cells,we used cBioPortal tools.We checked the frequencies of each genetic alteration using TCGA pancancer (70655 samples from 217 non-redundant studies) and LIHC cohort (1829 samples from eleven studies) and their correlation withFABP5mRNA expression and HCC cancer stages.
Expression of FABP5 in tumor and non-tumor cells and its correlation with patient survival
We compared mRNA expression of FABPs in tumorvsnon-tumor tissues using TCGA pan-cancer (9664 tumor tissue samples and 711 non-tumor tissue samples) and LIHC cohort (360 tumor tissue samples and 50 non-tumor tissue samples) and checked if high expression correlates with poor patient outcomes using the GEAIP2 platform.
Correlation of FABP5 expression with cancer hallmarks and oncogenes in HCC cells
We assessed correlation ofFABP5expression with cancer hallmarks enriched in HCC cells (G2M checkpoint and TP53 signaling) and highlighted genes within the gene sets using the GEAIP2 platform.
Cell viability assay
The cell viability assay was performed as described in our earlier study[12].Briefly,2000 HepG2 or Huh7 cells per well were pre-seeded into white 96-well plates overnight.The cells were then treated with a 2-fold serial dilution of SBFI-26 (0-100 μmol/L).The cell viability was measured using a VICTOR Nivo Multimode Microplate Reader (PerkinElmer Health Sciences Inc,Shelton,CT,United States) at 72 h post treatment by using CellTiter-Glo 2.0 Reagent.
Statistical and computational analysis
We performed Pearson's or Spearman's correlation test to determine whether there was a significant link between the two variables.The log-rank test was used to determine the statistical significance of gene expression in correlation with patient outcome.aP< 0.05;bP< 0.01;cP< 0.001;dP< 0.0001.
RESULTS
FABP4, FABP5, FABP8, FABP9 and FABP12 are frequently co-amplified in cancer
We discovered that a cluster of FABP genesFABP4,FABP5,FABP8,FABP9andFABP12at the adjacent loci of chromosome 8,but not other FABPs located at different chromosomes,showed high frequencies of genetic alteration (3%) in the pan-cancer cohort (Figure 1A).The frequency of genetic alterations of these FABPs reaches 5%-6% in the LIHC cohort (Figure 1B).The prevalent type of alteration is amplification and co-occurrence of amplification of these genesFABP4,FABP5,FABP8,FABP9andFABP12is highly significant (P< 0.001) (Figure 1C).Other genetic alterations such as mutations,structural variants and homo-deletion also occur but at much lower frequencies (Figure 1A and B).

Figure 1 Genetic alternation of FABP family members in various cancer types.A-B: Genetic alternation of each FABP family member (FABP1,FABP2,FABP3,FABP4,FABP5,FABP6,FABP7,PMP2,FABP9 and FABP12) in pan-cancer types (70655 samples from 217 non-redundant studies) (A) and in various liver cancer subtypes (1829 samples from eleven studies) (B) using The Cancer Genome Atlas datasets.The mutation frequency for each FABP gene was shown next to the gene name;C: Co-occurrence of genetic alternation of the gene loci of FABP family members in liver cancer.
FABP5 and FABP4 amplifications occur at the highest frequencies in patients with prostate adenocarcinoma or HCC
Interestingly,when we checked genetic alterations of FABPs in different cancer types,we discovered that patients with prostate adenocarcinoma (PRAD) or HCC have the highest genetic alteration frequencies among others and again with genetic amplification as the prevalent type.FABP4showed the highest frequency in PRAD (7.9%) followed by HCC (7.8%) using TCGA pan cancer cohort,whileFABP5showed the highest frequency in HCC (8.1%) followed by prostate PRAD (Figure 2A and C).Consistent with these,when we checked their genetic status in various liver cancer cohorts,we observed highFABP4alteration frequencies up to 12.5% andFABP5up to 12.2% in the aggressive subtype HCC (Figure 2B and D).Importantly,FABP5mRNA expression is much higher in the patients withFABP5amplification than those withFABP5gain,diploid,or deletion (P< 0.0001) (Figure 2E).Moreover,HCC cases at stage II-IV showed higher expression ofFABP5(P=0.071),but notFABP4(P=0.179) (Figure 2F).
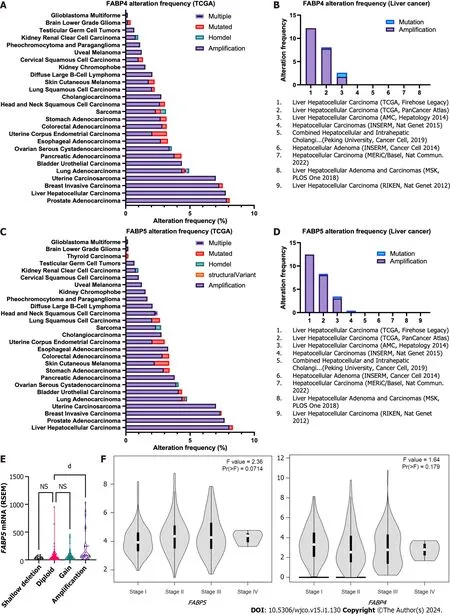
Figure 2 Frequencies of genetic alternations and expression of FABP4 and FABP5 in various cancer types.A-B: Frequencies of genetic alternations of FABP4 in major cancer types using The Cancer Genome Atlas (TCGA) datasets via cBioportal (A) and in various liver cancer subtypes (B) using TCGA datasets via cBioportal;C-D: Frequencies of genetic alternations of FABP5 in pan-cancer types (C) and in various liver cancer subtypes (D) using TCGA datasets via cBioportal;E: FABP5 expression in liver cancer with different FABP5 genetic alteration types using TCGA datasets via cBioportal;F: Expression of FABP5 (left panel) and FABP4 (right panel) in liver cancer at stage I-IV using TCGA datasets via GEPIA2 portal.dP < 0.0001;NS: Not significant.
FABP5 and FABP4 are expressed at much higher levels in tumor tissues compared to non-tumor counterparts in HCC
To find out whether amplification and resulting high expression of FABP family members has clinical significance in cancer,we compared their expression in tumor tissues and non-tumor controls for various cancer types (Figure 3A).Among all family members,FABP3,FABP4,andFABP5are expressed ubiquitously across various cancer types (Figure 3A),whileFABP1,FABP6,FABP7,andFABP8are expressed selectively in restricted cancer types.In contrast,FABP2,FABP9,andFABP12are expressed at extremely low levels,if any (Figure 3A).FABP1,FABP3,FABP4,andFABP5,but not other family members,are selectively expressed in LIHC patients (Figure 3A and Supplementary Figure 1A),whileFABP3,FABP4andFABP5are also selectively expressed in PRAD patients (Figure 3A and Supple-Smentary Figure 1A).When we checked the expression of FABPs in tumor tissues compared to non-tumor controls,we found thatFABP5is significantly upregulated in adrenocortical carcinoma (ACC),glioblastoma multiforme (GBM),kidney renal clear cell carcinoma (KIRC),brain lower-grade glioma (LGG),liver hepatocellular carcinoma (LIHC),lung adenocarcinoma (LUAD),skin cutaneous melanoma (SKCM),and uveal melanoma (UVM),whileFABP4is significantly upregulated only in colon adenocarcinoma (COAD),lung squamous cell carcinoma and stomach adenocarcinoma (Figure 3B).In patients with LIHC,among the selectively expressed FABPs,onlyFABP5andFABP4showed significant upregulation of mRNA expression in tumor tissues compared to non-tumor controls.Other FABPs,such asFABP1(restricted expression in normal liver),did not show differential expression in tumor and non-tumor tissues,even though it is expressed at prominent levels in patients with LIHC (Figure 3C and Supplementary Figure 1A-C).More importantly,highFABP5expression significantly correlates with overall patient survival (P=6.6e-5) and disease-free survival (P=6.6e-5) (Figure 3D-E).We found that highFABP5expression significantly correlates with overall survival not only in LIHC but also in other cancer types including ACC,GBM,KIRC,LGG,LUAD,SKCM,and UVM,whereFABP5is found to be upregulated in tumor tissues compared to non-tumor control (Figure 3B and Supplementary Figure 1D).In contrast,highFABP4expression showed less significant correlation with poor overall survival (P=0.047) (Figure 3E).In addition,expression of other FABPs (FABP1andFABP3) expressed in LIHC,does not significantly correlate with overall survival (P=0.21 and 0.091,respectively) (Supplementary Figure 1E-G).Together,these data indicate thatFABP5,among other FABPs,is selectively upregulated in tumor tissues and its expression correlates well with poor clinical outcomes in patients with HCC.
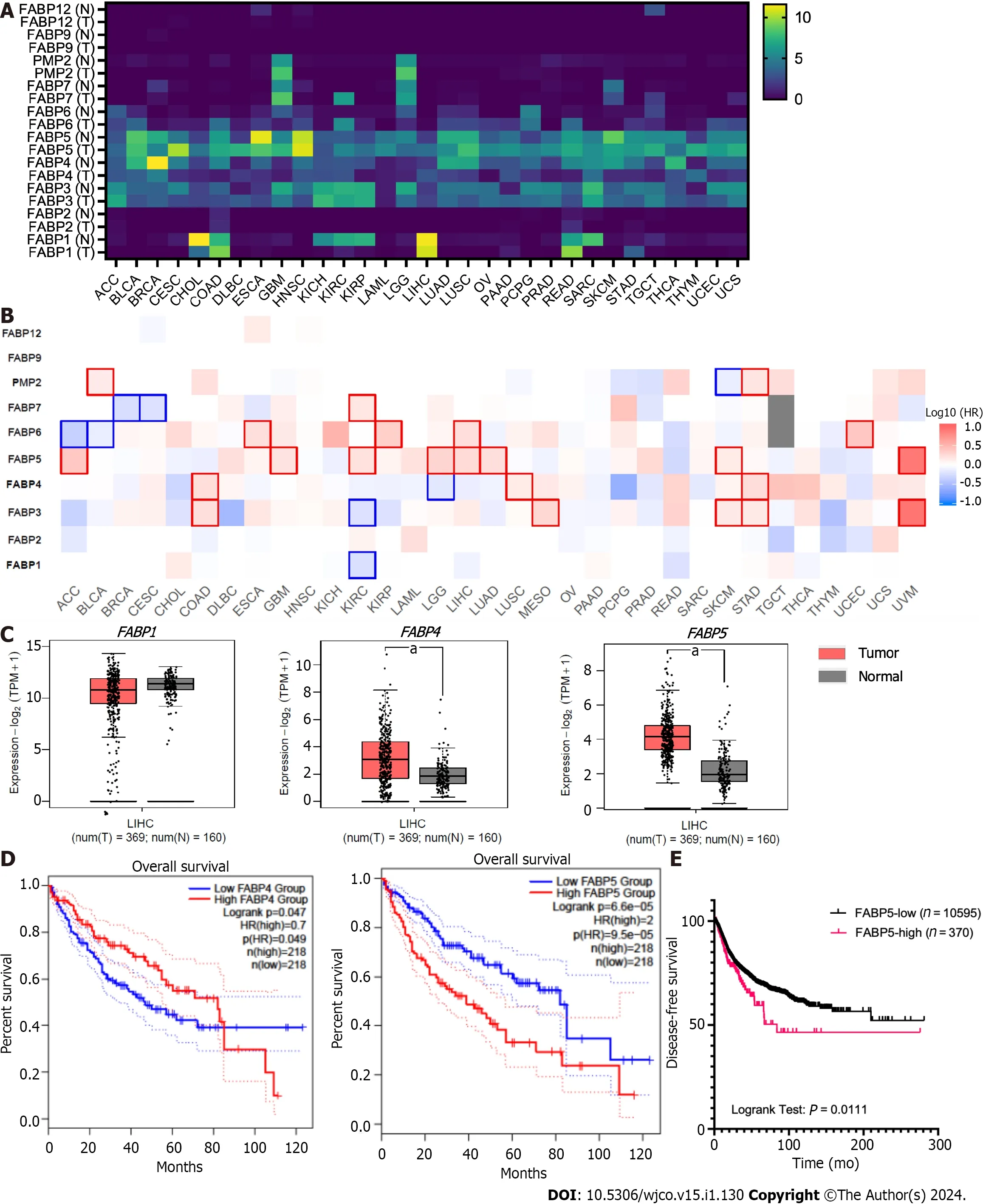
Figure 3 Expression of FABP family members in tumor tissues and non-tumor tissue control in various cancer types. A-B: Expression of each FABP family member (FABP1,FABP2,FABP3,FABP4,FABP5,FABP6,FABP7,PMP2,FABP9 and FABP12) in tumor tissues and non-tumor tissue control in various cancer types (A) and statistical significance when comparing their expression in tumor tissues with non-tumor tissue control (B) using GEPIA2 portal;C: Higher expression of FABP4 and FABP5,but not FABP1,was observed in liver tumor tissues comparing to non-tumor tissue control;D-E: High expression levels of FABP5,but not FABP1 or FABP4,in liver cancer correlate with overall patient survival (D) and disease-free survival (E).aP < 0.05.
High FABP5 expression selectively associated with enrichment of cancer hallmarks G2M checkpoint and TP53 signaling
E2F targets,G2M checkpoint and DNA repair have been identified to be the top three cancer hallmarks enriched in HCC tumor cells compared to non-tumor controls[13],which supported that cell cycle and DNA repair signaling networks are critical for HCC cancer malignancies and progression.Therefore,we are interested in checking ifFABP5overexpression correlates with enrichment of these important cancer hallmarks in HCC.We found thatFABP5expression correlates well with G2M checkpoint gene signature (r=0.33,P=1.1e-10) (Figure 4A and B),and most of the genes in the dataset,includingATR(r=0.35,P=3.9e-12)[13],BRCA1(r=0.23,P=8.1e-6),CCNB1(r=0.61,P=0),CDK1(r=0.56,P=0),CDKN2D(r=0.37,P=121e-13),CHEK1(r=0.46,P=0),CHEK2(r=0.17,P=0.001),PI4KA(r=0.29,P=9.3e-9),PRKDC(r=0.43,P=1.4e-11),RPS6K1(r=0.3,P=3.6e-9),andYWHAH(r=0.38,P=5.7e-14) (Figure 4C).Among these,expressions ofCCNB1andCDK1showed the highest correlation withFABP5expression (Figure 4C).The strongCCNB1-FABP5correlation is observed in LIHC but also in many other cell types including COAD (r=0.43,P=6.5e-14),DLBCL (r=0.54,P=6.5e-14),KICH (r=0.8,P=6.7e-16),KIRC (r=0.46,P=0),READ (r=0.36,P=0.00043),tenosynovial giant cell tumor (r=0.43,P=1.3e-7),and UVM (r=0.75,P=1.3e-15) (Supplementary Figure 2A and B).
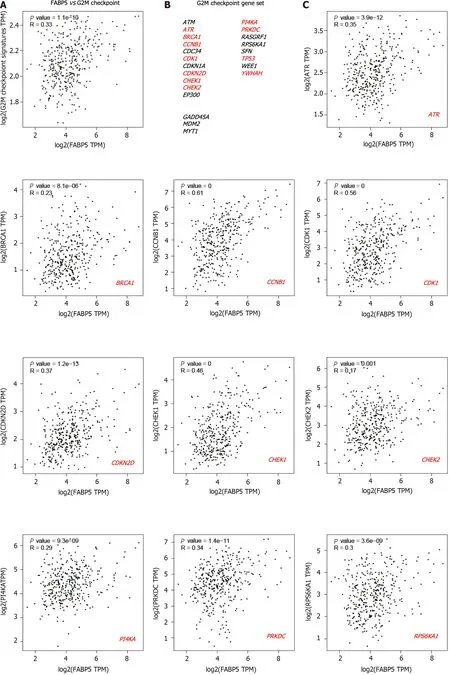
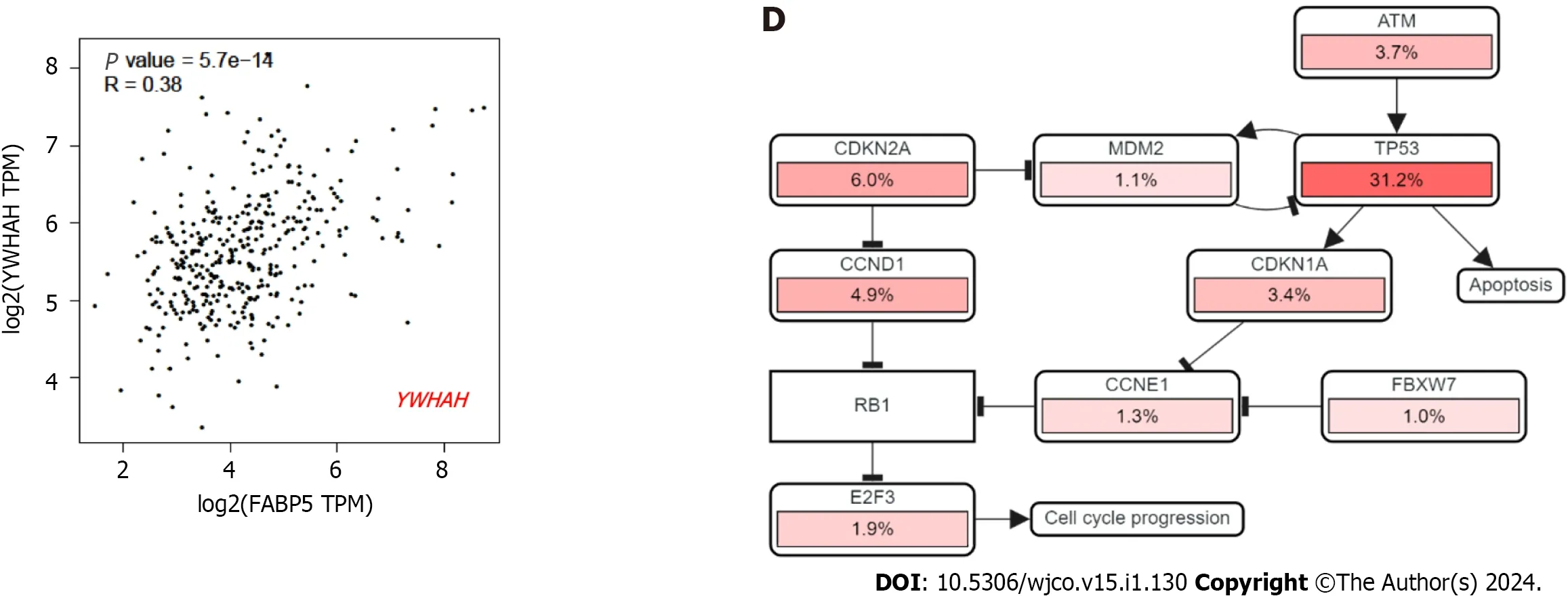
Figure 4 Expression of FABP5 correlates with cancer hallmark G2M checkpoint and genes within. A: FABP5 positively correlated with enrichment of cancer hallmark G2M checkpoint in patients with liver hepatocellular carcinoma (LIHC);B: Gene list included in the cancer hallmark G2M checkpoint.The genes that showed positive correlation with FABP5 are highlighted in red (P < 0.01);C: Highlighted genes in the G2M checkpoint gene set positively correlate with FABP5 expression in patients with LIHC.P values show the statistical significance of the correlation;R values indicate the correlation coefficient;D: Altered expression of individual genes in the G2M checkpoint gene set showed in the context of signaling network.Pink bars show upregulation of gene expression.
In an independent analysis using cBioportal platform,we found that most genes involved in cell cycle network are dysregulated withTP53as the top gene (Figure 4D).Therefore,we checked ifFABP5expression is associated with expression ofTP53and p53 signaling gene signature.We found thatFABP5expression correlates well withTP53expression (r=0.22,P=1.6e-5),its signaling gene signature (r=0.33,P=1.1e-10) (Figure 5A and B),and almost half of the genes in the dataset,includingBCL2(r=0.15,P=0.0044),CDK2(r=0.28,P=6.6e-8),CDK4(r=0.49,P=0),E2F1(r=0.15,P=0029),PCNA(r=0.31,P=2.2e-9),andRB1(r=0.21,P=5.2e-5) (Figure 5C).In an independent analysis using cBioportal platform,we found that most genes involved in TP53 signaling network are dysregulated withTP53as the top gene (Figure 5D).
High FABP5 expression correlated well with PLK1 and BIRC5 expression
In our previous study,we demonstrated that thatPLK1,a master regulator of cell cycle,andBIRC5,a multifunctional gene only expressed at G2M phase,are two important oncogenes that are highly co-expressed in HCC and the cotargeting of PLK1 and BIRC5 synergistically inhibited tumor growth of HCC preclinical modelsinvitroandinvivo[12].To investigate the relationship betweenFABP5expression andPLK1-BIRC5co-expression in cancer,we first checked expression of selected FABPs together withCCNB1,TP53,BIRC5andPLK1(Figure 6A).Expression ofFABP5,but not other FABPs includingFABP1,FABP3,FABP4andFABP6appeared to be well-correlated with expression ofPLK1andBIRC5across cancer types,in addition toCCNB1andTP53(Figure 6A).Consistent with our previous findings[12],expression ofPLK1andBIRC5showed remarkable correlation in pan cancer (r=0.73,P=0) and even higher in LIHC (r=0.83,P=0) (Figure 6B and C).Expression ofFABP5also showed some correlation withPLK1(r=0.19,P=0) orBIRC5(r=0.15,P=0) in cancer and much better correlation in LIHC patient cohort (r=0.58,P=0;r=0.58,P=0,respectively) (Figure 6D-G and Supplementary Figure 3A and B).These data demonstrate thatFABP5is highly correlated to the expression ofPLK1-BIRC5co-expression,which is selectively in patients with HCC.
HCC cells with high FABP5 expression are sensitive to FABP5 inhibition
The stabilizingTP53mutation Y220C in Huh7 cells resulted in overexpression of p53,which is much higher than that in HepG2 cells harboring wild typeTP53gene.Interestingly,FABP5 protein expression is also expressed at a much higher level in Huh7 cells than HepG2 cells (Figure 7A).BIRC5,CDK1,CDK2,and CDK4,but not MCL-1,are also expressed at much higher levels in FABP5highHuh7 cells than FABP5lowHepG2 cells (Figure 7A).Huh7 cells are more sensitive to FABP5 inhibition by SBFI-26,a specific inhibitor of FABP5,than in HepG2 cells (IC50=89 and 145 µmol/L) at 6 d upon treatment (Figure 7B and C).Long treatment for 6 d led to further inhibition of cell viability of Huh7 cells than shorter treatment for 3 d (IC50=89 and 749 µmol/L,respectively) (Figure 7D and E).This demonstrated that like many other compounds targeting regulators of cellular mechanism,the anti-tumor effect of FABP5 inhibitor SBFI-26 is a slow process,which requires time to show the anti-tumor effect.Together,these data indicate that HCC cells with high FABP5 expression are sensitive to FABP5 inhibition.
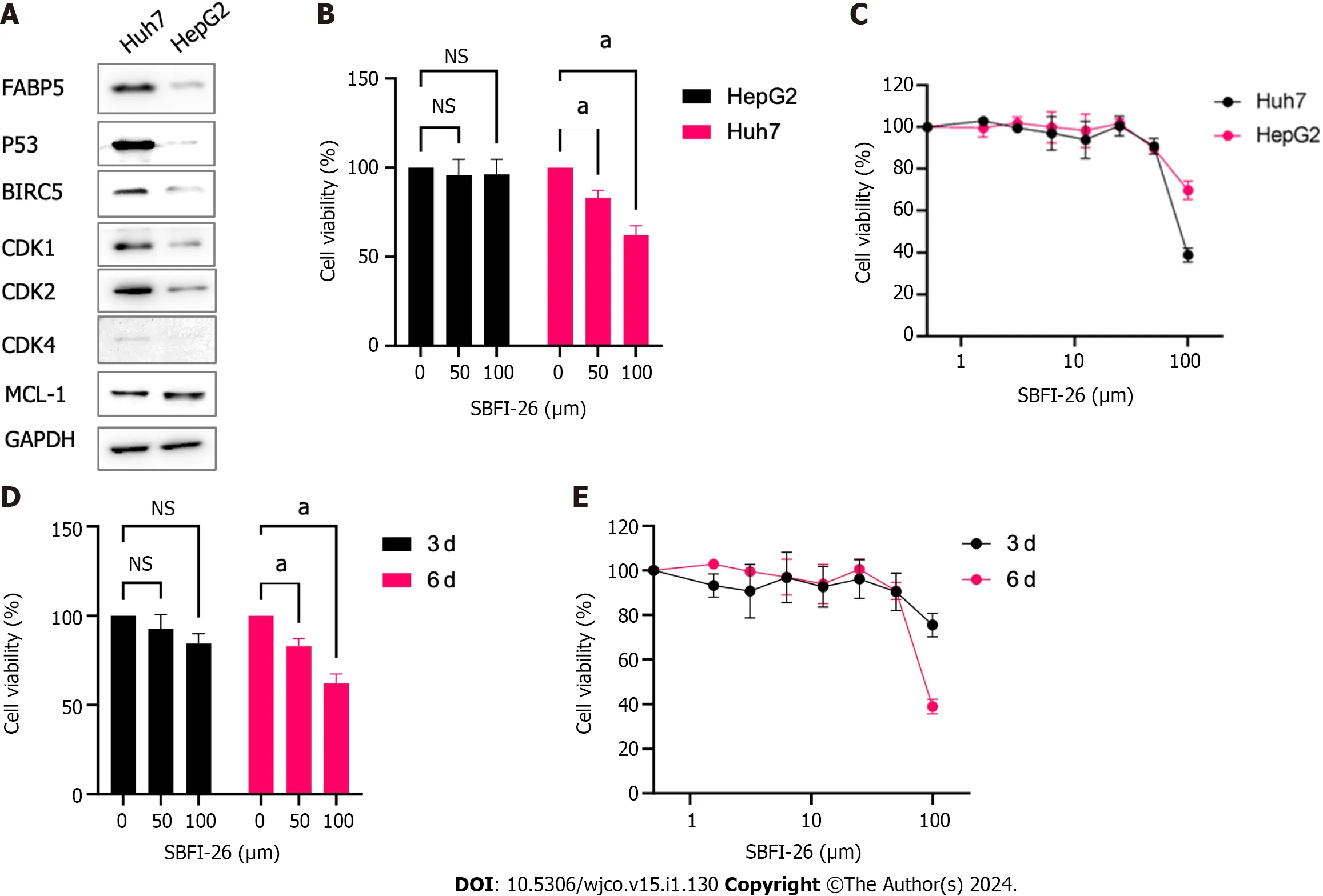
Figure 7 Huh7 cells are sensitive to FABP5 inhibition by SFBl-26. A: Western blot shows the expression of FABP5,p53,and other proteins in Huh7 and HepG2 cells;B: 3-dose (0,50 and 100 µmol/L) cell viability assay shows SBFI-26 inhibited the cell viability of hepatocellular carcinoma (HCC) cell lines Huh7 and HepG2 cells post treatment for 6 d;C: 8-dose (2-fold serial doses up to 100 µmol/L) cell viability assay showed SBFI-26 inhibited the cell viability of HCC cell lines post treatment for 6 d;D: 3-dose (0,50 and 100 µmol/L) cell viability assay shows SBFI-26 inhibited the cell viability of Huh7 cells in time-dependent manner;E: 8-dose cell viability assay showed SBFI-26 inhibited the cell viability of Huh7 cells at 3-and 6-d post treatment;NS: Not significant.
DlSCUSSlON
In this study,we discovered theFABP5gene is frequently amplified together with other adjacent family membersFABP4,FABP8,FABP9andFABP12as a gene cluster.However,onlyFABP5andFABP4are highly expressed in HCC patients and are significantly upregulated in tumor cells compared to non-tumor controls.Compared toFABP4,FABP5is expressed at higher levels in cancer including LIHC and more differentially expressed in tumor cells compared to nontumor controls.Consistent with our data,Ohataetal[14] performed immunohistochemical staining of FABP5 for 243 paired HCC and adjacent non-tumor liver tissue samples.The study confirmed that all normal liver cells were stained negatively,while liver tumor cells can be divided into two groups,FABP5 positive (57.2%) and FABP5 negative (42.8%).Therefore,this data supports that FABP5 is overexpressed in 57.2% of patients with liver tumors.Our data of FABP5 expression in HepG2 and Huh7 cells is consistent with published data in this study as well.This study showed a positive correlation of high FABP5 expression with distant metastasis and invasion.However,Huh7 and HepG2 are both considered to be low metastatic.Our data showed that high expression ofFABP5mRNA correlated well with G2M checkpoint (P=1.1e-10,r=0.33) and TP53 signaling in liver cancer cells (P=1.7e-5,r=0.22) (Figures 4 and 5).We confirmed some of these gene expression differences involved in these two signaling networks including CDK1,CDK2,CDK4,and BIRC5 by western blotting in FABP5 low expressing HepG2 cells and FABP5-high expressing Huh7 cells (Figure 7A).The hotspot mutation Y220C ofTP53gene results in its decreased DNA binding and reduced p53 tumor suppressor function,leading to cancer progression[15-17].This may explain that Huh7 cells carrying TP53 Y220C mutation grow much faster than hepG2 cells with wild type TP53 (cell doubling time,24 and 48 h,respectively).Therefore,it is possible that the TP53 genetic status affects the cell proliferation and expression of FABP5,which requires further validation.
Furthermore,the correlation ofFABP5expression with poor patient survival is more significant than that ofFABP4expression.These data suggestFABP5is the predominant gene across FABPs that are dysregulated in cancer,and it is the most important member that contributes significantly to cancer malignancy and progression,especially in patients with HCC.It is interesting to find out that highFABP5expression correlates well with top cancer hallmarks and two oncogenesPLK1andBIRC5that were identified in HCC patients in our and other studies earlier[12,13].These data strongly suggest thatFABP5is a novel therapeutic target for treating HCC and provides valuable insights for potential therapeutic development in treating patients with HCC.
Small molecule inhibitors targeting FABP proteins,especially FABP4,are currently under development by multiple efforts.Early preclinical data provide evidence that targeting FABP4 by BMS309403 is promising in treating cancer for multiple cancer disease models[2,7,18].In this study,our data suggests that FABP5 is a novel therapeutic target in patients with HCC and other cancer types.FABP5 inhibitors such as SFBI-26 are emerging and demonstrate that targeting FABP5 is feasible and promising in treating cancer[13].
We found that the gene cluster withFABP4,FABP5,FABP8,FABP9,andFABP12in adjacent loci in chromosome 8 are often co-amplified in many cancer types but with highest frequencies in PRAD and HCC.Amplification is the most common genetic alteration type for these FABPs.In contrast,other family membersFABP1,FABP2,FABP3,FABP6andFABP7also showed expression in cancer,ubiquitously or selectively,but are not frequently altered at genetic level.Interestingly,not all co-amplified family members are expressed in cancer due to amplification.OnlyFABP4andFABP5are expressed across various cancer types,suggesting that genetic amplification itself is necessary but not sufficient for their abnormal expression in cancer.Expression ofFABP4,as a major target of PPARγ[19],has been shown to be controlled by PPARγ[4],andFABP4has been shown to negatively regulate PPARγ expression level,likely through a negative feedback signaling loop.In contrast toFABP4,FABP5has been shown to facilitate fatty-acid induced PPARγ activation and downstream signaling,and activated PPARγ in turn upregulates FABP5 expression levels in prostate cancer[10].Whether this is also the case in HCC requires further investigation.In this study,we discovered thatFABP5is the one with greatest changes in mRNA expression across family members in patients with LIHC comparing tumor with non-tumor tissues and its expression highly correlates with poor patient outcome and enriched cancer hallmarks involved in cell cycle progression.
Dysregulated cell metabolism and cell cycle progression are key interconnecting events for cancer malignancy and progression[20].The top three cancer hallmarks previously reported in HCC patients are E2F targets,G2M checkpoint and DNA repair[13].All these lead to dysregulated cell cycle progression.Interestingly,we discovered thatFABP5expression highly correlates with top cancer hallmarks enriched in LIHC patients (G2M checkpoint),which likely drive cancer cell survival and proliferation[13].In our previous study,we demonstrated that two oncogenes,PLK1andBIRC5,are highly co-expressed in HCC and co-targeting of PLK1 and BIRC5 synergistically inhibited tumor growth of HCC preclinical modelsinvitroandinvivo[12].Both PLK1 and BIRC5 are master regulators in cell cycle,powerful in promoting cell cycle progression and inhibiting cell death[21,22].In this study,we confirmed thatPLK1andBIRC5are coexpressed in cancer including HCC.More interestingly,FABP5expression correlates very well with expression ofPLK1andBIRC5in multiple cancer types.The strong correlation ofFABP5with cell cycle hallmark and cell cycle master regulators suggests its critical role in contribution to cancer cell progression when overexpressed.
In addition to its upregulation and functions in tumor cells,FABP5 is also found to be dysregulated in multiple immune cell types and can serve as a novel immune-related prognostic marker and a target of immunotherapy[23].FABP5 was reported to regulate mitochondrial integrity and functions as cell-intrinsic checkpoint for Treg suppressive function in tumor microenvironment[24].However,how FABP5 is dysregulated and the underlying mechanism in antitumor immunity have not been fully understood and requires further investigation.
CONCLUSlON
FABP5is frequently amplified in HCC,leading to its abnormal expression.HighFABP5expression correlates well with worse patient outcome,enriched cancer hallmarks and oncogenes in HCC.Targeting FABP5 by SBFI-26 is more effective in FABP5-high expressing cells than FABP5-low expressing cells.
ARTlCLE HlGHLlGHTS
Research background
FABP5amplification can serve as a prognosis biomarker for the prediction of patient outcome and as a novel therapeutic target for treating hepatocellular carcinoma (HCC) withFABP5amplification.
Research motivation
FABP5is frequently amplified in cancer,especially in HCC,which leads to its abnormal expression in HCC.HighFABP5expression correlates well with worse patient outcome,enriched cancer hallmarks and oncogenes in HCC.Targeting FABP5 by SBFI-26 is more effective in FABP5-high expressing cells (Huh7) than FABP5-low expressing cells (HepG2).
Research objectives
FABP4,FABP5,FABP8,FABP9andFABP12as a gene cluster is frequently amplified in cancer,which is the most common genetic alteration for FABPs.FABP5is highly overexpressed in cancer and its expression correlates well with worse patient outcomes.FABP5expression highly correlates with enrichment of G2M checkpoint,TP53 signaling pathway,and many genes in the gene sets such asCDK1,CDK4,andTP53.Furthermore,FABP5also correlates well with two coexpressed oncogenesPLK1andBIRC5in liver hepatocellular carcinoma (LIHC) patients.FABP5-high expressing Huh7 cells also expressed higher protein levels of p53,BIRC5,CDK1,CDK2,and CDK4 than FABP5-low expressing HepG2 cells.Huh7 is more sensitive to FABP5 inhibition than HepG2 cells.
Research methods
In this study,we accessed the public available portal of The Cancer Genome Atlas datasets of pan cancer and liver hepatocellular carcinoma LIHC by using cBioPortal and GEPIA2 portal.Based on mutation and CNA (copy number variation) datasets,we investigated genetic alterations of FABP family members in various cancer types.Based on mRNA datasets,we investigated FABP expression and their correlation with patient clinical outcome,enriched cancer hallmarks and oncogenes.For validation,we determined the protein levels of FABP5 and its correlated genes in Huh7 and HepG2 and evaluated the potential of targeting FABP5 in treating HCC.
Research results
The present study aimed to understand the genetic alterations and expression of FABP family members and their associated cancer hallmarks and oncogenes in contributing to cancer malignancies,especially HCC.
Research conclusions
Several family members includingFABP4,FABP5,andFABP7are abnormally expressed in cancer cells beyond tissue expression restriction and play important roles in cancer malignancy and progression.However,the mechanism leading to their abnormal expression is not clear.
Research perspectives
Reprogrammed lipid metabolism plays crucial roles in cell survival,growth,and evolution in many cancer types including HCC,an aggressive subtype of liver cancer.Fatty acid binding protein (FABP) family members including FABP5 play important roles in contribution to cancer progression and metastasis;however,how these FABP members,especially FABP5,are dysregulated in HCC and their contribution to HCC cancer progression have not been well defined.
ACKNOWLEDGEMENTS
We would like to thank the patients and their families who contributed to this research study.We also appreciate the availability of the TCGA dataset and the built-in analysisviathe web-based bioinformatic platforms cBioportal and GEPIA2.
FOOTNOTES
Co-first authors:Yan Li and William Lee.
Author contributions:Li Y conceived and designed the study;Li Y,Lee W,Zhao ZG,Liu Y,Cui H,and Wang HY performed the experiments and analyzed the data;Li Y,Lee W,Zhao ZG,Liu Y,Cui H,and Wang HY interpreted the data;Li Y and Lee W drafted the manuscript;Liu Y,Cui H,and Wang HY revised the manuscript;All authors approved the final version of the article.Lee W contributed primarily to the bioinformatics analysis and therefore Lee W and Li Y contributed equally to the study.Li Y conceived and designed the study,performed the experiments,analyzed and interpreted the data,drafted and revised the manuscript.Lee W contributed to the bioinformatics analysis,data analysis and interpretation,as well as manuscript drafting and revision.Both authors have made substantial contributions to the study and are therefore qualified as co-first authors of the paper.
Supported byTianjin Key Medical Discipline Construction Project,No.TJYXZDXK-034A.
lnstitutional review board statement:This study was approved by the Institutional Review Board (IRB) of Tianjin Third Central Hospital.
Conflict-of-interest statement:All authors have seen and agreed with the contents of the manuscript and there is no conflict of interest to declare.
Data sharing statement:For this bioinformatics analysis,we acquired the TCGA data and LIHC data as well as the data for other cancer patients from cBioPortal (https://www.cbioportal.org/).The bioinformatic platform GEPIA2 also used the TCGA datasets for the builtin analysis.No additional data are available.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers.It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license,which permits others to distribute,remix,adapt,build upon this work non-commercially,and license their derivative works on different terms,provided the original work is properly cited and the use is non-commercial.See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:China
ORClD number:Yan Li 0000-0003-2602-7423;William Lee 0009-0008-2736-6025;Zhen-Gang Zhao 0000-0001-7249-4474;Yi Liu 0000-0001-8082-8868;Hao Cui 0000-0002-5629-7814;Hao-Yu Wang 0000-0001-6672-2733.
S-Editor:Liu JH
L-Editor:Filipodia
P-Editor:Zhang XD
杂志排行
World Journal of Clinical Oncology的其它文章
- Prognostic factors of breast cancer brain metastasis
- Radiotherapy for hyoid bone metastasis from lung adenocarcinoma: A case report
- What are the changes in the hotspots and frontiers of microRNAs in hepatocellular carcinoma over the past decade?
- Gene signatures to therapeutics: Assessing the potential of ivermectin against t(4;14) multiple myeloma
- Predicting colorectal cancer prognosis based on long noncoding RNAs of disulfidptosis genes
- ldentification of the key genes and mechanisms associated with transcatheter arterial chemoembolisation refractoriness in hepatocellular carcinoma
