Circulating RNA ZFR promotes hepatocellular carcinoma cell proliferation and epithelial-mesenchymal transition process through miR-624–3p/WEE1 axis
2024-03-04LiZhngSiHeHoGunYoZhoDiZhng
Li Zhng ,Si He ,Ho Gun ,Yo Zho ,Di Zhng ,∗
a Department of General Surgery, the Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an 710004, China
b Department of Breast Cancer, Shaanxi Provincial Cancer Hospital, Xi’an 710000, China
Keywords: Hepatocellular carcinoma circZFR miR-624–3p WEE1 Liver cancer
ABSTRACT Background: Hepatocellular carcinoma (HCC),the most common type of primary liver cancer,is the fourth leading cause of cancer-related deaths worldwide.Previous evidence shows that the expression of circulating RNA ZFR (circZFR) is upregulated in HCC tissues.However,the molecular mechanism of circZFR in HCC is unclear.Methods: Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) was employed to detect the expression of circZFR,microRNA-624–3p (miR-624–3p) and WEE1 in HCC tissues and cells.RNase R assay and actinomycin D treatment assay were used to analyze the characteristics of circZFR.For functional analysis,the capacities of colony formation,cell proliferation,cell apoptosis,migration and invasion were assessed by colony formation assay,5-ethynyl-2′-deoxyuridine (EdU) assay,flow cytometry assay and transwell assay.Western blot was used to examine the protein levels of WEE1 and epithelial-mesenchymal transition (EMT)-related proteins.The interactions between miR-624–3p and circZFR or WEE1 were validated by dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay.Xenograft models were established to determine the role of circZFR in vivo.Results: circZFR and WEE1 were upregulated,while miR-624–3p expression was reduced in HCC tissues and cells.circZFR could sponge miR-624–3p,and WEE1 was a downstream gene of miR-624–3p.Knockdown of circZFR significantly reduced the malignant behaviors of HCC and that co-transfection with miR-624–3p inhibitor restored this change.Overexpression of WEE1 abolished the inhibitory effect of miR-624–3p mimic on HCC cells.Mechanistically,circZFR acted as a competitive endogenous RNA (ceRNA)to regulate WEE1 expression by targeting miR-624–3p.Furthermore,in vivo studies have illustrated that circZFR knockdown inhibited tumor growth.Conclusions: circZFR knockdown reduced HCC cell proliferation,migration and invasion and promoted apoptosis by regulating the miR-624–3p/WEE1 axis,suggesting that the circZFR/miR-624–3p/WEE1 axis might be a potential target for HCC treatment.
Introduction
Hepatocellular carcinoma (HCC) is one of the leading cause of cancer-related death worldwide [1].HCC is featured by aggressive nature,poor prognosis and limited treatment opportunities,posing a serious threat to people’s health [2].The mainstream treatment for HCC includes liver resection,liver transplantation or percutaneous local ablation,and is usually complemented by radiotherapy and systemic drug therapy,but the recurrence rate is still close to 40% within 1 year after surgery,and the 5-year overall survival rate is only 15%-40% [3,4].Most HCC patients are diagnosed at advanced stages and are unable to be treated by surgical resection [5].Accordingly,there is an urgent need to investigate the mechanisms and development of HCC,as well as to identify new curative targets and specific biomarkers.
Circular RNAs (circRNAs) are novel non-coding RNAs,which are closed-loop structures formed by the variable shearing of specific terminal reverse complementary precursor mRNAs [6,7].circRNAs are found widely in eukaryotic cells and are characterized by structural stability,RNase R resistance and different expression in tissue and cell type,which give circRNAs potential as molecular diagnostic markers of disease [8,9].There is growing evidence that dysregulation of circRNA performs an essential part in the development and progression of a variety of tumors,including HCC [10–12].Yang et al.suggested that circZFR expression was enhanced in HCC tissues,and that the increased circZFR promoted cell proliferation and migration by inhibiting miR-511 [13].Moreover,circZFR knockdown substantially restrained the multiplication of HCC cells and epithelial-mesenchymal transition (EMT),and circZFR promoted HCC progression by regulating the expression of CTNNB1 via miR-3619–5p [14].There are many downstream binding targets of circZFR,but regulation of miR-624-3p by circZFR has not been reported.
MicroRNAs (miRNAs) are endogenous non-coding small RNAs with specific functions in the reverse transcriptional regulation of gene expression and are powerful regulators of a variety of cellular activities [15,16].miRNA has a pivotal position in enhancing cancer progression [17].Correlation study indicated that overexpressed miR-624-3p acted as a sponge for circRNA to target and regulate downstream genes in the development of esophageal squamous cell carcinoma (ESCC) [18,19].Moreover,Lin et al.found that brainderived neurotrophic factor (BDNF) promoted tumor lymphangiogenesis by downregulating miR-624-3p expression in chondrosarcoma [20].With the progress of bioinformatics,miR-624-3p has been predicted as the target of circZFR,but whether miR-624-3p participates in the regulation of HCC by circZFR needs further study.
The WEE1 kinase family is composed of three serine/threonine kinases that shares a conserved molecular structure and serves as part of the DNA damage response (DDR) pathway,which has a critical role in cell cycle regulation [21,22].It has been reported that WEE1 can be involved in regulating cancer progression.For example,Gao et al.[23]presented that HEIH expedited retinoblastoma progression via mediating miR-194-5p to regulate WEE1 expression,and Ma et al.[24]reported that circ_0013958 targeted miR-532-3p to promote WEE1 expression and consequently expedite HCC progression.Similar results were reported by Li et al.[25].However,whether WEE1 is regulated by circZFR in HCC is unclear.
The objective of this research was to explore the function and regulatory mechanism of circZFR in HCC and to provide evidence for circZFR as a potential biomarker in HCC.
Methods
Clinical samples collection
HCC tumor tissues and corresponding paracancerous tissues from HCC patients (n=57) were acquired from the Second Affiliated Hospital of Xi’an Jiaotong University.Samples were quickfrozen in liquid nitrogen and stored in a-80 °C refrigerator for subsequent experiments.The study was approved by the Ethics Committee of the Second Affiliated Hospital of Xi’an Jiaotong University,and informed consent was obtained from the patients.
Cell lines and culture
HCC cell lines (MHCC97-H,Huh-7) and normal cell line (THLE-3) were from Bena (Beijing,China) and American Type Culture Collection (ATCC,Manassas,VA,USA).All cell lines were cultured in Roswell Park Memorial Institute 1640 (RPMI-1640;BIOSUN,Shanghai,China) with 10% fetal bovine serum (FBS;Genetimes,Shanghai,China).The cells were cultured with 5% CO2at 37 °C.
Cell transfection
circZFR short-hairpin RNA (sh-circZFR) and its control group(sh-NC),miR-624-3p mimic (miR-624-3p) and its blank control group (miR-NC),miR-624-3p inhibitor (in-miR-624-3p) and its blank control group (in-miR-NC),and overexpression of WEE1 and matching control (pc-DNA) were obtained from RiboBio Ltd.(Guangzhou,China).MHCC97-H and Huh-7 cells (2×105cells/well) were inoculated in 6-well plates.Lipofectamine 2000(Invitrogen,Carlsbad,CA,USA) was then employed to guide the cell transfection.The transfection process was started when the cell density reached 70%-80%.The transfection effect was evaluated by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) assay 48 h after transfection.
qRT-PCR assay
Total RNAs from HCC tissues and cell lines were extracted by Trizol reagent (Invitrogen).And cDNA was generated from total RNA using PrimeScript RT Reagent Kit (TaKaRa,Beijing,China).The relative expression was processed by 2-ΔΔCtmethod,and GAPDH and U6 were perceived as internal reference.All sequences of primers were listed in Table 1.
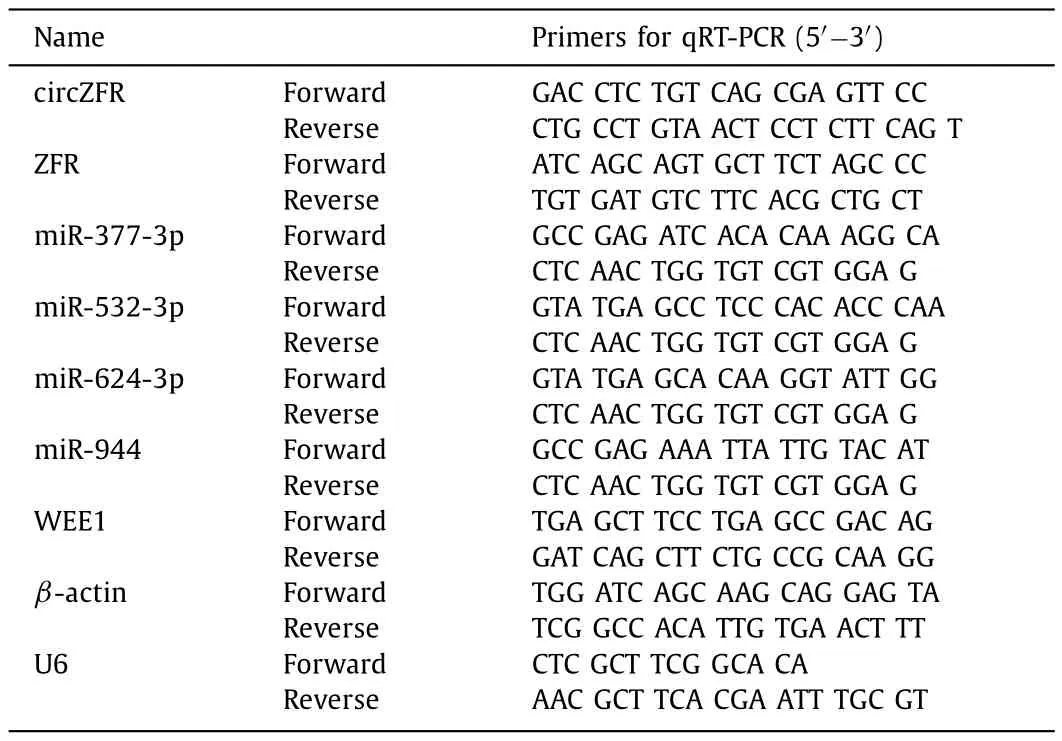
Table 1Primers sequences used for quantitative reverse transcriptase polymerase chain reaction (qRT-PCR).
RNase R assay and actinomycin D treatment assay
The structural stability of circRNAs was assayed with RNase R(Tiangen Biochemical Technology,Beijing,China).RNase R (3 U/μg)or the mock was incubated with total RNA (3 μg) for half an hour at 37 °C.Actinomycin D (2 mg/mL,Sigma-Aldrich,St.Louis,MO,USA) was added to MHCC97-H and Huh-7 cells culture mediums at 4,8,12 and 24 h to inhibit transcription.The corresponding expression level of circZFR and linear molecule in the treated cells was detected using qRT-PCR assay.
Colony formation assay
MHCC97-H and Huh-7 cells were seeded into 6-well plates (200 cells/well) and cultured in complete medium for half a month until the cell colonies were clearly visible.Paraformaldehyde (Beyotime,Shanghai,China) was utilized to immobilize colonies for 10 min,and then colonies were dyed with crystal violet (ComWin Biotech,Beijing,China).The stained colonies were observed and counted under an inverted microscope (Olympus,Tokyo,Japan).
5-Ethynyl-2′ -deoxyuridine (EdU) assay
EdU assay kit (RiboBio,Guangzhou,China) was employed to assess the proliferation of cells.MHCC97-H and Huh-7 cells were seeded in 96-well plates (1×104cells/well) and then incubated with 50 μmol/L EdU buffer.Subsequently,4% formaldehyde was used to immobilize the cells.Each well was incubated with penetrant for 10 min.Conclusively,EdU solution and DAPI (Ribobio)were used to incubate the cells at 37 °C for 20 min.Three fields were randomly selected for statistics,and fluorescence microscopy was used to photograph EdU-positive cells.
Flow cytometry assay
Annexin V-FITC/propidium iodide (PI) Cell Apoptosis Detection Kit (Beyotime) was applied to detect apoptosis.After 48-h culture,the transfected cells were washed by cold PBS.Annexin V-FITC and PI were successively added to stain cells for 15 min.The positive cells were detected and quantified by flow cytometry (Agilent,Santa Clara,CA,USA).
Transwell assay
The 24-well transwell chamber (Corning Incorporated,Corning,NY,USA) was adopted to estimate the capacities of invasiveness(with Matrigel,Corning Incorporated) and migration (without Matrigel) in HCC cells.The transfected MHCC97-H and Huh-7 cells were added into the upper chambers,and 700 μL of 20% FBS was appended to the bottom chamber.Subsequently,the cells were gently removed from the substrate with a few cotton swabs,and were immobilised with methanol and dyed with crystal violet.The number of cells was counted under a high magnification microscope (original magnification×100;Olympus).
Western blot assay
Total proteins were first extracted by radioimmunoprecipitation analysis buffer (RIPA;Millipore Corporation,Billerica,MA,USA).The BCA protein assay kit (Thermo Fisher Scientific,Waltham,MA,USA) was utilized to quantify proteins which were loaded onto 12% SDS-PAGE.The PVDF membranes which were immersed with Tris-HCl buffer solution were subsequently employed to transfer separated proteins.Then membranes were removed to a container containing a hermetic solution (5% skim milk).Following that,the membranes were incubated with primary and secondary antibodies purchased from Abcam (Cambridge,MA,USA),including anti-Bax (1:1000,ab32503,Abcam),anti-E-cadherin (1:5000,ab76319,Abcam),anti-Vimentin (1:1000,ab92547,Abcam),anti-PCNA (1:2000,ab92552,Abcam),anti-p21 (1:2000,ab109520,Abcam),anti-Bcl-2 (1:1000,ab32124,Abcam),anti-WEE1 (1:1000,ab137377,Abcam),anti-βActin (1:1000,ab8227 Abcam) and goat anti-rabbit immunoglobulin G (IgG) H&L (HRP) (1:2000,ab6721,Abcam).The protein bands in membranes were detected with the ECL kit (Thermo Fisher Scientific),and the density of all bands was analyzed using Image J software.
Dual-luciferase reporter assay
The bioinformatics prediction platform starBase (http://starbase.sysu.edu.cn/) was applied to identify the binding sites between miR-624-3p and circZFR or WEE1.The wild-type (WT) and mutanttype (MUT) fragments of circZFR or WEE1 were constructed into downstream part of firefly luciferase gene in pGL3 vectors (Realgene,Nanjing,China),named as circZFR WT,circZFR MUT,WEE1 3′ UTR WT and WEE1 3′ UTR MUT.MHCC97-H and Huh-7 cells were co-transfected with luciferase reporter plasmid and miR-624-3p or miR-NC.Dual-luciferase reporter assay kit (Realgene) was used to detect the luciferase activity after the cells were lysed.
RNA immunoprecipitation (RIP) assay
The immunoprecipitation assay was employed to validate the predicted target relationship between circZFE and miR-624-3p by Magna RIP RNA-binding protein immunoprecipitation kit (Millipore).After being revealed to RIP lysis,MHCC97-H and Huh-7 cells were incubated with magnetic beads coupled to Argonaute 2 (Ago2) antibody or IgG antibody which was regarded as a negative control.After washing the unbinding,the immunoprecipitation RNA was isolated and purified for qRT-PCR.
Xenograft mouse model
Huh-7 cells (5×106cells/200 μL) transfected with sh-NC or shcircZFR were injected into nude mice (n=5 mice/group).Thirtyfive days later,mice were sacrificed and the tumor weight and volume were measured.All animal experiments were approved by the Animal Care and Use Committee of the Second Affiliated Hospital of Xi’an Jiaotong University.
Immunohistochemistry (IHC) assay
Paraffin-embedded HCC tissue slides were dewaxed and hydrated,and then antigen was extracted with sodium citrate buffer.After that,tissue sections were quenched in 0.3% H2O2and sealed with 5% goat serum.The slides were hatched with anti-Ki67 (1:1000,ab15580,Abcam),anti-Bax (1:250,ab32503,Abcam),anti-E-cadherin (1:8000 0,ab76319,Abcam),anti-Vimentin (1:500,ab92547,Abcam),anti-WEE1 (1:1000,ab137377,Abcam) at 4 °C overnight and subsequently incubated with a ready-to-use secondary antibody (goat anti-rabbit IgG H&L (HRP),1:1000,ab6721,Abcam) at room temperature.Afterwards,the protein signaling was observed under 5 fields using DAB (Vector Laboratories,Peterborough,UK) solution under light microscope.
Statistical analysis
GraphPad Prism 7 (GraphPad,San Diego,CA,USA) was utilized for data analysis.Student’st-test and one-way analysis of variance(ANOVA) followed by Tukey’s test were employed to compare the variables between groups.Pearson’s correlation analysis was applied to measure the correlation between two groups treated differently.And all of data were exhibited as mean ± standard deviation based on ≥3 replicates.P<0.05 was considered statistically significant.
Results
circZFR was upregulated in HCC tissues and cells
The association of clinicopathological features and circZFR expression in HCC tissues was presented in Table 2.Based on the median value (2.08) of circZFR expression,the patients were divided into two groups: low circZFR expression group (n=28)and high circZFR expression group (n=29).High circZFR expression was associated with the malignant manifestations,including lager tumor size,late TNM stage and tumor metastasis (Table 2).The ring structure of circZFR was presented first,showing that circZFR was derived fromZFRgene,located on chromosome 5,and formed by exon 13-exon 17 end to end (Fig.1 A).qRT-PCR confirmed that the expression of circZFR was significantly higher in HCC tissues and cancer cells (MHCC97-H and Huh-7) than that in adjacent tissues and non-cancer cell line (THLE-3) (Fig.1 B and C).RNase R method and actinomycin D treatment assay showed that circZFR expression was almost not affected by RNase R digestion or actinomycin D treatment,indicating its high stability,while linear molecule ZFR was greatly degraded by RNase R digestion or actinomycin D (Fig.1 D-G).
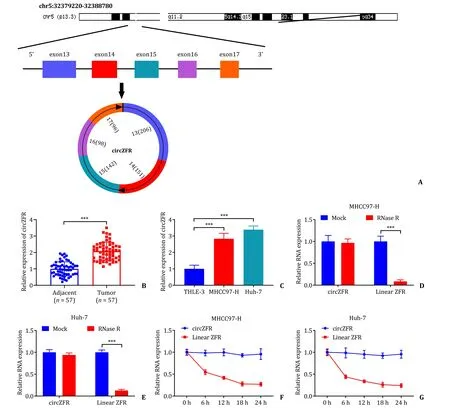
Fig.1. circZFR was upregulated in HCC tissues and cells.A: The structure of circZFR;B and C:the expression of circZFR in tumor tissues (n=57) and adjacent normal tissues,cancer cells (MHCC97-H and Huh-7) and non-cancer cells (THLE-3) detected by qRT-PCR;D and E:the stability of circZFR detected by RNase R degradation;F and G:the stability of circZFR detected by actinomycin D.∗∗∗: P < 0.001.HCC: hepatocellular carcinoma;qRT-PCR: quantitative reverse transcriptase polymerase chain reaction.

Table 2Relationship between clinicopathological characteristics and circZFR expression in patients with hepatocellular carcinoma (n=57).
circZFR knockdown repressed the proliferation and enhanced the apoptosis of HCC cells
The efficacy of circZFR knockdown was shown in Fig.2 A,and the expression of linear ZFR was not affected in MHCC97-H and Huh-7 cells after sh-circZFR transfection compared to sh-NC (Fig.S1).Colony formation assay showed that sh-circZFR significantly decreased the number of colonies in MHCC97-H and Huh-7 cells(Fig.2 B).Similarly,the EdU assay revealed that sh-circZFR suppressed the proliferation of HCC cells as the transfection of shcircZFR obviously reduced the EdU positive cell rate in comparison to sh-NC (Fig.2 C).Flow cytometry showed that sh-circZFR promoted apoptosis in MHCC97-H and Huh-7 cells (Fig.2 D).Transwell assay found that the numbers of migrated and invaded cells transfected with sh-circZFR were significantly reduced (Fig.2 E and F).Bax protein was known to promote apoptosis and E-cadherin and Vimentin proteins were associated with invasive metastasis of tumor cells.Western blot assay indicated that sh-circZFR elevated the protein expression levels of Bax,E-cadherin and p21,and that sh-circZFR declined the protein expression level of Vimentin,PCNA and Bcl-2 (Fig.2 G-J).These data underlined the fact that circZFR downregulation inhibited the proliferation of HCC cells.
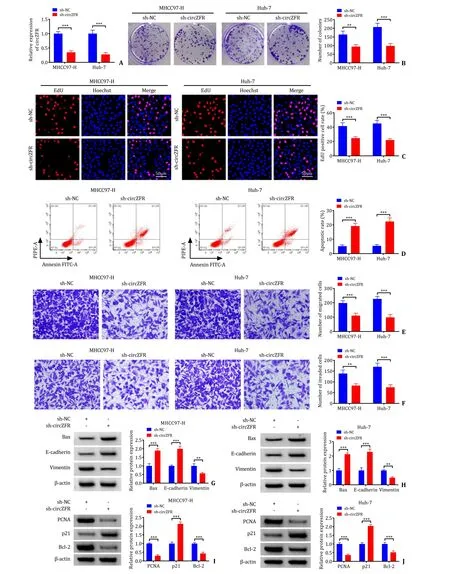
Fig.2. Effect of knockdown circZFR on the proliferative and apoptotic functions of HCC cells.A: The expression level of circZFR determined by qRT-PCR in MHCC97-H and Huh-7 cells transfected with sh-NC or sh-circZFR;B: colony formation assay was used to assess proliferation of transfected MHCC97-H and Huh-7 cells;C: EdU proliferation assay was performed in transfected MHCC97-H and Huh-7 cells;D: apoptosis of MHCC97-H and Huh-7 cells was detected by flow cytometry;E and F: the role of circZFR on cell migration and invasion was monitored by transwell assay;G-J: the protein expression levels of Bax,E-cadherin,Vimentin,PCNA,p21 and Bcl-2 were determined by Western blot.∗∗: P < 0.01;∗∗∗: P < 0.001.qRT-PCR: quantitative reverse transcriptase polymerase chain reaction;EdU: 5 Ethynyl 2′ deoxyuridine.
circZFR regulated miR-624-3p expression
The circBank (http://www.circbank.cn/),starBase (https://starbase.sysu.edu.cn/),and circInteractome (https://circinteractome.nia.nih.gov/) tools predicted that four miRNAs bind to circZFR,including miR-377-3p,miR-532-3p,miR-624-3p and miR-944(Fig.3 A).The qRT-PCR assay revealed that circZFR knockdown hardly affected miR-377-3p expression but upregulated miR-532-3p,miR-624-3p and miR-944 expression,among which miR-624-3p expression was the most affected,we therefore selected miR-62-–3p for the follow-up experiment (Fig.3 B and C).miR-624-3p was predicted to be a target of circZFR using bioinformatics tools and the specific binding site was shown in the Fig.3 D.Subsequently,the efficacy of miR-624-3p mimic in MHCC97-H and Huh-7 cells was detected,showing that miR-624-3p expression was greatly elevated after miR-624-3p transfection (Fig.3 E).Dual-luciferase reporter assay displayed that co-transfection of miR-624-3p and circZFR WT significantly inhibited luciferase activity in MHCC97-H and Huh-7 cells,while co-transfection of miR-624-3p and circZFR MUT produced no significant changes (Fig.3 F and G).RIP experiments confirmed that circZFR and miR-624-3p could be immunoprecipitated by Ago2 antibody (Fig.3 H and I),further suggesting an interaction between circZFR and miR-624-3p.Furthermore,the results from qRT-PCR assay demonstrated that miR-624-3p expression was clearly lower in HCC tumor tissues and cell lines (Fig.3 J and K).And Pearson’s correlation analysis revealed a negative correlation between circZFR and miR-624-3p (Fig.3 L).Therefore,the above data showed that miR-624-3p was the target of circZFR.

Fig.3. circZFR targeted miR-624-3p.A: Venn diagram illustrated miRNAs that could be combined with circZFR,and miRNAs were predicted from different sites;B and C:detection of miRNAs expression in MHCC97-H and Huh-7 cells transfected with sh-circZFR by qRT-PCR assay;D: the starBase predicted the candidate targets of circZFR and showed the mutant binding sites between miR-624-–3p and circZFR;E: the efficiency of miR-624-3p mimic in MHCC97-H and Huh-7 cells was detected by qRT-PCR;F and G: dual-luciferase reporter assays were performed to confirm the association between miR-624-3p and circZFR;H and I: the relationship between miR-624-3p and circZFR was verified by RIP assay;J: the expression of miR-624-3p in HCC tissues (n=57) and adjacent normal tissues was tested by qRT-PCR;K: miR-624-3p expression in THLE-3,MHCC97-H and Huh-7 cells was tested by qRT-PCR;L: Pearson’s correlation analysis showed the correlation between miR-624-3p expression and circZFR expression.∗∗∗: P< 0.001.qRT-PCR: quantitative reverse transcriptase polymerase chain reaction.
The in-miR-624-3p alleviated the inhibition of cell malignant behavior caused by sh-circZFR
qRT-PCR showed that miR-624-3p inhibitor significantly decreased the miR-624-3p (Fig.4 A).Colony formation assay revealed that in-miR-624-3p restored the inhibitory effect on the colony formation caused by sh-circZFR (Fig.4 B and S2A).Furthermore,EdU assay indicated that transfection of sh-circZFR significantly reduced the EdU positive cells,meaning that sh-circZFR inhibited the proliferation of HCC cells,while co-transfection with in-miR-624-3p alleviated this inhibition caused by sh-circZFR (Fig.4 C,S2B and S2C).Co-transfection with in-miR-624-3p suppressed the promotion of apoptosis induced by sh-circZFR as detected by flow cytometry analysis (Fig.4 D and S2D).Transwell assay demonstrated that cotransfection with in-miR-624-3p restored the inhibitory effect of sh-circZFR on cell migration and invasion (Fig.4 E and F).Western blot results disclosed that transfection of sh-circZFR in MHCC97-H and Huh-7 cells increased Bax,E-cadherin and p21 protein expression and declined the protein expression of Vimentin,PCNA and Bcl-2,while co-transfection of sh-circZFR and in-miR-624-3p eliminated the effects (Fig.4 G-J).These data suggested that circZFR regulated the proliferation,apoptosis,migration,and invasion of HCC cells by targeting miR-624-3p.
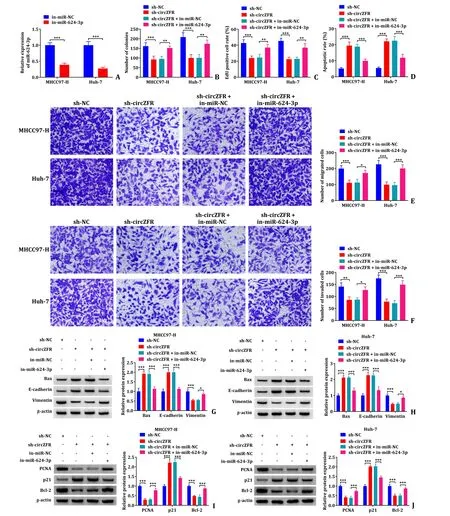
Fig.4. The in-miR-624-3p alleviated the inhibition of cell behavior caused by sh-circZFR.A: Detection of transfection efficiency of miR-624-3p inhibitor was performed using qRT-PCR assay;B: colony formation assay was used to assess proliferation of transfected MHCC97-H and Huh-7 cells;C: EdU proliferation assay was performed in transfected MHCC97-H and Huh-7 cells;D:apoptosis of MHCC97-H and Huh-7 cells was detected by flow cytometry;E and F: the cell migration and invasion were monitored by transwell assay;G-J: the protein expression levels of Bax,E-cadherin,Vimentin,PCNA,p21 and Bcl-2 were determined by Western blot.∗: P < 0.05;∗∗: P <0.01;∗∗∗: P < 0.001.qRT-PCR: quantitative reverse transcriptase polymerase chain reaction;EdU: 5 Ethynyl 2′ deoxyuridine.
WEE1 was a target of miR-624-3p
As shown in the Fig.5 A,starBase predicted that miR-624-3p has a target binding site to WEE1.Dual-luciferase reporter assay found that co-transfection of miR-624-3p and WEE1 3′ UTR WT significantly inhibited luciferase activity in MHCC97-H and Huh-7 cells,while co-transfection of miR-624-3p and WEE1 3′ UTR MUT had no significant changes (Fig.5 B and C).Furthermore,the results from qRT-PCR assay and Western blot indicated that WEE1 mRNA expression and protein expression levels were significantly higher in HCC tumor tissues and cell lines (Fig.5 D-G).Therefore,the above data implicated that WEE1 was the target of miR-624-3p.
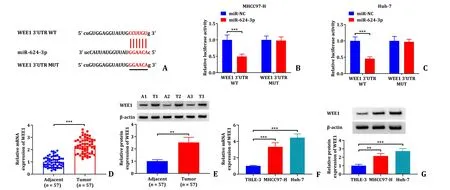
Fig.5. The validation of the relationship between miR-624-3p and WEE1 on targeting.A: The complementary sequences between miR-624-3p and WEE1;B and C: dualluciferase reporter assays were performed to confirm the association between miR-624-3p and WEE1 3′ UTR;D-G: WEE1 mRNA expression and protein expression level in HCC tumor tissue and cell lines were detected by qRT-PCR assay and Western blot.∗∗: P < 0.01;∗∗∗: P < 0.001.HCC: hepatocellular carcinoma;qRT-PCR: quantitative reverse transcriptase polymerase chain reaction.
circZFR regulated the miR-624-3p/WEE1 axis
The associations of circZFR,miR-624-3p and WEE1 were further investigated on the basis of the previous results.Western blot results disclosed that transfection of sh-circZFR significantly reduced the protein expression levels of WEE1 in MHCC97-H and Huh-7 cells,while co-transfection of sh-circZFR and in-miR-624-3p recovered their levels (Fig.6 A and B).Pearson’s correlation analysis exhibited a negative correlation between WEE1 expression and miR-624-3p expression (r=-0.6171,P<0.001),and a positive correlation between WEE1 expression (r=0.6330,P<0.001) and circZFR expression in tumor tissues (Fig.6 C and D).The data verified that circZFR positively enhanced WEE1 expression via targeting miR-624-3p.
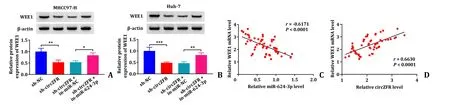
Fig.6. Relationship between circZFR,miR-624-3p and WEE1.A and B: The protein expression level of WEE1 in MHCC97-H and Huh-7 cells was determined by Western blot;C and D: Pearson’s correlation analysis showed the correlation between WEE1 expression and miR-624-3p expression or circZFR expression.∗: P < 0.05;∗∗: P < 0.01;∗∗∗: P< 0.001.
Overexpression of WEE1 recovered HCC cell malignant behaviors suppressed by miR-624-3p mimic
As shown in Fig.7 A,WEE1 level was increased in MHCC97-H and Huh-7 cells transfected with overexpressed WEE1.Subsequent Western blot indicated that miR-624-3p reduced the protein expression level of WEE1,while co-transfection with overexpressed WEE1 recovered its level (Fig.7 B).Colony formation assay showed that WEE1 overexpression alleviated the inhibitory effect on the colony formation caused by miR-624-3p (Fig.7 C and S3A).What’s more,EdU assay proved that transfection of miR-624–3p significantly reduced the EdU-positive cells of HCC,meaning that miR-624-3p inhibited the proliferation of HCC cells,however co-transfection of WEE1 increased the number of EdU-positive cells (Fig.7 D,S3B and S3C).Co-transfection with WEE1 suppressed the apoptotic effect of miR-624-3p as detected by flow cytometry analysis (Fig.7 E and S3D).Transwell assay demonstrated that co-transfection with WEE1 alleviated the inhibitory effects of miR-624-3p on cell migration and invasion (Fig.7 F-H).Furthermore,Western blot results disclosed that transfection of miR-624-3p in MHCC97-H and Huh-7 cells increased Bax,E-cadherin and p21 protein expressions and declined the protein expression levels of Vimentin,PCNA and Bcl-2,while co-transfection of WEE1 and miR-624-3p eliminated the effects (Fig.7 I-L).These data suggested that miR-624-3p regulated the proliferation,apoptosis,migration,and invasion of HCC cells by targeting WEE1.
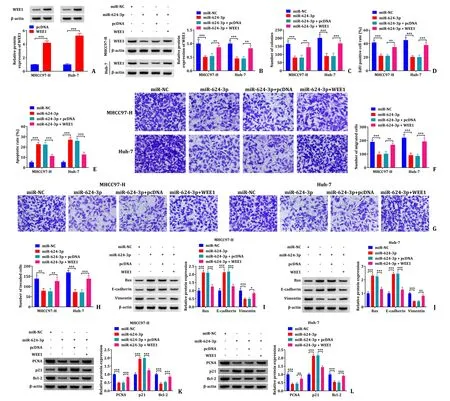
Fig.7. Overexpression of WEE1 alleviated the inhibition of cell malignant behavior caused by miR-624-3p mimic.A: Detection of transfection efficiency of WEE1 overexpression in MHCC97-H and Huh-7 cells by Western blot;B: the protein expression level of WEE1 was determined by Western blot;C: colony formation assay was used to assess proliferation of transfected MHCC97-H and Huh-7 cells;D: EdU proliferation assay was performed in transfected MHCC97-H and Huh-7 cells;E: apoptosis of MHCC97-H and Huh-7 cells was detected by flow cytometry;F-H: the cell migration and invasion were monitored by transwell assay;I-L: the protein expression levels of Bax,E-cadherin,Vimentin,PCNA,p21 and Bcl-2 determined by Western blot.∗∗: P < 0.01;∗∗∗: P < 0.001.
circZFR knockdown restricted tumorigenicity of HCC cells in vivo
Tumor volume and weight in the sh-circZFR group were smaller in comparison to the sh-NC group (Fig.8 A-C).The qRT-PCR results showed that the expression of circZFR and WEE1 was significantly lower in the sh-circZFR group and that miR-624-3p expression was significantly higher than that in the sh-NC group (Fig.8 D).Western blot results disclosed that transfection of sh-circZFR significantly reduced the protein expression level of WEE1 in subcutaneous tumor tissues (Fig.8 E).The data from IHC assay also showed that the expression level of WEE1 in the sh-circZFR group was lower than that in the sh-NC group (Fig.8 F).Western blot indicated that sh-circZFR repressed the protein expression of Vimentin,and increased the protein expression of Bax and E-cadherin (Fig.8 G).Furthermore,IHC results showed that the expression levels of Vimentin and Ki67 were significantly decreased in the sh-circZFR group and the expression of Bax and E-cadherin were significantly increased in the sh-circZFR group (Fig.8 H).The data strongly supported that circZFR knockdown restrained tumor growth in animal models via regulation of miR-624-3p and WEE1 expressions.
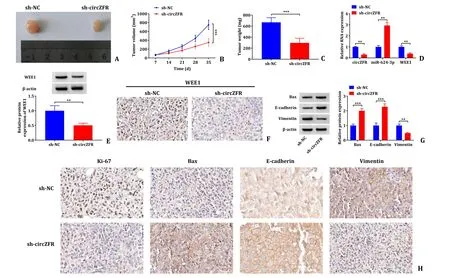
Fig.8. circZFR knockdown restricted tumorigenicity of HCC cells in vivo.A-C: Tumor volume and weight after sh-circZFR in vivo ;D: the expression levels of circZFR,miR-624-3p and WEE1 mRNA were determined by qRT-PCR;E: the protein expression level of WEE1 determined by Western blot;F: the positive rate of WEE1 analyzed by IHC;G: the protein expression levels were determined by Western blot;H: the positive rate of Ki-67,Bax,E-cadherin and Vimentin analyzed by IHC.∗∗: P < 0.01;∗∗∗: P < 0.001.qRT-PCR: quantitative reverse transcriptase polymerase chain reaction;IHC: immunohistochemistry.
Discussion
Competitive endogenous RNA (ceRNA) is a natural decoy for libraries of common miRNAs [26,27].ceRNA regulates a variety of biological processes,including cancer,through shared miRNA interactions [25].Here we showed,for the first time,the role of circZFR in HCC cell functional properties through regulating the miR-624-3p/WEE1 axis.
Analysis of the functions and mechanisms of circRNA in tumor is a current hot topic of research.For example,hsa_circ_0091581 expression was enhanced in HCC tissues and cells and promoted cell proliferation via the miR-526b/c-Myc axisinvitroandinvivo[28].Interestingly,it has been confirmed that circZFR was upregulated in HCC tissues,and that circZFR regulated HMGA2 expression by sponging miR-375,thus acting as a facilitator in the progression of HCC [29].Besides,silencing circZFR significantly reduced the expression of the target geneAKT1through upregulating miR-511,thereby inhibiting malignant behavior of HCC cells [13].Consistent with these previous findings,our data also confirmed that circZFR was abnormally upregulated in HCC tissues and cell lines.In function,circZFR knockdown retarded the multiplication,invasion and migration of HCC cells and promoted apoptosisinvitro.Ourinvivoexperiments demonstrated that circZFR downregulation also inhibited tumor growth in nude mice.It is recommended that circZFR may have a facilitating function in HCC progression.
Bioinformatics analysis identifies a number of downstream binding targets for circZFR,among which the binding site between miR-624-3p and circZFR is highly conserved,but the mechanism of action between them has not yet been reported.A previous study reported that miR-624 was poorly expressed in HCC cells [30].In accordance with this study,our data verified that miR-624-3p was downregulated in HCC tissues and cells.A subsequent negative regulatory role between miR-624-3p and circZFR was verified by Pearson’s correlation analysis.Furthermore,functional analysis indicated that miR-624-3p inhibition restored the inhibited malignant behaviors of HCC cells caused by circZFR deficiency.It confirmed that circZFR promoted HCC cell proliferation by targeting miR-624-3p.miRNAs regulate tumor progression and metastasis by binding to target genes to regulate mRNA degradation or translational suppression [31].In this experiment,we confirmed WEE1 as a downstream target of miR-624-3p.Previous studies have revealed that WEE1 was highly expressed in HCC,and its overexpression enhanced EMT and cell migration in HCC cells and also promoted HCC tumourigenesisinvivo[25,32].Consistent with these findings,our data demonstrated that WEE1 was highly expressed in HCC tissues and cells.Interestingly,WEE1 expression in HCC was negatively associated with miR-624-3p expression but positively correlated with circZFR expression.Overexpression of WEE1 eliminated the blocking effect of miR-624-3p mimic on the malignant behavior of HCC cells,further demonstrating the action mechanism of circZFR/miR-624-3p/WEE1 axis in HCC development.
In conclusion,our study demonstrated that circZFR expression was upregulated in HCC tissues and cells,and that abnormal circZFR expression was associated with adverse clinical outcomes in HCC patients.circZFR positively regulated WEE1 through sponging miR-624-3p,thereby promoting HCC progression.These data may provide new targets for subsequent research and treatment of HCC.
Acknowledgments
None.
CRediT authorship contribution statement
Li Zhang:Conceptualization,Formal analysis,Writing– original draft,Writing– review &editing.Sai He:Investigation,Methodology,Writing– original draft.Hao Guan:Data curation,Software,Writing– original draft.Yao Zhao:Software,Supervision.Di Zhang:Project administration,Writing– review &editing.
Funding
None.
Ethical approval
This study was approved by the Ethics Committee of the Second Affiliated Hospital of Xi’an Jiaotong University.Written informed consent was obtained from all participants.Animal study was approved by the Animal Care and Use Committee of the Second Affiliated Hospital of Xi’an Jiaotong University.
Competing interest
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
Supplementary materials
Supplementary material associated with this article can be found,in the online version,at doi:10.1016/j.hbpd.2023.07.003.
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Recent advances in promising drugs for primary prevention of gastroesophageal variceal bleeding with cirrhotic portal hypertension
- Stereotactic body radiotherapy in pancreatic adenocarcinoma
- Application of ultrasonography-elastography score to suspect porto-sinusoidal vascular disease in patients with portal vein thrombosis
- Polydatin ameliorates hepatic ischemia-reperfusion injury by modulating macrophage polarization
- Hypomethylation of glycine dehydrogenase promoter in peripheral blood mononuclear cells is a new diagnostic marker of hepatitis B virus-associated hepatocellular carcinoma
- AGK2 pre-treatment protects against thioacetamide-induced acute liver failure via regulating the MFN2-PERK axis and ferroptosis signaling pathway
