A novel pathogen Fusarium cuneirostrum causing common bean(Phaseolus vulgaris) root rot in China
2024-01-17DongDengWenqiWuCanxingDuanSuliSunZhendongZhu
Dong Deng,Wenqi Wu,Canxing Duan,Suli Sun,Zhendong Zhu
Institute of Crop Sciences,Chinese Academy of Agricultural Sciences,Beijing 100081,China
Abstract Several fungal pathogens cause root rot of common bean,among which Fusarium spp.are the most common pathogens causing Fusarium root rot (FRR) worldwide.FRR has been becoming an increasingly severe disease of common bean in China,but the species of Fusarium spp.have remained unclear.Thus,this study was performed to identify the pathogen causing common bean root rot in Liangcheng County,Inner Mongolia,China.Nineteen Fusarium-like isolates were obtained after pathogen isolation and purification.The pathogenicity test indicated that eight isolates caused severe disease symptoms on common bean,while 11 other isolates were not pathogenic.The eight pathogenic isolates,FCL1-FCL8,were identified as Fusarium cuneirostrum by morphological characterization and phylogenetic analysis using partial sequences of EF-1α,ITS,28S,and IGS regions.Host range test showed that the representative F.cuneirostrum isolate FCL3 was also pathogenic to mung bean,while not pathogenic to adzuki bean,chickpea,cowpea,faba bean,pea,and soybean.Moreover,50 common bean and 50 mung bean cultivars were screened for resistance to FRR,and seven highly resistant or resistant cultivars of common bean were identified,while no resistant cultivars of mung bean were screened.This study revealed that F.cuneirostrum was one of common bean FRR pathogens in Inner Mongolia and it could induce mung bean root rot as well.To our knowledge,this is the first report of F.cuneirostrum causing FRR of common bean in China.
Keywords: Fusarium cuneirostrum,fusarium root rot,pathogenicity,molecular phylogenetic analysis,Phaseolus vulgaris
1.Introduction
Common bean,also known as dry bean (Phaseolus vulgaris),is one of the most important food legumes in the world,which are planted in more than 115 countries and the annual output accounts for more than 50% of the total production of edible legumes (FAO 2022).However,disease is a serious problem restricting common bean production (Singh and Schwartz 2010).More than 20 diseases of common bean were documented in China,but only a few diseases were researched in more detail,such as common bacterial blight,anthracnose,and Fusarium wilt(Wangetal.2009;Chenetal.2012;Xueetal.2012).
Root rot is a serious disease in most common bean production regions in the world.Many fungal and oomycete pathogens have been reported to cause root rot on common bean,includingAphanomyceseuteiches,Fusariumspp.,Macrophominaphaseolina,Phymatotrichum omnivorum,Plectosphaerellacucumerina,Pythiumspp.,Rhizoctoniasolani,andThielaviopsisbasicola,and they result in symptoms such as poor stands,chlorosis,root rot,and stunting (Schwartzetal.2005;Nzungizeetal.2011;Jacobsetal.2018;Yangetal.2018).Among the root rots,Fusarium root rot (FRR) occurs widespread in common bean production areas worldwide,which cause yield losses up to 84% (Abawi and Pastor 1990;Park and Tu 1994;Macedoetal.2017).Initially,the causal agent of FRR was identified asF.solanif.sp.phaseoli(Snyder and Hansen 1941).However,based on morphological and molecular phylogenetic analyses,F.solanif.sp.phaseoliwas re-identified asF.phaseoliby Aokietal.(2003).Fusariumphaseoliwas assigned toF.solanispecies complex (FSSC) clade 2 with two otherFusariumspecies,F.brasilienseandF.cuneirostrum,which have been reported to cause common bean root rot in the United States,Brazil,Canada,and Uganda (O’Donnell 2000;Henriquezetal.2014;Jacobsetal.2018;Sangetal.2018).Additionally,there are otherFusariumspecies not belonging to the FSSC also can cause FRR,such asF.equiseti,F.graminearum,F.redolens,andF.sporotrichioides(Bilgietal.2011;Adesemoyeetal.2018).Fusariumoxysporum,documented as a vascular wilt pathogen in more than 100 plants species,has been found to cause root rot on common bean in some cases (Abawi 1989;Aokietal.2014;Adesemoyeetal.2018;Husainietal.2018).
Fusarium root rot of common bean has been becoming severe in major production regions in China,and the pathogens of FRR were generally regarded asF.solaniandF.oxysporumusing basic morphological characteristics in previous studies (Yu 1955;Wangetal.2010;Liuetal.2017).However,with application of multilocus molecular phylogeny,more cryptic species in FSSC andF.oxysporumspecies complex (FOSC) have been resolved and described as new species (Aokietal.2003,2005,2014;Lombardetal.2019).Thus,the objective of this study was to identify pathogen species inciting FRR of common bean using pathogenicity test,morphological characterization,molecular phylogenetic analysis,and host range test,and screening for resistance cultivars.
2.Materials and methods
2.1.Disease survey and pathogen isolation
During a disease survey of common bean in August 2019,severe root rot was observed in some common bean fields in Liangcheng County (40°53´N,112°50´E),Inner Mongolia,China.To identify the causal agents,the diseased plants were collected from these fields for pathogen isolation and identification.
The taproots of diseased plants were washed under tap water and cut into several 2-3 mm thick pieces.Each piece was sterilized in 2% sodium hypochlorite for 3 min,rinsed with sterile distilled water three times,and dried on sterilized filter paper.Every three to four pieces were cultivated on a potato dextrose agar (PDA) (Becton Dickinson,USA) medium containing 0.1% lactic acid at 25°C with a 12 h light period.Fusarium-like isolates were obtained and purified by single-spore isolation (Summerelletal.2003).All single-spore isolates were stored at-80°C on PDA for future use.
2.2.Pathogenicity test
The inoculum preparation for the pathogenicity and host range tests was performed with slight modifications according to the method described by Sunetal.(2019).Several pieces of each isolate were cut from the edge of colony cultured for 14 days,placed in 100 mL mung bean broth (mung bean 40 g,distilled water 1 L,boiled for 60 min,filtered,sterilized at 121°C for 30 min),and cultured for 4 days in an incubation shaker (27°C,100 r min-1).After filtering through four layers of gauze,the conidial suspension was adjusted to a final concentration of 1.0×106spores mL-1to inoculate the plants.
The common bean cultivars Tianzhenhuang and Pinjinyun 3 were used for the pathogenicity test in this study.Fifteen seeds were planted in three duplicate paper cups (600 mL) filled with fresh vermiculite and the planted cups were placed in the greenhouse for 12 days at 25°C.The seedlings were gently pulled out.After washing the roots,the bottom 1/3 sections of the roots were cut off,the remaining roots were immersed in the spore suspension for 3 min,and then planted in a new paper cup with fresh vermiculite.The plants inoculated with sterile water after cutting roots were used as controls.The inoculated plants were placed in a greenhouse at 25°C with natural light (Sunetal.2019).
The symptoms were investigated at 4-wk after inoculation.Disease severity was scaled based on the evaluation method described by Aokietal.(2005) as follows: 1=no symptoms;2=light symptom development with mottling and mosaic (1-20% foliage affected);3=moderate symptom development with interveinal chlorosis and necrosis (21-50% foliage affected);4=heavy symptom development (51-80% foliage affected);and 5=severe symptom development with interveinal chlorosis and necrosis and/or dead plants (81-100% foliage affected).Moreover,at the end of the above evaluation,all evaluated plants were removed from the vermiculite and the roots were cleaned under running water before assessment for root rot using 1-5 scale according to Aokietal.(2005): 1=healthy roots and tap root;2≤25%of lateral roots and tap root with necrosis;3=25-50% of lateral roots and tap root with visible necrosis;4=51-90%of lateral roots and tap roots with necrosis;and 5≥90% of root system with necrosis,plants dead.These data were used to calculate the foliage and root disease index (DI)of each cultivar by using the formula:
DI=[∑(n×s)/(N×5)]×100
where n=the number of plants at that scale,s=the scale of the disease severity,and N=the total number of plants tested.As long as the DI of an isolate was reached 50.0 or the average disease severity of foliage (ADSF) or root(ADSR) attained 3.0,it could be confirmed that the isolate was pathogenic (Table 1) (Aokietal.2005;Banietal.2012).The pathogenicity test was repeated twice.
2.3.Growth rate and morphological characteristics
The 7-mm-diameter mycelial plugs of each pathogenic isolate were cut from the edge of an active colony by a puncher.The plugs were transferred to the center of a 90-mm-diameter plate containing PDA,and each isolate was plated onto three different PDA dishes and incubated at 25°C in the dark.Colony diameters were measured every 3 days by calculating from two measurements taken at right angles to each other until the 28th day or the colonies covered the whole dishes.The morphological characteristics of these isolates were observed by subculturing at 25°C for 20 days on PDA and carnation leaf agar (CLA) plates (Aokietal.2005).The conidia were observed and measured under a light microscope(Olympus 31X,Japan).
2.4.Molecular phylogenetic analysis
Eight pathogenic isolates were sub-cultured on cellophane-covered PDA for 14 days at 25°C.Mycelia of each isolate were scraped from the PDA plates with a sterilized blade.Total genomic DNA of all isolates was extracted from mycelium by using the Fungi Genomic DNA Extraction Kit (Solarbio,Beijing,China),according to the manufacturer’s instruction.PCR amplifications were conducted using the primer pair EF-1/EF-2 for the translation elongation factor 1-alpha (EF-1α) region(O’Donnelletal.1998),primer pair ITS4/ITS5 for the nuclear ribosomal internal transcribed spacer (ITS) region(Whiteetal.1990),primer pair ITS5/NL4 for domains D1 and D2 at the 5’-end of the nuclear large subunit rDNA(28S) region (Aokietal.2005),and primer pair NL11/CNS1 for the entire nuclear ribosomal intergenic spacer(IGS) region (Aokietal.2005) to confirm the identity of these pathogenic isolates (Appendix A).PCR reactions were carried out using a Gene Amp 9700 thermocycler(Applied Biosystems,Foster City,CA) in 40 μL reaction mixtures containing: 4 μL DNA (5 ng μL-1),2 μL of each primer,20 μL of 2×TaqPCR Mastermix (Vazyme,Nanjing,China),and 14 μL ddH2O.The PCR reaction program included 1 cycle of 4 min at 94°C;38 cycles of 30 s at 94°C,1 min at 55°C,and 2 min at 72°C;followed by 1 cycle of 10 min at 72°C and 4°C hold.All PCR products were analyzed using gel electrophoresis in 1.5% agaroses gel with the Gelgreen Nucleic Acid Gel Stain (Biotium,USA),and photographed over a UV transilluminator.
PCR products were purified with TIAN quick Midi Purification Kit (TIANGEN,Beijing,China),and submittedto Sangon Biotech (Shanghai,China) Co.,Ltd.for cloning and sequencing using the aforementioned primers.SeqMan 7.1.0 from DNAStar (Madison,WI,USA) were used to contig and edit the DNA sequences.The resulting sequences were blasted and aligned in NCBI database(http://www.ncbi.nlm.nih.gov),and then deposited in the GenBank database under isolate numbers (Appendix B).Phylogenetic analysis was conducted with sequences generated in this study and high similarity sequences of isolates in Aokietal.(2005) study and GenBank (Appendix B).Multiple sequence alignments were performed using MUSCLE method in MEGA11 with default parameters.The alignments were edited to remove trimmed areas and discarded incomplete sequences.Phylogenetic tree for EF-1α,ITS,28S,and IGS regions and their tandem sequences were constructed respectively using the Maximum Parsimony method with 1,000 bootstrap replications and the pairwise deletion option (Aokietal.2005).
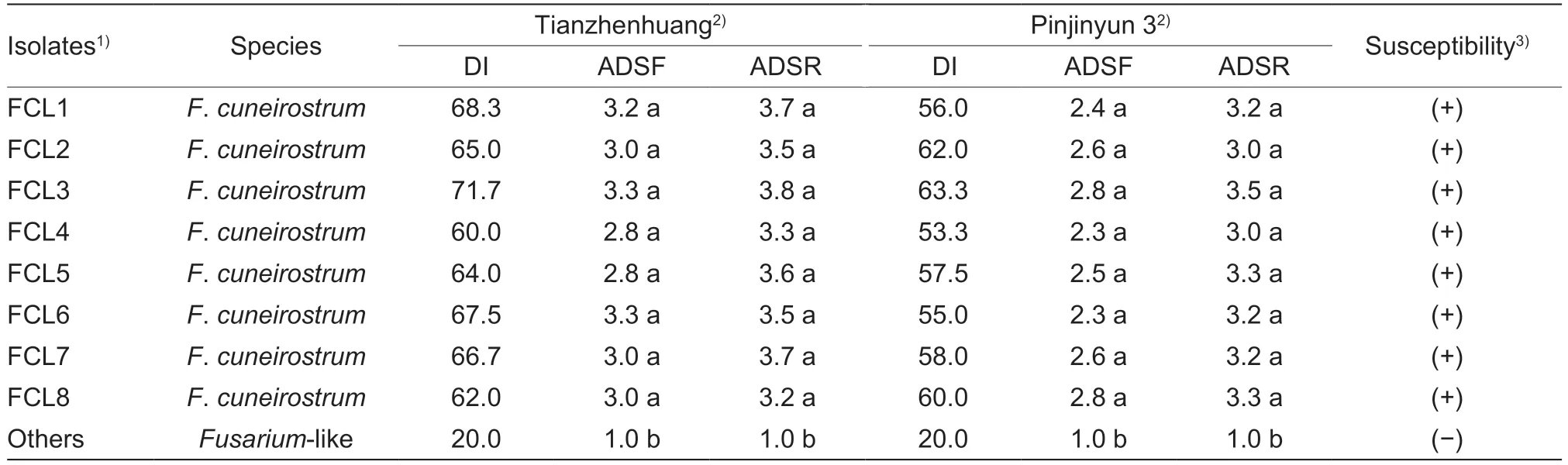
Table 1 Pathogenicity of 19 Fusarium species isolates on common bean cultivars Tianzhenhuang and Pinjinyun 3
2.5.Host range test
The representative pathogenic isolate FCL3 was selected to conduct the host range test.The tested hosts were constituted of several legume crops,including mung bean(Vignaradiatacvs.Jilv 7 and Zhenglv 8),pea (Pisum sativumcvs.Longwan 1 and Zhongwan 4),chickpea(Cicerarietinumcvs.Muying 1 and A-1),cowpea (Vigna unguiculatacvs.Yujiang and Zhongjiang 1),adzuki bean(Vignaangulariscvs.Jihong 11 and Baihong 4),faba bean (Viciafabacvs.Yundou 690 and Lincan 8),and soybean (Glycinemaxcvs.Williams and Zhonghuang 13).In addition,to further determine the pathogenicity of the isolates to soybean,another 10 soybean cultivars were tested as well (Table 2).The inoculation procedure and evaluation method were the same as in the pathogenicity test.The host range test was also repeated twice.
2.6.Resistance evaluation of common bean and mung bean cultivars
A total of 50 common bean and 50 mung bean cultivars were screened for the resistance to root rot,and their information is described in Table 3 and Appendix C.One representative pathogenic isolate FCL3 was used for resistance evaluation.The inoculation procedure was same as preceding pathogenicity test.The resistance criterion was evaluated according to DI and disease severity as follows: highly resistant (HR),20≤DI≤30,ADSF or ADSR≤2.0;resistant (R),30<DI≤40,ADSF or ADSR≤2.5;intermediate (I),40<DI≤50,ADSF or ADSR≤3.0;susceptible (S),50<DI≤75;highly susceptible(HS),75<DI≤100.For those cultivars identified as highly resistant or resistant to root rot,repeated identification was performed.
2.7.Statistical analysis
To analyze the significance of the differences in DI,ADSF,and ADSR in the different common bean or mung bean cultivars,respectively,all data obtained from the disease severity of foliage and root were subjected to an analysis of variance (one-way ANOVA) (Aokietal.2005;Banietal.2012).Whenever the ANOVA was statistically significant(P≤0.05) for a specific variable,a Duncan’s multiple range test was executed to evaluate the differences of the means between each cultivar (Banietal.2012).The coefficient of correlation existing between the different disease parameters was calculated using the nonparametric Spearman’s rank correlation coefficient analysis.All statistical analyses were performed with IBM SPSS v26.0 (IBM Corporation,Armonk,NY,USA).
3.Results
3.1.Disease symptoms and pathogen isolation
During the field surveys in Liangcheng County,Inner Mongolia,many diseased common bean plants,which displayed similar symptoms of Fusarium root rot (FRR)with chlorosis,stunting,and taproots with brown lesions or necrosis,were be found in common bean fields (Fig.1).The incidence rate of diseased plants was nearly 30% in the severely affected fields.

Fig.1 Symptoms of root rot on common bean caused by Fusarium cuneirostrum in the field.A,common bean suffering from Fusarium root rot showing wilt in the field.B,the discolored and root rot of infected common bean.C,the discolored vascular system of infected common bean.
NineteenFusarium-like isolates were obtained after pathogen isolation and purification.Out of the 19 isolates,eight isolates grew slowly and their colonial morphology,such as color and fluffy mycelium,were similar as one ofF.cuneirostrumandF.phaseoli(Aokietal.2005;Schwartzetal.2005;Wangetal.2019).
3.2.Pathogenicity test
Four-weeks after inoculation,the pathogenicity test indicated that eight isolates (FCL1 to FCL8) were pathogenic to the common bean cultivars Tianzhenhuang and Pinjinyun 3,among the 19 isolates (Table 1).The DI of eight pathogenic isolates was varied from 53.3 for FCL4 to 71.7 for FCL3 in the two cultivars,and the values for ADSF and ADSR ranged from 2.3 in FCL4 and FCL 6 to 3.8 in FCL3.All inoculated common bean plants showed typical FRR symptoms similar to those in the field,including foliar chlorosis and defoliation,lateral root and tap root necrosis,and reddish-brown lesions on base stems and tap roots (Fig.2-A and B).The diseased plants were dwarfed,wilted,and easily pulled out.No symptoms were observed in the control plants or nonpathogenic isolates.The same pathogens were re-isolated from the diseased plants to confirm identity and Koch’s postulates.

Fig.2 Symptoms of common bean cultivar Pinjinyun 3 and mung bean cultivar Jilv 7 inoculated with Fusarium cuneirostrum.The whole plants wilt status of common bean (A) and mung bean (B).The root rot status of common bean (C) and mung bean (D).
3.3.Growth rate and morphological characteristics
The eight pathogenic isolates had similar colony with undulate margins,radial cream-grey mycelia and greygreen conidia pustules on PDA (Fig.3-A and B).The mycelia were milky white or slightly purplish in the early stage,while the color deepened to purple with the age.The average diameter of colonies cultured on PDA for 28 days were 71.8 mm,and the average growth rate was 2.1-2.8 mm d-1.The macroconidia produced on PDA or CLA medium were falcate,three to five septate,mostly four septate (46.6 to 53.6×4.7 to 6.8 μm on average),with obviously a wedge shaped and slightly protruding basal foot cell (Fig.3-C and D).The morphological characteristics of these isolates were similar asF.cuneirostrumreported by Aokietal.(2005).

Fig.3 Culture morphology and molecular characteristics of Fusarium cuneirostrum isolated from common bean in Inner Mongolia,China.Front (A) and back (B) of colony on PDA plate after 25 days incubation.Conidiospore on PDA (C) and CLA (D) plates(Bars=10 μm).
3.4.Molecular phylogenetic analysis
Approximately 540 bases were determined for ITS and 28S,685 bases for EF-1α,and 2,578 bases for IGS.The four nuclear loci sequences of the eight pathogenic isolates were blasted in GenBank,which showed 100%identical to respective those of knownF.cuneirostrumexcept the EF-1α gene of FCL7 was 99.71%.The trimmed sequences of the EF-1α,ITS,28S,and IGS were cascaded,and a total sequence length about 4,340 bp from the eight isolates and other referenced isolates was contained in the final dataset.The similar topology was carried in the phylogenetic tree that constructed by the concatenated sequence dataset and maximum parsimony method.The phylogenetic analyses revealed that the eight isolates were clustered in a single phylogenetic group withF.cuneirostrumisolates,which were the cause of FRR on common bean and mung bean (Fig.4).This result was consistent with the study of Aokietal.(2005)and inferred the host range tests.

Fig.4 Phylogenetic tree showing relationships between eight Fusarium cuneirostrum isolates and other species within the F.solani species complex from NCBI GenBank based on Maximum Parsimony analysis of the concatenated nuclear loci the translation elongation factor 1-alpha,nuclear ribosomal internal transcribed spacer,domains D1 and D2 at the 5´-end of the nuclear large subunit rDNA,and the entire nuclear ribosomal intergenic spacer sequences.F.venezuelense NRRL 22395 and F.cryptoseptatum NRRL 22412 were used as outgroups.
Based on morphological characterization and molecular phylogenetic analysis of the eight isolates,they were identified asF.cuneirostrum.
3.5.Host range test
In the host range test ofF.cuneirostrumisolate FCL3 on seven other legumes,the isolate showed no pathogenicity to adzuki bean,faba bean,chickpea,cowpea,pea,and soybean with no obvious symptoms.The three disease ratings were all in the lowest range with DI of 20.0 and ADSF and ADSR of 1.0 (Table 2).However,the isolates were strongly pathogenic to mung bean,and caused the similar or more serious symptoms as those on common bean,with chlorosis,plant wilted,defoliation,root rot,and reddish-brown stems and tap roots (Fig.2-C and D).The highest disease ratings,including DI of 100.0 and ADSF and ADSR of 5.0 were obtained from two mung bean cultivars (Table 2).Thus,these results indicated that theF.cuneirostrumisolate FCL3 from common bean was also pathogenic to mung bean,which verified the result of phylogenetic analyses and were in keeping with the study of Aokietal.(2005).
3.6.Resistance evaluation of common bean,mung bean,and soybean cultivars
A total 50 common bean and 50 mung bean cultivars were inoculated for screening resistance sources to the representativeF.cuneirostrumisolate FCL3.
Large variations in DI,ADSF,and ADSR exhibited among the common bean cultivars.The DI varied from 20.0 to 100 and ADSF and ADSR ranged 1.0 to 5.0 in the common bean cultivars (Table 3).To confirm the suitable and convenient method of disease scoring for screening the resistance common bean cultivars to FRR,the correlation among the different disease ratings was examined using a non-parametric Pearson correlation coefficient analysis.The results showed that three disease ratings were significantly correlated(Table 4).The most significant correlation was obtained between DI and ADSF (r=0.975,P<0.01).Similarly,the high and same significant correlations were observed between the ADSR,DI (r=0.956,P<0.01),and ADSF(r=0.920,P<0.01).Combining three disease ratings,five common bean cultivars,Longyundou 5,Longyundou 12,Longyundou 17,Longyundou 27,and Pinyun 2,and two common bean cultivars,Longyundou 26,and YD154,were highly resistant and resistant toF.cuneirostrumisolate FCL3,respectively.Cultivars Longyundou 4,Longyundou 13,and Longyundou 22 were intermediate,including heterozygous.Among the remaining 40 cultivars,34 and six cultivars showed susceptible and high susceptible,respectively.

Table 4 Correlation of the three disease ratings assessed calculated according to the Pearson correlation coefficient1)
For mung bean cultivars,hardly any variations could be found in the three disease ratings (Appendix C).All inoculated cultivars were highly susceptible toF.cuneirostrumwith the maximal or almost maximal DI,ADSF,and ADSR.Even if one or more seedlings of some cultivars showed no symptom in leaves,their roots have been severely rotted.Thus,statistical analysis was not applicable to the result of mung bean cultivars.
4.Discussion
Fusariumcuneirostrumwas formally described by Aokietal.(2005).Up to now,it has been found in Africa,America,and Asia (Aokietal.2005;Henriquezetal.2014;Sangetal.2018).Oudman (2018) recently investigated causal agents of Fusarium root rot (FRR) on dry bean in Michigan,United States,and revealed 50% of the pathogenic isolates to beF.cuneirostrum,suggesting it was a prevailing pathogen causing FRR on common bean.
In China,several pathogens,such asP.ultimum,R.solani,F.equiseti,F.oxysporum,andF.solani,are documented to cause common bean root rot,generally causing an average of 10-30% yield loss (Wangetal.2010;Liuetal.2017).Among them,FRR caused byFusariumspp.is the most serious,which can reduce yield over 80% (Wangetal.2010;Liuetal.2017).But these pathogens were identified only by morphological observation and pathogenicity test (Wangetal.2010).In this study,a field disease survey was conducted on common bean in Liangcheng County,Inner Mongolia.A total of 19Fusarium-like isolates were isolated from symptomatic plants.Pathogenicity tests indicated that eight slow-growth isolates,whose morphological characteristics were similar withF.brasilienseandF.cuneirostrum,were pathogenic to the common bean cultivars Tianzhenhuang and Pinjinyun 3.The isolates caused typical FRR symptoms such as yellow and wilted leaves,reddish-brown lesion on taproot,and dwarfing.
To accurately identify the pathogen,multilocus genetic analysis was used to identity phylogenetic species.(O’Donnelletal.1998,2008,2010;Lombardetal.2019).In this study,DNA sequence analysis showed that the EF-1α,ITS,28S,and IGS sequences of the eight isolates were consistent withF.cuneirostrumisolates currently available in GenBank.Molecular phylogenetic analysis based on tandem EF-1α,ITS,28S,and IGS sequences classified the eight isolates into one distinct clade withF.cuneirostrumisolates,which caused FRR of common bean (Fig.4).Therefore,based on the morphological characteristics and molecular phylogenetic analysis,the isolates FCL1-FCL8 was identified asF.cuneirostrum.
Host range test revealed theF.cuneirostrumcommon bean isolates also displayed strong pathogenic to mung bean cultivars Jilv 7 and Zhenglv 8,but no pathogenic to other six legumes cultivars including 12 soybean cultivars.The symptoms on mung beans were even more severe as those on common beans.In previous study,F.cuneirostrumhad been reported to cause root rot on common bean,mung bean,and soybean sudden death syndrome (SDS) (Aokietal.2005).However,Aokietal.(2012) subsequently describedF.cuneirostrumstrain NRRL 31949 causing soybean SDS as a newFusariumspecies,F.crassistipitatum.Moreover,our inoculation results for 12 soybean cultivars revealed thatF.cuneirostrumwas not pathogenic to soybean as well,which supported the result of Aokietal.(2005,2012).
FRR is a soil-borne disease.Planting resistant cultivars is the most effective and economical strategy to control FRR (Singh and Schwartz 2010;Wangetal.2010).In this study,resistant phenotypes of 50 common bean cultivars and 50 mung bean cultivars toF.cuneirostrumisolate FCL3 were assessed with the scoring of DI,ADSF,and ADSR.The high correlation coefficient obtained when comparing measurements with each other confirmed that the three disease ratings were adequate to classify common bean resistance toF.cuneirostrum.Five highly resistant and two resistant common bean cultivars were obtained (Table 3),while no resistant mung bean cultivars were screened (Appendix C).
In this study,it was revealed thatF.cuneirostrumwas the primary causal agent of FRR in Liangcheng County,Inner Mongolia.To date,this is a first report thatF.cuneirostrumcaused FRR on common bean in China.The findings are important for Fusarium root rot control and resistance breeding of common bean and other legumes.Moreover,mung bean is also a major and native crop in China,so it is important to investigate whetherF.cuneirostrumcauses root rot on mung bean in the field and to screen more mung bean and common bean cultivars to findF.cuneirostrum-resistant cultivars in further.
5.Conclusion
In this study,we isolated eightF.cuneirostrumisolates from common bean in Inner Mongolia.These pathogens were identified through morphology,pathogenicity and host range tests,and molecular characterization.Seven highly resistant or resistant common bean cultivars toF.cuneirostrumwere screened by artificial inoculation method.This study provides important information forF.cuneirostrumcausing Fusarium root rot on common bean in China,which will guide the disease control and common bean breeding and warn that mung bean may also be suffering fromF.cuneirostrum.
Acknowledgements This study was supported by the China Agriculture Research System of MOF and MARA (CARS-08),and the Scientific Innovation Program of the Chinese Academy of Agricultural Sciences.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.01.010
杂志排行
Journal of Integrative Agriculture的其它文章
- Advances in DNA methylation and its role in cytoplasmic male sterility in higher plants
- Assessment of molecular markers and marker-assisted selection for drought tolerance in barley (Hordeum vulgare L.)
- 3D genome organization and its study in livestock breeding
- SUPER WOMAN 2 (SPW2) maintains organ identity in spikelets by inhibiting the expression of floral homeotic genes OsMADS3,OsMADS58,OsMADS13,and DROOPING LEAF
- Genetic dissection and validation of a major QTL for grain weight on chromosome 3B in bread wheat (Triticum aestivum L.)
- Coupling of reduced inorganic fertilizer with plant-based organic fertilizer as a promising fertilizer management strategy for colored rice in tropical regions
