SUPER WOMAN 2 (SPW2) maintains organ identity in spikelets by inhibiting the expression of floral homeotic genes OsMADS3,OsMADS58,OsMADS13,and DROOPING LEAF
2024-01-17HuiZhuangJinsongLanQiuniYangXiaoyuZhaoYuhuanLiJingyaZhiYalinShenGuanghuaHeYunfengLi
Hui Zhuang ,Jinsong Lan ,Qiuni Yang ,Xiaoyu Zhao,Yuhuan Li,Jingya Zhi,Yalin Shen,Guanghua He,Yunfeng Li
Rice Research Institute,Key Laboratory of Application and Safety Control of Genetically Modified Crops,Academy of Agricultural Sciences,Southwest University,Chongqing 400715,China
Abstract Flower organ identity in rice is mainly determined by the A-,B-,C-and E-class genes,with the majority encoding MADS-box transcription factors.However,few studies have investigated how the expression of these floral organ identity genes is regulated during flower development.In this study,we identified a gene named SUPER WOMAN 2 (SPW2),which is necessary for spikelet/floret development in rice by participating in the regulation of the expression of pistil identity genes such as OsMADS3,OsMADS13,OsMADS58 and DL.In the spw2 mutant,ectopic stigma/ovary-like tissues were observed in the non-pistil organs,including sterile lemma,lemma,palea,lodicule,and stamen,suggesting that the identities of these organs were severely affected by mutations in SPW2.SPW2 was shown to encode a plant-specific EMF1-like protein that is involved in H3K27me3 modification as an important component of the PRC2 complex.Expression analysis showed that the SPW2 mutation led to the ectopic expression of OsMADS3,OsMADS13,OsMADS58,and DL in non-pistil organs of the spikelet.The ChIP-qPCR results showed significant reductions in the levels of H3K27me3 modification on the chromatin of these genes.Thus,we demonstrated that SPW2 can mediate the process of H3K27me3 modification of pistil-related genes to regulate their expression in non-pistil organs of spikelets in rice.The results of this study expand our understanding of the molecular mechanism by which SPW2 regulates floral organ identity genes through epigenetic regulation.
Keywords: rice (Oryza sativa),spikelet,organ identity,H3K27me3
1.lntroduction
Rice has been recognized as one of the most important agronomic crops in the world,and it has been selected as the model plant for functional genomics studies of crops.The development of its spikelet/floret greatly affects the formation of rice yield and quality.Therefore,investigating the identities of floral organs is of great significance.
Recent molecular genetic studies inArabidopsishave made considerable progress in our understanding of floral organ development.The identities of the four whorls of floral organs (sepal,petal,stamen,and carpel)are regulated by four groups of genes that are known as the “ABCE model” (Bowmanetal.1991;Coen and Meyerowitz 1991;Weigel and Meyerowitz 1994;Pelazetal.2000;Theissen and Saedler 2001;Dittaetal.2004;Soltisetal.2007;Litt and Kramer 2010).These genes primarily encode the MADS-box transcription factors and control the homeotic transformation of floral organs inArabidopsis(Theissenetal.2016).In contrast to dicots likeArabidopsis,rice has a different floral morphology.The spikelet,which is the last inflorescence unit in rice,consists of degenerative sterile glumes and a floret with four whorls of floral organs (lemma,palea,lodicules,stamens,and pistil) (Zhuangetal.2020).Due to the differences between dicots and monocots,the ABCE model ofArabidopsisand its regulatory network are not fully applicable to monocots such as rice (Kyozukaetal.2000;Fornaraetal.2003;Kateretal.2006;Zhuangetal.2020).
In rice,OsMADS14,OsMADS15,andOsMADS18are A-class (AP1/FUL-like) genes involved in lemma and palea formation (Jeonetal.2000;Kyozukaetal.2000;Pelucchietal.2002;Fornaraetal.2004;Wangetal.2010).The B-class (AP3-like) genesSUPERWOMAN1(SPW1)/OsMADS16,OsMADS2,andOsMADS4are associated with lodicule and stamen development (Kangetal.1998;Leeetal.2003;Nagasawaetal.2003;Prasad and Vijayraghavan 2003;Yadavetal.2007).OsMADS3andOsMADS58are homologs of the C-class geneAGAMOUS(AG) and play a major role in regulating stamen and pistil development (Yamaguchietal.2006).Pistil identity in rice is also regulated byDROOPING LEAF(DL),a homologous gene ofCRABSCLAW(CRC)inArabidopsis(Alvarez and Smyth 1999;Yamaguchietal.2004).OsMADS13is classified as a D-class gene in rice that plays a key role in specifying ovule identity (Drenietal.2007,2011;Dreni and Kater 2014).The E-class genesLEAFYHULLSTERILE1(LHS1)/OsMADS1,OsMADS5,OsMADS7,OsMADS8,andPANICLE PHYTOMER2(PAP2)/OsMADS34control the overall process of flower organ development,following a distinct regulation pattern compared to the A-,B-,C-,and D-class genes (Jeonetal.2000;Pelucchietal.2002;Agrawaletal.2005;Bommertetal.2005;Prasadetal.2005;Chenetal.2006;Lietal.2009).
Recent studies have reported the involvement of additional factors,beyond MADS-box transcription factors,in regulating the development of rice flower organs.These factors ensure the precise spatial and temporal control of mRNA or protein expression of the A-,B-,C-,and E-class genes,which are crucial for the initiation and positioning of floral organs.Two key regulators in rice flower development areABERRANT PANICLEORGANIZATION1(APO1) andAPO2,which share homology with theUNUSUALFLORAL ORGANS(UFO) andLEAFY(LFY) genes inArabidopsis,respectively.The interaction between UFO and LFY results in the activation of the B-class geneAPETALA3(AP3) and the C-class geneAG(Siriwardana and Lamb 2012).In a similar manner,the concerted action of APO1 and APO2 regulates floral meristem and organ identity in rice,and theapo1-1apo2-1mutant displays the development of glume-like organs that undergo homeotic transformation from lodicules positioned on the palea side (Ikeda-Kawakatsuetal.2012).Thesl1mutant shares similarities with thespw1mutant,as both mutants exhibit homeotic transformation of lodicules into glume-like organs and stamens into carpels.SL1positively regulates the expression ofSPW1to control the identities of lodicules and stamens (Horigomeetal.2009;Xiaoetal.2009).CHIMERICFLORALORGANS1(CFO1)/OsMADS32,which encodes a MIKC-type MADSbox protein,acts as a negative regulator ofDLand plays a crucial role in maintaining the identity of floral organs.Thecfo1mutant exhibits developmental abnormalities,such as defects in the margin region of the palea (mrp),the formation of chimeric flower organs,and the presence of ectopic floral organs (Sangetal.2012;Wangetal.2015).MOSAICFLORALORGANS1(MFO1)/OsMADS6is another significant MADS-box gene involved in flower development.Functional deficiency ofMFO1/OsMADS6results in a phenotype similar to that ofcfo1,and it also negatively regulates the expression ofDL(Lietal.2010;Duanetal.2012;Huetal.2021).
Polycomb group (PcG)-mediated repression is a common mechanism for maintaining the epigenetic inhibition of target genes in plants (Schwartz and Pirrotta 2007,2008;Köhleretal.2008).PcG proteins form two major complexes: Polycomb Repressive Complex 1(PRC1) and PRC2.PRC2 is primarily responsible for histone H3 Lys27 methylation (H3K27me3),while PRC1 is recruited to the modified site to maintain and stabilize the methylation (Caoetal.2002;Mülleretal.2002;Schubertetal.2005;Turcketal.2007;Calonjeetal.2008;Schatlowskietal.2008;Mozgovaetal.2015).InArabidopsis,mutations in the PcG protein have been shown to reduce the level of H3K27me3 modification of the target gene,thus affecting flower morphogenesis.CURLYLEAF(CLF) encodes a histone methyltransferase of PRC2 for H3K27me3 that restricts the expression ofAGduring vegetative growth (Goodrichetal.1997;Schubertetal.2006).Another gene,WAVYLEAVES ANDCOTYLEDONS(WLC),also contributes to downregulatingAGexpression through aberrant methylation in vegetative organs (Levy and Dean 1998).EMBRYONIC FLOWER 1 (EMF1) interacts with EMF2 to recruit PRC2,facilitating the deposition of repressive histone modifications (e.g.,H3K27me3) inAG’s regulatory regions,leading to the repression ofAGexpression(Calonjeetal.2008;Kimetal.2010).In rice,OsEMF1,also known asDEFORMEDFLORALORGAN1(DFO1),CURVEDCHIMERICPALEA1(CCP1),orDS1,is a homolog of theEMF1gene ofArabidopsis(Aubertetal.2001;Yanetal.2015;Zhengetal.2015;Liuetal.2018).OsEMF1 has been found to interact with the rice PcG proteins OsMSI1 and OsiEZ1 to regulate H3K27me3-mediated epigenetic repression ofOsMADS58,thereby maintaining rice floral organ identity.TheOsEMF1mutation results in abnormal palea development that is characterized by the presence of ectopic stigmatic tissues and other pleiotropic phenotypes (Yanetal.2015;Zhengetal.2015;Liuetal.2018).
In this study,we report a new allele ofOsEMF1,namedSUPERWOMAN2(SPW2),which is involved in maintaining spikelet development in rice.We show thatSPW2can inhibit the expression of pistil identity genesOsMADS3,OsMADS13,OsMADS58,andDLin non-pistil organs by participating in the PcG complex-mediated H3K27me3 modification.The results of this study expand our understanding of the molecular mechanisms by whichOsEMF1regulates flower organ identity genes through epigenetic modifications.
2.Materials and methods
2.1.Plant materials and growth conditions
Thespw2mutant was isolated from an ethyl methanesulfonate (EMS)-inducedindicarice (Oryza sativa) XIDA 1B (wild type (WT)) mutant bank.In 2014,spw2mutants were crossed to the sterile line 56S (indica),and the F1and F2populations were generated for genetic analysis and gene mapping.Rice (Oryzasativa) plants were planted in the Xiema,Rice Research Institute of Southwest University (Chongqing,China) under natural conditions in summer.The transgenic plants were grown in a greenhouse under standard WT growing conditions.
2.2.Microscopy
Spikelets of WT andspw2were fixed with formalinaceto-alcohol (FAA) solution at 4°C overnight,followed by dehydration steps and then embedding in paraffin(Paraplast Plus,Sigma).The tissues were sliced into 8 μm sections,affixed to microscope slides,and stained with Safranin O and Fast Green (ThermoFisher,America).Sections were observed and photographed using a light microscope (Nikon E600,Japan).For scanning electron microscopy (SEM),panicles of WT plants andspw2mutant plants were selected and the outer bracts of the spikelets were stripped by using anatomical needles and tweezers,and then they were directly observed using a scanning electron microscope (SU3500,Japan) at -20°C under a low-vacuum environment.
2.3.Map-based cloning of spw2
The 216 F2plants with typicalspw2mutant phenotypes were selected from aspw2/56S F2population for gene mapping.A screen for molecular markers linked toSPW2was performed using genetic markers from publicly-available rice databases,including Gramene(http://www.gramene.org) and Rice Genomic Research Program (http://rgp.dna.affrc.go.jp/publicdata/caps/index.html).Candidate genes predicted using the Rice Genome Brower website (http://rice.plantbiology.msu.edu)were amplified from bothspw2and XIDA 1B (WT),and sequenced directly.The primers used in the mapping and candidate gene analysis are listed in Appendix A.
2.4.Vector construction and transformation
To construct the fusion expression vector of SPW2P:SPW2:GFP,a 6,605-bpSPW2genome sequence with terminator,coupled with the~5,000 bp upstream,was amplified.The fragment was inserted into the binary vector pCAMBIA1300 using the pEASY-Uni Seamless Cloning and Assembly Kit (Transgene,China).The recombinant plasmids were transformed into thespw2(mutant-type) using theAgrobacteriumtumefaciensmediated transformation method as described by Xiaoetal.(2009).The primer sequences used are listed in Appendix A.
2.5.RNA extraction and qRT-PCR analysis
Total RNA of the WT andspw2was extracted using an RNAprep Pure Plant RNA Purification Kit (Tiangen,China),and cDNA was synthesized from 1 μg total RNA using Superscript III reverse transcriptase (TaKaRa,Japan).qRT-PCR analysis was performed with a 7500 Real-Time PCR System (Applied Biosystems,America)and a SYBR PremixExTaqII Kit (TaKaRa),andACTINwas used as the endogenous control.At least three replicates were performed,from which the mean values were used to represent the expression levels.The primers used are listed in Appendix A.
2.6.Protein sequence and phylogenetic analysis
Amino acid sequences homologous toSPW2were downloaded from the National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov/).The phylogenetic tree was constructed using the maximum likelihood method based on the JTT matrix-based model with the lowest Bayesian information criterion scores(MEGA 5.0 software).
2.7.In situ hybridization
RNAinsituhybridization was performed as described previously (Sangetal.2012),and the gene-specificSPW2probe was amplified and labeled using the DIG RNA Labeling Kit (SP6/T7,Roche,Germany).Probes for known floral organ genes (DL,OsMADS3,OsMADS13andOsMADS58) were prepared using the same method.The primer sequences are listed in Appendix A.
2.8.Y2H assays
The full-length coding regions ofSPW2andLF1were amplified and ligated into the yeast expression vector pGBKT7 to detect autoactivation of the SPW2 protein.The constructs pGBKT7-SPW2,pGBKT7-LF1 (used as a positive control) (Zhangetal.2017) and pGBKT7 (used as a negative control) were transformed separately into the yeast strain Y2HGold.Transformants were selected on synthetic defined (SD)/-Trp or SD/-Ade/-His/-Trp media (Clontech,America).Detailed procedures are described in the manufacturer’s instructions (Yeast Protocols Handbook,PT3024-1;Clontech).The primers used are listed in Appendix A.
2.9.Transcriptional activity analysis
Transcriptional activity of the full-length SPW2 protein was analyzed in rice protoplasts using the dual luciferase reporter assay system.A Glomax 20-20 luminometer(Promega,America) was used to measure the relative luciferase activity.Experimental procedures for rice protoplast extraction were performed as previously described by Lietal.(2020).
2.10.ChlP-qPCR
The WT andspw2panicles were used in the ChIP examination.Young panicles (<5 cm) were collected for the isolation of nuclear extracts.The EpiQuik Plant ChIP Kit (P-2014-48,Epigentek,America) and anti-Histone H3(tri methyl K27) antibody (ChIP grade;ab6002,Abcam,England) were used for the ChIP assays.All PCR experiments were conducted using 40 cycles of 95°C for 5 s,60°C for 30 s,and 72°C for 30 s in a reaction mixture containing 10 pmol of each primer and 1 μL of DNA from ChIP or a control of 1 μL of input DNA diluted 20-fold (per biological replicate) as the template.More than three technical replicates of each were used to produce data for statistical analysis.Experimental procedures for ChIPqPCR were performed as previously described by Xuetal.(2010).The primers used are listed in Appendix A.
3.Results
3.1.Characterization of the spw2 mutant phenotype
Thespw2mutant exhibited typical pistilloid phenotypes in non-pistil organs,including sterile lemmas,lemmas,paleas,lodicules and stamens (Fig.1).In the WT,the spikelet is composed of one pair of extremely degraded rudimentary glumes,one pair of sterile lemmas (also called empty glumes) and one terminal fertile floret.The floret comprises two additional bracts,the lemma and palea,which encase three whorls of floral organs: two lodicules,six stamens,and a central carpel (Fig.1-AH;Hoshikawa 1989).At the flowering stage,the sterile lemmas exhibited elongation and/or widening in 26.5% ofspw2sipikelets,and the stigmatic tissues were observed on the tips of the sterile lemma in 43% of thespw2spikelets (Fig.1-I-J,N,P and S).Stigmatic tissues were also observed on the tips of thespw2lemmas,and some lemmas exhibited curvature,accounting for 38 and 37%of cases,respectively (Fig.1-I,K,O and S).The paleas of the majority ofspw2spikelets exhibited curvature and formed stigmatic tissues both at the top and in the margin region (mrp),with the basal area of mrp showing an ovule-like structure (Fig.1-Q-S).Furthermore,due to the curvature of both the lemma and palea inspw2spikelets,they could not hook together (Fig.1-N-O).Approximately half of the lodicules inspw2spikelets had undergone transformation into a pistil-like structure,with the base enlarging like an ovary and exhibiting two stigmas(Fig.1-L,N and S).We also observed degeneration of the anther and filament in thespw2stamens,and stamenpistil chimeras were observed in whorl 3,displaying a stigma on an anther (Fig.1-M,N and S).In about 20% of thespw2spikelets,stamens were completely transformed into pistil/pistil-like organs (Fig.1-M and S).

Fig.1 Phenotypes of spikelets at the flowering stage in the wild type (WT) and spw2 mutant.A-H,spikelets of the WT at the inflorescence development stage In9 (heading and flowering).A and B,the WT spikelet,the lemma was removed in B.C-G,lemma,palea,sterile lemma,lodicule,stamen and pistil of WT.H,transverse sections of the WT spikelet showing a phenotype similar to those in A-G.I-R,spikelets of the spw2 mutant at stage In9.I,J and O,the spw2 spikelet,the lemma was removed in J.K,lemma of the spw2 spikelet.L,palea and lodicule of the spw2 spikelet.M,stamen and pistil of the spw2 spikelet.N,transverse sections of the spw2 spikelet showing a phenotype similar to those in I-M.P,sterile lemma of the spw2 spikelet.Q and R,palea of the spw2 spikelet.S,percentage of abnormal organs in spw2 spikelets.White text indicates the normal organs,and red text indicates the abnormal organs.Scale bars 1,000 μm.bp,body of palea;ch-st,chimera of stamen and pistil;cu-le,curved lemma;cu-pa,curved palea;le,lemma;le-sl,lemma-like sterile lemma;lo,lodicule;mrp,margin of palea;pa,palea;pi,pistil;pi-le,pistil-like lemma;pi-pa,pistil-like palea;pi-sl,pistil-like sterile lemma;pi-st,pistil-like stamen;sl,sterile lemma;st,stamen.
The morphology of WT andspw2spikelets was observed using SEM.In the WT spikelet,the sterile lemma had a smooth abaxial surface with a few trichomes,while the lemma had a silicified abaxial epidermis with trichomes and protrusions,including an elongated awn on the tip (Appendix B-a-e).The palea consisted of a main body (bp) and a margin region (mrp).The bp resembled the lemma,while the mrp had a smooth abaxial epidermis without trichomes or protrusions (Appendix B-f-h).The lodicule had a smooth abaxial epidermis with round or elliptical cells arranged irregularly (Appendix B-i-j).The stamen consisted of an anther and a filament,and the pistil’s stigma had stigmatic papillae cells (Appendix B-k-l).SEM analysis revealed that thespw2spikelets exhibited ectopic stigmatic tissues in various non-pistil organs,including sterile lemmas,lemmas,paleas,lodicules,and stamens (Appendix B-m-y).Somespw2sterile lemmas showed a rough abaxial epidermis that was similar to the WT lemma,with an orderly parallel arrangement of tubercles and trichomes (Appendix B-o).The basal cells of the mrp inspw2had a square shape and vertical distribution,resembling the abaxial epidermal cells of the ovary (Appendix B-t).Similar transformations were also observed in the lodicules and stamens ofspw2spikelets(Appendix B-u-x).
Compared with the WT,thespw2plants also exhibit pleiotropic defects during vegetative growth,which are mainly reflected in reduced plant height and increased tiller number (Appendix C-a).During the reproductive growth phase,developmental defects in the inflorescence were also found in thespw2mutant with a reduction in panicle length and a significant reduction in the numbers of primary and secondary branches and spikelets per panicle (Appendix C-b).Moreover,the abnormal floral organ development inspw2spikelets also resulted in abnormal seed morphology.The palea and lemma ofspw2cracked to varying degrees,and even failed to enclose the grain properly,resulting in an abnormal grain shape (Appendix C-c).
3.2.Histological analysis of organs in spw2 spikelets
In accordance with these observations,almost all the nonpistil organs (sterile lemmas,lemmas,paleas,lodicules,and stamens except for the pistil) inspw2spikelets tended to be partially transformed into pistil-like organs (Fig.1).Therefore,we further explored the histologic structures of these organs (Fig.2).In a transverse section of the sterile lemma of the WT,three layers of structures were observed: the abaxial epidermis,the adaxial epidermis,and several layers of spongy parenchyma cells sheath the vascular bundles (Fig.2-A1 and A2).Regarding the lemma and bp of the WT,numerous vascular bundles and a four-layered structure consisting of a silicified abaxial epidermis,several layers of fibrous sclerenchyma cells,several layers of spongy parenchyma cells,and a vacuolated adaxial epidermis were observed(Fig.2-A3 and A4).The mrp of the WT consisted of a nonsilicified abaxial epidermis,several layers of spongy parenchyma cells,several fibrous sclerenchyma cells,and a nonvacuolated adaxial epidermis (Fig.2-C1).The lodicule of the WT consisted of parenchymatous cells and interspersed tracheal elements (Fig.2-C2),while the filament of the WT consisted of one central vascular bundle and several layers of parenchymatous cells(Fig.2-C3).The structure of the ovule was histologically well-defined,clearly visible in nuclear staining,and displayed regular cell shapes (Fig.2-C4).
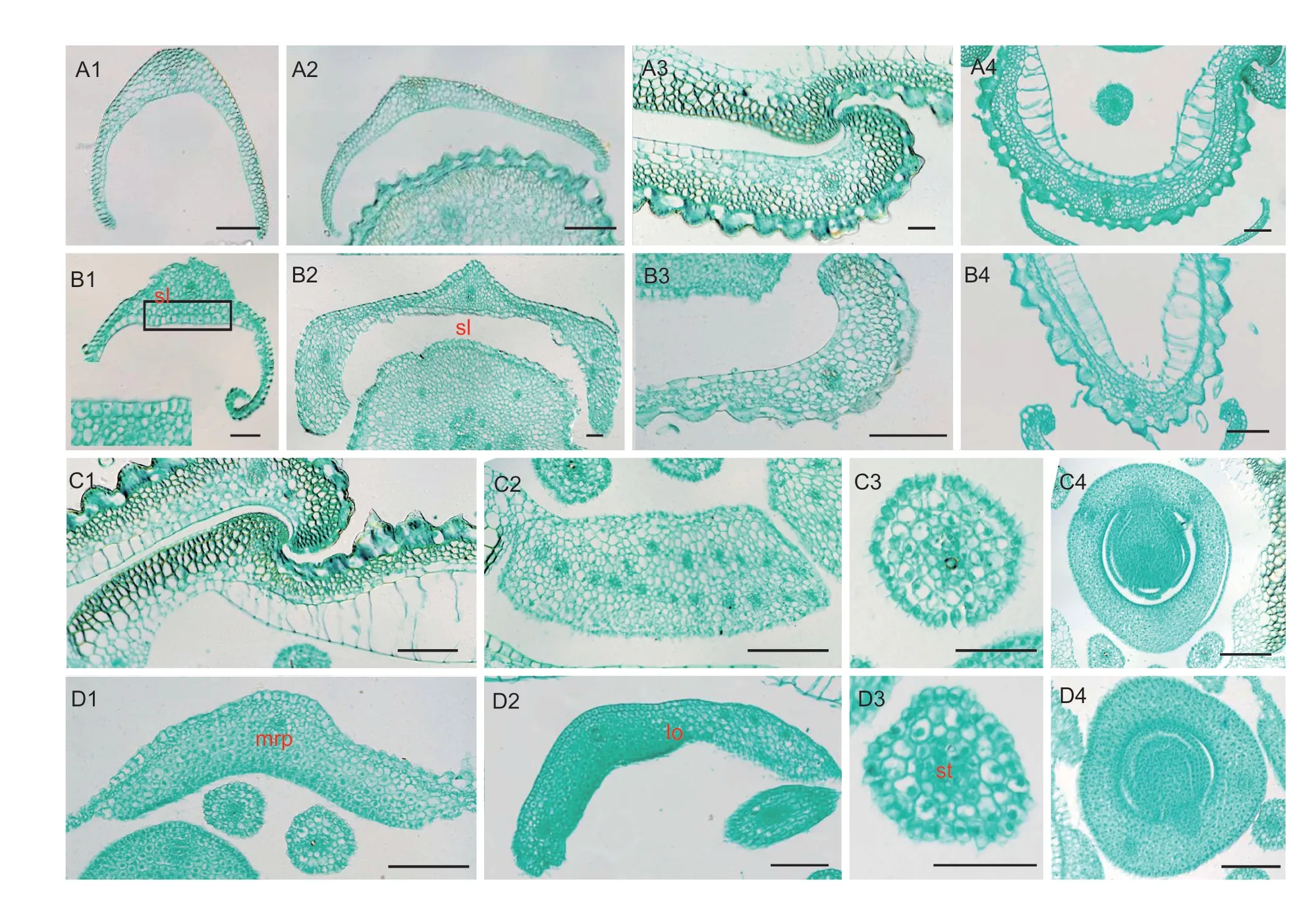
Fig.2 Histological analysis of organs in the spikelets of the wild type (WT) and spw2 mutant.A-D,transverse sections of organs in a WT spikelet and a spw2 spikelet.A and C,transverse sections of organs in a WT spikelet.B and D,transverse sections of organs in a spw2 spikelet.A1 and A2,inner sterile lemma (isl) and outer sterile lemma (osl) of the WT.B1 and B2,lemma-like isl and osl of spw2 spikelets.A3,lemma of the WT.B3,lemma of spw2.A4,bp (body of palea) of the WT.B4,abnormal bp in spw2 spikelets.C1,mrp of the WT.D1,pitilloid mrp in spw2 spikelets.C2,lodicule of the WT.D2,abnormal lodicule in spw2 spikelets.C3,stamen of the WT.D3,pistilloid stamen in spw2 spikelets.C4,ovule of the WT.D4,ovule of spw2.White text indicates the normal organs,and red text indicates the abnormal organs.bp,body of palea;mrp,margin of palea;lo,lodicule;sl,sterile lemma;st,stamen.Scale bars,100 μm.
In the elongated and/or widenedspw2sterile lemmas,several layers of ectopic fibrous sclerenchyma cells,along with cells resembling ovule cells in form,were observed between the abaxial epidermis and parenchyma cell layers,and the number of vascular bundles was randomly increased to three (Fig.2-B1 and B2).The transverse section of thespw2lemma and bp showed high spatial and compositional similarities compared to those of the WT (Fig.2-B3 and B4).However,the cells comprising thespw2mrp were significantly different from those of the WT,as they were composed of spongy parenchyma cells and fibrous sclerenchyma cells with distinct morphological features resembling ovule cells (Fig.2-D1).In somespw2lodicules and filaments,the parenchymatous cells were partially or completely replaced by ectopic ovule cells (Fig.2-D2 and D4).Taken together,these morphological features suggest that the lateral organs other than the pistil inspw2spikelets have acquired a partial pistil-like identity.Consequently,we conclude thatSPW2is involved in the specification of organ identity in the spikelet.
3.3.Abnormal early spikelet development in the spw2 mutant
In rice,spikelet development is categorized into Sp1(formation of a pair of rudimentary glume primordia) stage to Sp8 (formation of ovule and pollens) stage (Ikedaetal.2004).The spikelets were analyzed using SEM with the same imaging conditions for each set of WT andspw2mutants.During the Sp4 (formation of palea primordium)stage,rudimentary glume primordia and sterile lemma primordia were formed and the lemma and palea primordia were developing (Fig.3-A).Subsequently,the lodicule primordia,stamen primordia and carpel primordia developed in an orderly manner during the Sp5-Sp7(formation of lodicule primordia,stamen primordia and carpel primordium) stages (Fig.3-B-D).By the Sp8 stage,the lemma primordia and palea primordia had hooked together,enclosing the inner floral organs(Fig.3-E).In thespw2spikelets,except for the slightly larger size of the sterile lemma primordia compared to the WT,there were no significant differences during the Sp4-Sp6 stages (Fig.3-F-G).However,during the Sp6-Sp8 stages,the development process of thespw2palea primordia was much slower than that of the WT (Fig.3-H-I).Moreover,the degenerated palea primordia lost the ability to hook with the lemma at the Sp8 stage,resulting in some of the inner floral organ primordia being exposed(Fig.3-J).At the Sp8 stage,punctiform prominence could be observed on the tips of some lemma primordia in thespw2spikelets (Fig.3-K).These results suggest that the sterile lemma and palea identities were altered during early floral development in thespw2mutant.
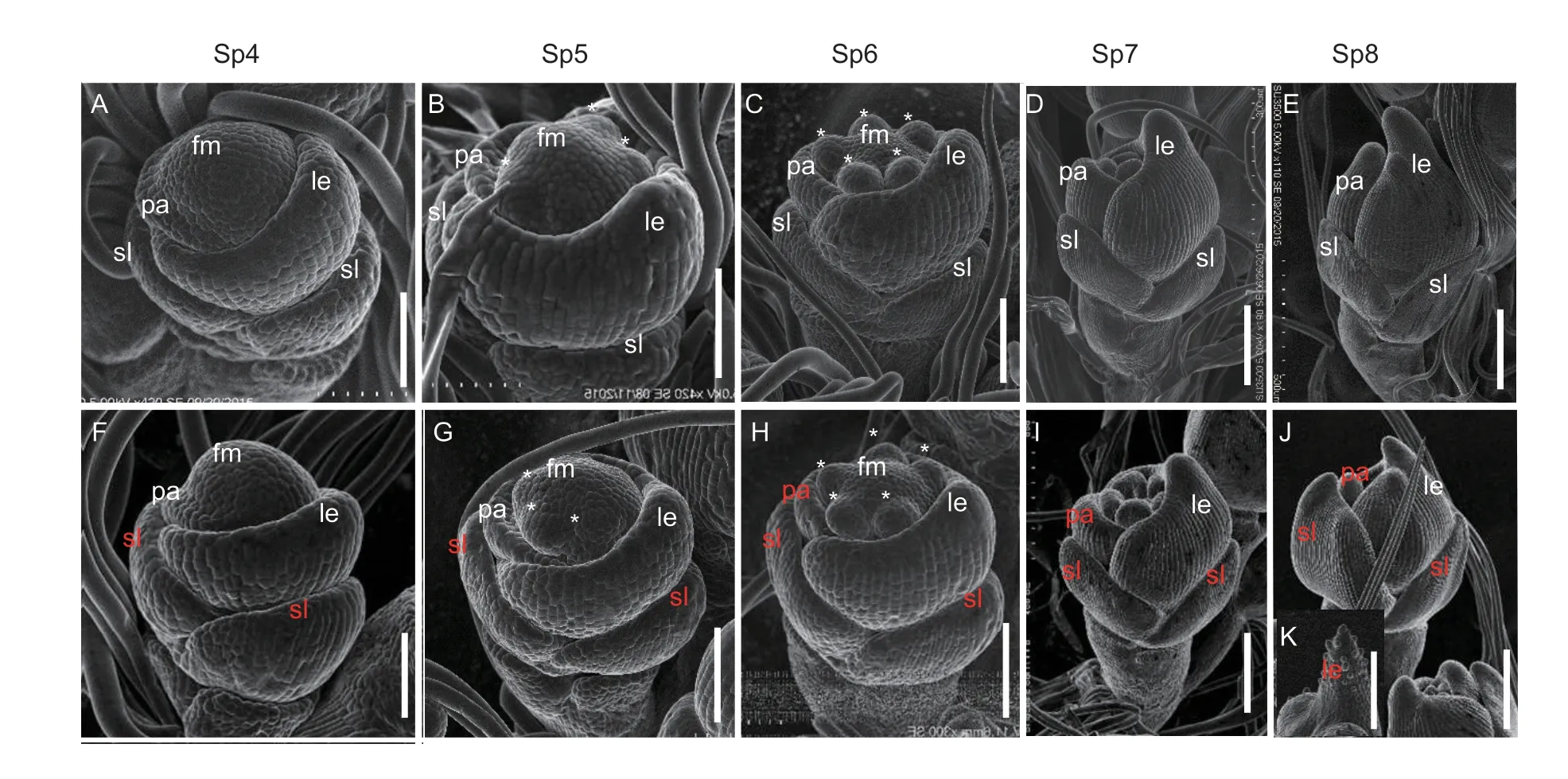
Fig.3 Scanning electron micrographs of spikelets at early developmental stages in the wild type (WT) and spw2 mutant.A-E,spikelets of the WT.F-K,spikelets of the spw2 mutant.K,the top of the lemma in spw2 spikelets.White text indicates the normal organs,and red text indicates the abnormal organs.Asterisks indicate the stamen.fm,floral meristem;le,lemma;pa,palea;sl,sterile lemma.Sp4-Sp8,staging of floral organ development in rice.Scale bars,100 μm.
3.4.Molecular cloning of SPW2
Thespw2mutant was crossed with the WT (XIDA 1B)and all F1progeny exhibited the WT phenotype.In the F2segregated population,692 plants displayed normal phenotypes,while 216 plants exhibited mutant phenotypes.The segregation data were consistent with the expected Mendelian ratio of 3:1,suggesting that thespw2trait is governed by a single recessive gene.
To elucidate the molecular function ofSPW2,we performed map-based cloning using the 216 individuals that displayed the characteristic mutant phenotype.Initially,theSPW2locus was mapped to the short arm of chromosome 1,specifically between the markers RM283 and RM581.Through further fine mapping,we narrowed down the mapping interval to a 112-kb genomic region delimited by the markers RM10425 and RM10440.Within this region,we identified 18 annotated open reading frames (ORFs) (Fig.4-A).A sequence comparison betweenspw2and the WT uncovered a single nucleotide substitution,where an A was replaced by a T in the third exon ofLOC_Os01g12890.This substitution resulted in a codon change from AGA to the termination codon TGA,leading to premature translation termination (Fig.4-B).

Fig.4 Map-based cloning of SPW2.A,mapping of the SPW2 gene.B,schematic illustration of the genomic structure of SPW2.The site of the mutation in SPW2 is shown.C,structure of the SPW2P:SPW2:NOS complementary vector.D,phenotype of the independent transgenic lines.Scale bars,100 μm.
Through a BLAST search and protein function analysis,we determined thatSPW2encodes a plantspecific EMF1-like protein consisting of 1,057 amino acids (Appendix D).InArabidopsis,EMF1 is known to be involved in H3K27me3-mediated silencing ofAG,acting downstream of EMF2,and likely interacts with both PRC1 RING-finger proteins and PRC2 component MULTICOPY SUPRESSOR OF IRA (MSI1) (Aubertetal.2001;Calonjeetal.2008).In rice,studies have demonstrated thatOsEMF1,a homolog ofEMF1,plays a crucial role in maintaining palea identity by collaborating with PcG proteins to regulate the epigenetic repression of H3K27me3 onOsMADS58(Yanetal.2015;Zhengetal.2015;Liuetal.2018).
To verify whetherLOC_Os01g12890was responsible forspw2,a genomic fragment of about 11 kb covering the promoter region and the entire ORF ofLOC_Os01g12890was cloned into the vector pCAMBIA1300.Subsequently,we performed Agrobacterium-mediated genetic transformation to introduce this construct into thespw2mutant (Fig.4-C).As expected,the functional complementation ofSPW2resulted in the restoration of the mutant to the WT phenotype.Notably,the flowers displayed a recovered morphology,providing strong evidence that the functional mutation ofSPW2is indeed responsible for the observed mutant phenotypes(Fig.4-D).These results confirm thatLOC_Os01g12890is theSPW2gene.
3.5.The expression pattern of SPW2
To investigate the spatial and temporal expression patterns of theSPW2gene in WT plants,we conducted both qRT-PCR analysis andinsituhybridization analysis.The qRT-PCR analysis provided insights into the transcript levels ofSPW2in various organs including roots,stems,leaves,sheaths and young panicles.Among these,the highest expression levels were observed in panicles measuring 2-5 cm in length.Stems exhibited a relatively strong expression,while roots,leaves,sheaths,and young panicles (<2 cm) displayed relatively weaker expression levels (Fig.5-A).During floral organ development,SPW2transcripts were detected in the sterile lemma as well as in all other floral organs,including the lemma,palea,lodicules,stamens,and pistil.Notably,high expression levels ofSPW2were observed in the sterile lemma and lodicule (Fig.5-A).To further confirm these expression patterns,we performedinsituhybridization,which provided visual evidence of gene expression.Consistent with the qRT-PCR results,theinsituhybridization analysis demonstrated the presence ofSPW2transcripts in the sterile lemma and all other floral organs (Fig.5-B).Taken together,these results indicate that theSPW2gene exhibits constitutive expression in both vegetative and reproductive organs of rice plants.
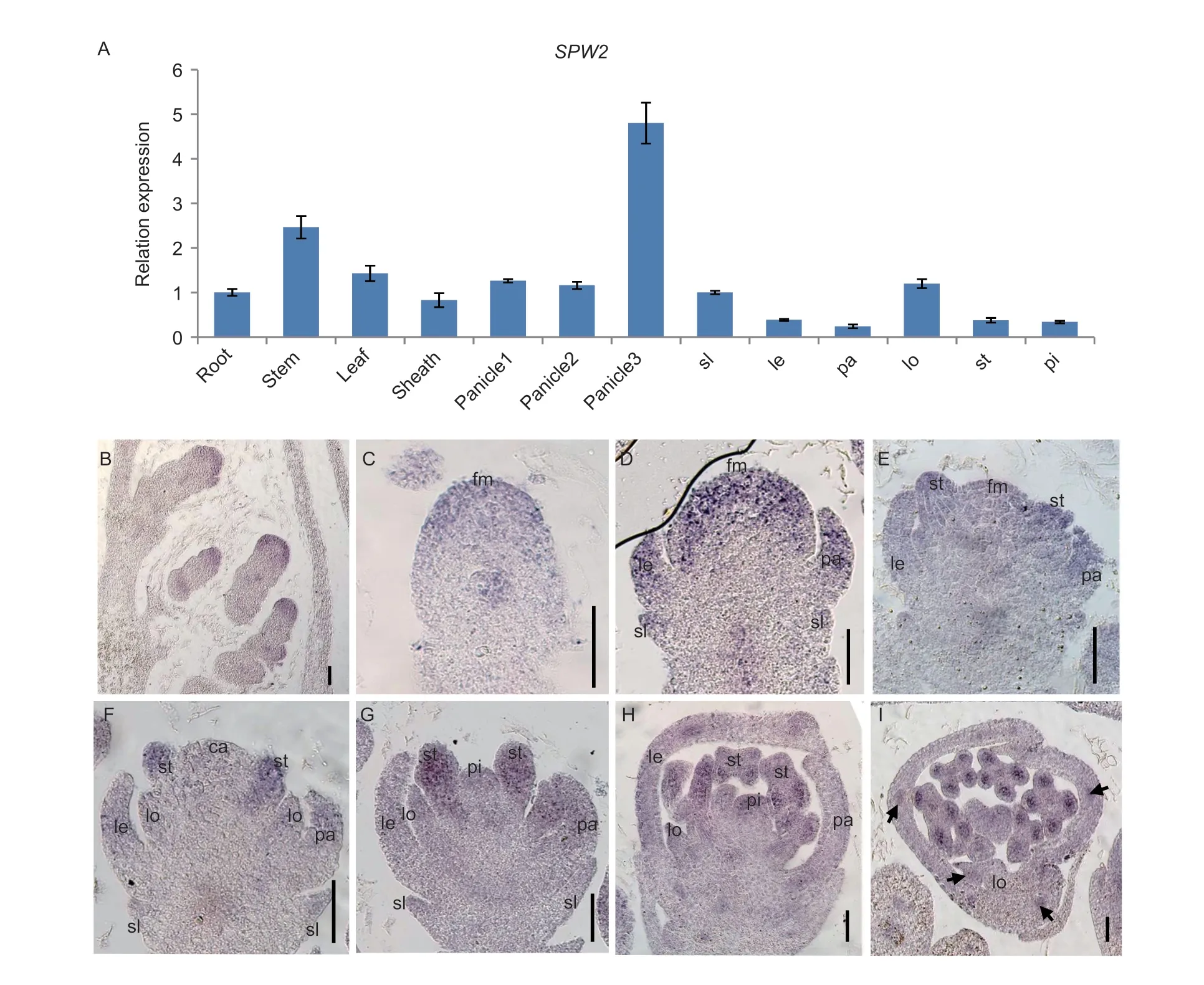
Fig.5 Expression analysis of SPW2.A,expression of the SPW2 gene by qPCR in wild type (WT).ACTIN was used as a control.Error bars indicate SD (n=3).At least three replicates were performed,from which the mean values were used to represent the expression levels.Panicle1,spikelets less than 0.5 cm;panicle2,0.5-2 cm spikelets;panicle3,2-3 cm spikelets;sl,sterile lemma;le,lemma;pa,palea;lo,lodicule;st,stamen;pi,pistil.B-I,in situ hybridization in WT panicles and spikelets,using an SPW2 antisense probe.B,spikelets at Sp1-Sp2,formation of rudimentary glume and empty glume primordia;C,a spikelet at Sp3;D-H,spikelets at Sp4-Sp8;I,transverse sections of organs in a WT spikelet.fm,floral meristem;lo,lodicule;st,stamen.Scale bars,50 μm.
3.6.Ectopic expression of pistil identity genes in mature spw2 spikelets
InArabidopsisthaliana,EMF1plays a crucial role in determining floral organ properties and morphogenesis.Theemf1-2mutant,which is a severe mutant ofEMF1,exhibits the formation of only carpelloid structures without the development of leaves,petals,or stamens.This phenotype is primarily attributed to the loss of function of a Polycomb Group (PcG)-mediated gene inhibition mechanism involvingEMF1,leading to the de-repression ofAG(Yangetal.1995;Aubertetal.2001;Moonetal.2003).Considering that thespw2mutant spikelets also display pistilloid organs,in order to determine whether theSPW2gene in rice also has a role in repressing the expression of pistil identity genes,we conducted a qRTPCR analysis to examine the expression levels ofDL,C-class genes (OsMADS3andOsMADS58),and D-class gene (OsMADS13) in both the WT andspw2mutant spikelets.
As expected,the RNA levels of theDL,OsMADS3,OsMADS13andOsMADS58genes in the mutant panicles were significantly higher than those in the WT (Appendix E).In rice,the developmental course of the inflorescence is categorized into nine stages (In1-In9) (Ikedaetal.2004).During stage In4,when the length of the inflorescence is less than 0.6 cm,the branching meristem undergoes elongation,and no lateral organs have differentiated.As the inflorescence continues to grow and reaches a length greater than 0.9 cm,the spikelet meristem and flower organ meristem form sequentially after stage In5.The ectopic expression of pistil identity genes,such asDL,OsMADS3,andOsMADS13,that was observed inspw2panicles at approximately 0.5-2 cm,could be attributed to the development of flower organs.However,sincespw2panicles that are less than 0.5 cm in length do not contain flower organ primordia,the early ectopic expression of theDL,OsMADS3,andOsMADS13genes observed as early as stage In4 is not a result of flower development.
Furthermore,the expression levels ofDL,C-class genes (OsMADS3andOsMADS58),and a D-class gene (OsMADS13) were found to be altered in nonpistil organs ofspw2spikelets.Specifically,in the sterile lemma,lemma,palea and lodicule,the expression levels of all these genes were significantly higher compared to the WT,which correlated with the observed pistilloid phenotypes inspw2spikelets (Appendix F).In the stamens,there was a substantial up-regulation ofDL,OsMADS13,andOsMADS58expression,which was consistent with the presence of pistil-like stamens in thespw2spikelets.These findings suggest thatSPW2may function as a negative regulator of the expression patterns of pistil identity genes.Mutations in theSPW2gene result in the ectopic expression of these genes,leading to the development of a severe pistilloid phenotype.
3.7.Ectopic expression of pistil identity genes in young spw2 spikelets
To further evaluate the genetic regulation among the relevant genes,we performedinsituhybridization to examine the expression patterns ofDL,OsMADS3,OsMADS13,andOsMADS58in thespw2mutant at an early stage of flower development.
In the WT,the expression ofDLwas first detected in the lemma primordia,and it subsequently became strongly detected in the carpel primordia (Fig.6-A1-A4).During the Sp8 stage,theDLexpression signal was observed in the peripheral domain of the medial vascular bundle of the lemma (Fig.6-A5).In thespw2mutant spikelet,a strongDLsignal was detected in the primordia of lemmas,similar to that of the WT (Fig.6-B1-B5).However,ectopic expression signals were detected in the abnormal sterile lemma primordia (triangles in Fig.6-B2 and B4),andDLtranscription signals inspw2flowers were found in the peripheral domains of lateral vascular bundles of the lemma-like sterile lemma,similar to those of the WT lemma (triangles in Fig.6-B5).Additionally,DLexpression was observed in the abnormal primordia of the mrp,lodicule and stamens inspw2flowers(triangles in Fig.6-B2-B5).The signal ofOsMADS3was first detected in the third-whorl founder cells positioned laterally to the flower meristem before the differentiation of stamen primordia (Fig.6-C1).Later,OsMADS3expression became restricted to the stamen and the carpel primordia of the WT (Fig.6-C2-C5).Inspw2spikelets,the extent of theOsMADS3signal gradually extends to the entire flower meristem before the emergence of stamen primordia(Fig.6-D1).Ectopic expression signals were detected in the lemmas,paleas,and abnormal lodicules ofspw2spikelets at the Sp6-sp8 stages (triangles in Fig.6-D2-D5).The expression ofOsMADS58was initially observed in the floral meristem when the lemma and palea primordia were just initiating (Fig.6-E1).Then,OsMADS58was expressed in the primordia of stamens,carpels,and ovules from their inception,and its expression persisted during the development of these floral organs (Fig.6-E2-E5).Inspw2mutant spikelets,strong ectopic signals ofOsMADS58were observed in the primordia of lemmas and palea at the Sp5 stage (Fig.6-F1-F2).As carpel primordia initiated,the ectopic transcripts ofOsMADS58were detected not only in the lemma primordia but also in the primordia of the abnormal stamens inspw2spikelets (Fig.6-F3-F5).The D-class geneOsMADS13showed high expression in whorl 4 just before carpel primordia initiation in the WT.Its expression continued in the ovule primordium and the inner layer of the developing carpel wall (Fig.6-G1-G4).At stage Sp8,detectable expression ofOsMADS13was observed in the ovule (Fig.6-G5).In comparison to WT flowers,ectopic expression ofOsMADS13could be detected in the primordia of sterile lemma,inner floral organs including lodicules,and ectopic pistils inspw2spikelets (Fig.6-H1-H5).
The observed ectopic expression of pistil identity genes in the primordia of sterile lemmas,mrp,lodicule and stamen inspw2spikelets suggests that these primordia have acquired hull and/or pistil identities during their developmental stages.Additionally,the ectopic expression ofOsMADS3,OsMADS13,andOsMADS58inspw2lemmas indicates the occurrence of stigma-like tissues on the tips of the lemmas during the early stages of flower development.These findings further support the notion that thespw2spikelet phenotype is partially caused by the ectopic expression ofDL,OsMADS3,OsMADS13,andOsMADS58,and that theSPW2gene plays a negative regulatory role in controlling the expression patterns of these genes.
3.8.SPW2 interferes with the transcription of pistil identity genes by mediating the H3K27me3 modification of chromatin
Previous reports indicated thatOsEMF1could represent a new class of molecules that plays a role in transcriptional regulation during different developmental periods in rice(Aubertetal.2001).To further explore the molecule mechanism by whichSPW2suppresses pistil identity genes,we performed transcriptional activity assays using both the Yeast Two-Hybrid (Y2H) System and the Dual-Luciferase Reporter (DLR) Assay System.
The ORFs ofSPW2and the known transcriptional activatorLF1were fused to pGBKT7.pGBKT7-LF1 and empty pGBKT7 were used as the positive control and negative control,respectively (Zhangetal.2017).pGBKT7,pGBKT7-LF1,and pGBKT7-SPW3 were transformed into yeast cells for growth on the selection medium.Yeast cells transformed with pGBKT7-LF1 were able to survive on the selection medium,whereas yeast cells transformed with pGBKT7-SPW2 or the empty pGBKT7 vector could not (Appendix G-a).This suggests that SPW2 does not have transcriptional activation activity in yeast cells.Next,we analyzed the transcriptional activity using the DLR System in rice protoplasts.We constructed dual-luciferase vectors for the SPW2 protein.The VP16 encoding a transcriptional activator was used as the positive control,and GAL4-BD was used as the negative control.The transcriptional activities of these proteins were measured in rice protoplasts.The SPW2 protein showed a slight enhancement of luciferase activity,approximately 2-fold,compared to control cells transfected with the empty vector,but the difference was not significant (P>0.05) (Appendix G-b).These results demonstrate that SPW2 does not exhibit significant transcriptional activation or repression activity as a transcription factor.
InArabidopsis,EMF1 has been shown to interact with specific regions ofAGinvivo,maintaining H3K27me3-mediated epigenetic silence and regulating shoot development (Chenetal.1997;Moonetal.2003;Calonjeetal.2008).Similarly,OsEMF1 in rice interacts with the PcG proteins OsMSI1 and OsiEZ1 to maintain floral organ identity by repressingOsMADS58through H3K27me3-mediated epigenetic regulation (Yanetal.2015;Zhengetal.2015).To investigate the potential regulatory role ofSPW2on pistil identity genes,we performed ChIP-qPCR analysis using an anti-H3K27me3 antibody to detect the levels of H3K27me3 in the promoters ofDL,OsMADS3,OsMADS13,andOsMADS58.
The results indicated that the levels of H3K27me3 inDL(P3 and P5 sites),OsMADS3(P1,P3 and P5 sites),OsMADS13(P2,P3 and P4 sites) andOsMADS58(P1,P2,P3,P4 and P6 sites) were significantly reduced in thespw2mutant compared with the WT (Fig.7).These results suggested thatSPW2regulates the transcription ofDL,OsMADS3,OsMADS13andOsMADS58by the modification of histone methylation.

Fig.7 Trimethylation of Lysine 27 of Histone H3 (H3K27me3) analysis of OsMADS3,OsMADS13,OsMADS58 and DL chromatin.A,C,E,and G,genomic structures of DL,OsMADS3,OsMADS13,and OsMADS58 and the regions tested in ChIP assays.B,D,F,and H,trimethylation of lysine 27 of histone H3 (H3K27me3) analysis of DL,OsMADS3,OsMADS13,and OsMADS58 chromatin.P1-P6,distribution of potential histone modifcation site(s) in the promoter of DL,OsMADS3,OsMADS13,and OsMADS58.Error bars indicate SD (n=3).At least three replicates were performed,from which the mean values were used to represent the expression levels.
4.Discussion
4.1.SPW2 is an important factor regulating flower organ identity in rice
In rice,the lack of function of certain floral organ identity genes leads to the transformation of floral organs into pistils or pistil-like structures,indicating their important role in rice floral development.Mutation of the B-class geneSPW1results in the conversion of stamens to carpels in rice (Kangetal.1998;Leeetal.2003).TheSL1gene in rice encodes a protein homologous toArabidopsisJAG.Mutants ofSL1exhibit the transformation of lodicules and stamens into glume-like organs and carpels,respectively(Horigomeetal.2009;Xiaoetal.2009).CFO1belongs to a monocot-specific clade in the MIKC-type MADSbox gene family,and its mutation leads to a defective palea,stamen-pistil chimeras,and ectopic floral organs(Sangetal.2012;Wangetal.2015).These studies indicate that the above-mentioned genes can suppress the ectopic expression of pistil identity in the second and third whorls,thus maintaining the normal development of non-pistil organs in the spikelets.Two reportedOsEMF1alleles in rice,ccp1anddfo1,exhibited defects in floral organ identity,with the mutants showing homeotic transformations of the margin region of the palea into pistil-like structures (Yanetal.2015;Zhengetal.2015).In accordance with the phenotypic analysis of these floral organ mutants,the most frequently observed transformation is the pistil-like stamen.Occurrences of pistilloid transformations in the palea and lodicule are rare,and there have been no reports of the transformation of sterile lemma or lemma into pistils.However,it is worth noting that organs closer to the pistil exhibit a higher resemblance to the pistil.
In this study,we identified a novel allelic mutant ofOsEMF1,calledspw2.In comparison to the reported mutants,spw2exhibited a remarkable super-pistilloid phenotype that affected a wide range of non-pistil organs in the spikelets,including the previously unreported sterile lemma and lemma.Previous studies revealed that the mutations inCCP1introduced a PscI restriction enzyme site,resulting in different digestion patterns,while premature stop codons indfo1-1anddfo1-2caused the absence of the DFO1 C-terminal domain(Yanetal.2015;Zhengetal.2015).Through mapbased cloning ofSPW2,we found that the premature translation termination causes a severe absence of the SPW2 structure,leading to null alleles.Based on these findings,SPW2can be regarded as a more representative complete loss-of-function mutant of theOsEMF1gene compared to thedfo1andccp1mutants.Compared to previously reported mutants,the pistil transformation in thespw2sterile lemma and lemma may be attributed to premature termination of the SPW2 protein or differences in the genetic background of the allelic variant.
Studies have confirmed that theOryzaancestor spikelet can be classified as a “three-floret spikelet”,where the sterile lemma is believed to originate from the degeneration of the lemma in the lateral florets (Arber 2010;Zhangetal.2017).For instance,PANICLE PHYTOMER2(PAP2) encodesOsMADS34,a regulator of sterile lemma development,and thepap2/osmads34mutant exhibits an elongated sterile lemma with lemma identity (Gaoetal.2010;Kobayashietal.2010).Similarly,ABERRANTSPIKELETANDPANICLE1(ASP1),which encodes a TOPLESS-like protein,results in a lemmalike sterile lemma in theasp1mutant (Yoshidaetal.2012).LONGSTERILELEMMA/ELONGATEDEMPTY GLUME1(G1/ELE1),a member of a plant-specific gene family that encodes a DUF640 domain protein,induces lemma-like organ formation in theg1/ele1mutant (Yoshidaetal.2009;Hongetal.2010).Furthermore,OsMADS1/LHS1determines the identities of the palea and lemma.Although it is not expressed in the sterile lemma,its ectopic expression leads to an enlarged sterile lemma resembling the lemma (Wangetal.2017).Collectively,these findings suggest that the sterile lemma is derived from the lemma of the lateral floret,and the sterile lemmas and lemma are homologous organs in rice.In this study,we observed varying degrees of pistilization in the non-pistil organs ofspw2spikelets.Interestingly,the sterile lemma and lemma ofspw2spikelets exhibited a similar phenotype,with the emergence of stigmatic tissue at their tips,which is a unique characteristic not previously reported in floral organ mutants.These findings indicate thatSPW2plays a crucial role in preserving the identities of the sterile lemma and lemma by preventing incorrect cell differentiation.Furthermore,our results provide additional support for the hypothesis proposing the homology between sterile lemmas and lemma in the“three-floret spikelet” theory.
4.2.SPW2 is responsible for the repression of pistil identity genes (OsMADS3,OsMADS13,OsMADS58,and DL) through epigenetic modification
In flowering plants,the regulation of the A-,B-,C-and E-class genes,along with their interactions,determines floral organ identity,resulting in proper floral patterning(Theissen and Saedler 2001;Soltisetal.2007;Litt and Kramer 2010;Theissenetal.2016).Moreover,various transcription factors and enzymes can individually or collectively regulate the spatio-temporal expression of these floral organ identity genes,facilitating the proper initiation of primordia.InA.thaliana,during the vegetative growth phase,H3K27me3-mediated gene repression involves PRC1 complex recruitment (including LIKE HETEROCHROMATIN PROTEIN 1[LHP1] and EMF1),compacting chromatin and inhibiting transcription at loci such asAP3andAG(Calonjeetal.2008;Winteretal.2011).Meanwhile,during flower patterning,the expression ofAP3andAGis directly promoted by the floral meristem identity geneLEAFY(LFY),which physically interacts with chromatin-remodeling ATPases SPLAYED (SYD) and BRAHMA (BRM),recruiting SYD to the regulatory regions ofAP3andAG,thereby triggering the reversal of polycomb repression (Wuetal.2012).The F-box protein UFO acts as a co-activator of LFY,enhancing the transcriptional activity of downstream B-class genesAP3andPI(Lohmann and Weigel 2002).APETALA2 (AP2) interacts with the transcriptional corepressor TOPLESS/TOPLESS RELATED (TPL),which further interacts with the histone deacetylase HDA19 to form the AP2-TPL-HDA19 complex,leading to repression of theAGgene in the two outer whorls (Kroganetal.2012).In contrast,AP2expression levels are not regulated byAG,as evidenced by the absence ofAP2transcripts in the central region of ag mutant flowers(Wollmannetal.2010).Instead,this control is mediated by miR172,a microRNA that inhibits the accumulation ofAP2mRNA and protein in the third and fourth whorls(Chen 2004;Wollmannetal.2010).
Our understanding of the regulatory mechanisms governing floral organ identities in rice is currently limited,with a scarcity of reports on the regulation of pistilrelated genes.In rice,four genes,namelyOsMADS3,OsMADS58,OsMADS13,andDL,are known to play a role in pistil development (Nagasawaetal.2003;Yamaguchietal.2006;Drenietal.2007).SPW1has been reported to genetically suppress the ectopic expression of theDLgene in the third whorl,ensuring normal development of the stamen.However,the current lack of biochemical evidence undermines the support for this viewpoint (Nagasawaetal.2003;Xiaoetal.2003).TheSL1also indirectly regulates the identity of stamens by maintaining the expression ofSPW1,which inhibits the expression ofDLin the third whorl (Nagasawaetal.2003;Yamaguchietal.2004;Xiaoetal.2009).Furthermore,CFO1negatively regulatesDLexpression in whorl 2,thereby inhibiting the formation of ectopic lodicule-pistil chimeras (Sangetal.2012;Wangetal.2015).As a component of the Polycomb group (PcG)complex,studies have verified thatOsEMF1regulates the expression of the C-class geneOsMADS58through histone modification with H3K27me3,thereby maintaining the proper morphology of the palea,lodicule and stamen in rice (Yanetal.2015;Zhengetal.2015;Liuetal.2018).
In this study,we found that all the pistil identity genes,includingOsMADS3,OsMADS58,OsMADS13,andDL,were ectopically expressed in the non-pistil organs ofspw2spikelets.The H3K27me3 levels were significantly reduced in the promoter regions ofOsMADS3,OsMADS13,OsMADS58,andDLinspw2young panicles.These results,combined with the observed pistilloid phenotype ofspw2,further suggest thatSPW2/OsEMF1mediates the levels of histone H3K27me3 on pistil identity genes such asOsMADS3,OsMADS13,OsMADS58,andDLto suppress their ectopic expression in non-pistil organs of rice spikelets,and maintains normal spikelet development.The results of our study highlight the functionality ofOsEMF1in regulating multiple genes involved in rice floral organ patterning,thereby enhancing our understanding of the molecular mechanisms underlying floral organ development in rice.
5.Conclusion
In this work,we isolated a novel allele forOsEMF1,namedSPW2,which encodes an ortholog ofEMF1inA.thaliana.TheSPW2gene plays an important role in repressing the expression of pistil identity genes includingDL,OsMADS3,OsMADS13andOsMADS58in rice through H3K27me3 modification.Loss of function ofSPW2results in a variety of pistilloid phenotypes in the non-pistil organs of the spikelet.
Acknowledgements
This work was supported by the Chongqing Modern Agricultural Industry Technology System,China(CQMAITS202301),the National Natural Science Foundation of China (32100287 and 31971919),the Natural Science Foundation of Chongqing,China(cstc2020jcyj-jqX0020 and cstc2021ycjh-bgzxm0066),and the China Postdoctoral Science Foundation Funded Project (2020M683219),and the Fundamental Research Funds for the Central Universities,China (SWUXDJH202315).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.07.010
杂志排行
Journal of Integrative Agriculture的其它文章
- Advances in DNA methylation and its role in cytoplasmic male sterility in higher plants
- Assessment of molecular markers and marker-assisted selection for drought tolerance in barley (Hordeum vulgare L.)
- 3D genome organization and its study in livestock breeding
- Genetic dissection and validation of a major QTL for grain weight on chromosome 3B in bread wheat (Triticum aestivum L.)
- Coupling of reduced inorganic fertilizer with plant-based organic fertilizer as a promising fertilizer management strategy for colored rice in tropical regions
- Effect of chemical regulators on the recovery of leaf physiology,dry matter accumulation and translocation,and yield-related characteristics in winter wheat following dry-hot wind
