Host-induced silencing of MpPar6 confers Myzus persicae resistance in transgenic rape plants
2024-01-17QiZhangWenqinZhanChaoLiLingChangYiDongJiangZhang
Qi Zhang ,Wenqin Zhan ,Chao Li ,Ling Chang ,Yi Dong# ,Jiang Zhang,
1 State Key Laboratory of Biocatalysis and Enzyme Engineering,School of Life Sciences,Hubei University,Wuhan 430062,China
2 Oil Crops Research Institute,Chinese Academy of Agricultural Sciences,Wuhan 430062,China
3 Agricultural Genomics Institute at Shenzhen,Chinese Academy of Agricultural Sciences,Shenzhen 518000,China
Abstract Plant-mediated RNA interference (RNAi) has emerged as a promising technology for insect control.The green peach aphid,Myzus persicae,feeds on over 400 species of host plants.Brassica napus (rape) is the second most important oilseed crop worldwide.Myzus persicae is highly reproductive and causes severe damage to the rape plants due to its quite flexible life cycle.In this study,we tested the RNAi effects of transgenic rape plants on M.persicae.By in vitro feeding M.persicae with artificial diets containing double-stranded RNAs (dsRNAs) targeting seven aphid genes,we identified a new gene encoding the partitioning-defective protein 6 (Par6) as the most potent RNAi target.Tissue-and stage-expression analysis of Par6 suggested this gene is highly expressed in the embryo and adult stage of M.persicae.We next generated transgenic rape plants expressing dsPar6 by Agrobacteriummediated transformation and obtained nine independent transgenic lines.Compared to wild-type control plants,transgenic rape lines expressing dsPar6 showed strong resistance to M.persicae.Feeding assays revealed that feeding transgenic rape plants to M.persicae significantly decreased MpPar6 expression and survival rate and impaired fecundity.Furthermore,we showed that the resistance levels to M.persicae are positively correlated with dsPar6 expression levels in transgenic rape plants.Our study demonstrates that transgenic rape plants expressing dsPar6 are efficiently protected from M.persicae.Interfering with the genes involved in embryo development could be the effective RNAi targets for controlling aphids and potentially other insect pests.
Keywords: oilseed rape,pest control,aphid,double-stranded RNA,RNA interference
1.Introduction
The green peach aphid,Myzuspersicae,is a typical sapsucking insect belonging to the order Hemiptera.It is distributed worldwide except in extreme climatological regions (Barbagalloetal.2007).Myzuspersicaeis highly polyphagous and feeds on over 400 species of host plants,including potato,cabbage,radish,mustard,rape,pepper,and spinach (Larson and Whitham 1991;Chenetal.2020).In a favorable climate,it could reproduce in parthenogenetic viviparity on the host plants for over 20 annual generations with overlapping generations (Capinera 2020).Both nymphs and adults ofM.persicaecan suck the phloem sap from host plants and excrete large drops of honeydew on the leaves and fruits.This promotes the appearance of fungal pathogens(Cladosporiumspp.) and viruses,thus resulting in serious damage to crops (Yuetal.2014;Abdollahzadehetal.2020).Other factors such as overcrowding,plant wilting,and viral RNA interference could additionally speed upM.persicaepopulation to produce the alates,facilitate the migration ofM.persicaeto neighboring plants,and spread bacterial and viral pathogens (Vanetal.1969;Jayasingheetal.2021).Because of these physiological advantages,M.persicaehas become one of the most notorious pests responsible for the economic losses of global agriculture(Alvarez 2007;Valenzuela and Hoffmann 2015).At present,the control ofM.persicaemainly relies on traditional chemical insecticides.However,the abuse of chemical insecticides not only leads to environmental pollution and the evolution of resistance but also causes the reduction of natural enemies (Bassetal.2014).
RNA interference (RNAi) is a highly conserved mechanism of gene regulation and defense in the eukaryotic organism.Double-stranded RNAs (dsRNAs)can trigger gene silencing in insects in a sequencespecific manner (Gordon and Waterhouse 2007;Hung and Slotkin 2021).When internalized into insect cells,dsRNAs are cleaved by the Dicer enzyme to generate small interfering RNAs (siRNAs),which guide the Argonaute protein of the RNA-induced silencing complex(RISC) to disassemble complementary messenger RNAs(mRNAs) (Cooperetal.2019).Degradation of mRNAs of essential insect genes may lead to multiple deleterious phenotypes of insects,including growth retardation,impaired fecundity,and increased mortality.RNAimediated pest control is considered an environmentally friendly method and has great application potential.Recently,plant-mediated RNAi for aphid control has been demonstrated.Feeding on transgenic wheat plants expressing dsSaCHS1(dsRNA targetingChitinsynthase1ofSitobionavenae) resulted in an almost 50% decreased population ofS.avenae15 days after flowering when compared with the non-transgenic plants (Zhaoetal.2018).Similarly,interference with theSaZFP(a gene encoding zinc finger protein ortholog inS.avenae) mRNA ofS.avenaeby feeding on transgenic wheat brought about over 40% mortality within 9 days (Sunetal.2019).Silencing ofMpC002(a putative effector gene encoding a predicted signal peptide inM.persicae) ofM.persicaeled to a more than 60% reduction in the population of aphids on transgenicArabidopsisplants (Colemanetal.2015).In agreement with this,the number of offspring ofM.persicaefeeding on transgenicArabidopsisplants was reduced by nearly 40% by the downregulation of the CP gene,encoding a cuticular protein (Bhatia and Bhattacharya 2018).
Oilseed rape (Brassicanapus) is an important source of vegetable oil grown worldwide.It is primarily grown as a commercial crop for the production of vegetable oil from its seeds,and its young edible leaves can also be used as forage for livestock.Myzuspersicaeis one of the main pests on rape plants (Drizouetal.2018) and causes massive damage by not only directly absorbing plant sap but also transmitting plant viruses such as Turnip mosaic virus and Beet western yellows virus (Symptomology and Aphids 2010).
This study identified a new gene encoding the partitioning-defective protein 6 (Par6) as an efficient RNAi target inM.persicae.We found that the expression ofPar6was much high in the embryos and adults ofM.persicae.When thePar6sequence was introduced into the nuclear genome of rape plants byAgrobacteriummediated transformation,transgenic plants expressing dsPar6displayed significant resistance toM.persicae.
2.Materials and methods
2.1.In vitro synthesis of dsRNA and screening of lethal RNAi target genes in aphid
By feedingM.persicaewith artificial diets containing synthetic dsRNA,we screened potential lethal targets of RNAi forM.persicaecontrol (Fraseretal.2000;Rispeetal.2008).Seven genes ofM.persicaewere selected as RNAi target genes.The target gene fragments were amplified with gene-specific primers by using aphid cDNA as a template (Appendix A).DsRNA was synthesized with the T7 RiboMAX Express RNAi Kit (Promega,Madison,WI,USA) according to the manufacturer’s instructions.The concentration of RNA was measured with a NanoPhotometer N60-touch (IMPLEN,Munich,Bavaria,Germany),and the RNA integrity was assessed by agarose gel electrophoresis.The GFP sequence (NCBI Sequence ID: NZ_CP084280.1) was used as a negative control.
The concentration of dsRNA in the artificial diet was set as 10 ng μL-1to screen for lethal RNAi targets.Artificial diet (2.5-3.5% amino acids,0.2-0.3% ascorbic acid trace element chelate,0.1-0.2% B vitamins,17-20% sucrose,3.0-4.0% tobacco sap,0.4-0.6% KH2PO4,0.1-0.3%MgCl2) was prepared in DEPC-treated water to avoid the degradation of dsRNA.To synchronize the developmental stages of aphids,30 adult aphids were selected for parthenogenesis on tobacco plants and removed from the plants after 24 h.When the nymphs reached the second instar,they were placed in transparent grass tubes with open sides.The artificial diet was sealed between the two layers of Parafilm,and 15 nymphs of the second instar were raised in each tube.The tubes were placed in an incubator at 25°C,75-80% relative humidity,and a photoperiod of 16 h light/8 h dark.The survival rate of aphids was recorded,and the diet was changed daily.On the third and seventh days,six aphids were randomly collected for gene-silencing analysis.The experiments were repeated three times.
2.2.Plant material and growth conditions
The variety ofBrassicanapus(rape) used in this study is Zhongshuang 6 (ZS6).The seeds were obtained from the germplasm resource of the Oil Crops Research Institute,Chinese Academy of Agricultural Sciences.Wild-type and transgenic oilseeds were pre-germinated in an aqueous solution containing 300 mg L-1kanamycin at 37°C for 24 h and then moved into the soil.After seed germination,the white cotyledons (susceptive to kanamycin) were removed,and the green cotyledons (resistant to kanamycin) remained.Rape plants were grown under controlled greenhouse conditions (photoperiod of 16 h light/8 h dark at 22°C).
2.3.Construction of transformation vectors
For the vector construction,cDNA ofM.persicaewas used as the template for PCR amplification.A 290-bpMpPar6DNA fragment was PCR amplified with primer pair Par6-F1-XhoI/Par6-R1-BglII (Appendix A),digested with restriction enzymesXhoI andBglII,and ligated into a similarly cut plasmid pUC-RNAi,generating plasmid pUC-Par6-1.The antisense strands ofMpPar6DNA fragment were PCR amplified with primer pair Par6-F2-SalI/Par6-R2-BamHI,digested with restriction enzymesBamHI andSalI,and inserted into similarly cut plasmid pUC-Par6-1,producing plasmid pUC-hpPar6.Finally,theMpPar6sequence was cloned asXhoI/SalI fragment from pUC-hpPar6 into a binary vector (Coutuetal.2007)forAgrobacterium-mediated nuclear transformation,generating the nuclear transformation vector pnuPar6.
2.4.Plant transformation
Construct pnuPar6 was first introduced intoAgrobacterium tumefaciens(strain GV3101) by electroporation;positive clones were selected on Luria-Bertani medium supplemented with 50 mg L-1rifampicin and 50 mg L-1kanamycin.After the verification by PCR analysis,a single positive colony was used to transformB.napuscv.ZS6.Oilseeds were sterilized by soaking in 75% ethanol for 1 min and in a 1.5% mercuric chloride solution for 10-15 min and germinated under darkness.After 6 days,the etiolated hypocotyls were cut into 7 mm segments and infected withA.tumefacienscontaining pnuPar6 in 50 mL liquid Murashige and Skoog (MS) medium supplemented with 30 g L-1sucrose and 100 μmol L-1acetosyringone (pH 5.8) (OD=0.3) for 0.5 h.After air drying,the hypocotyls were first transferred to a cocultivation medium (MS supplemented with 30 g L-1sucrose,18 g L-1mannitol,1 mg L-12,4-D,0.3 mg L-1kinetin,and 100 μmol L-1acetosyringone) for 2 days,and then to a selection medium (MS supplemented with 30 g L-1sucrose,18 g L-1mannitol,1 mg L-12,4-D,0.3 mg L-1kinetin,20 mg L-1AgNO3,25 mg L-1kanamycin,and 250 mg L-1carbenicillin) for screening of antibioticresistant calli.After 3 weeks,the calli were transferred to a regeneration medium containing MS salts,10 g L-1glucose,0.25 g L-1xylose,0.6 g L-1MES hydrate,2 mg L-1zeatin,0.1 mg L-1indole-3-acetic acid,25 mg L-1kanamycin,and 250 mg L-1carbenicillin for 2 weeks.The regeneration medium was replaced every 2 weeks for 3-4 regeneration cycles.Regenerated shoots were transferred onto a rooting medium containing MS salt and 10 g L-1sucrose.Rooted shoots were transferred into the soil.The transgenic lines were initially tested for the presence of the transgene by PCR assays.
2.5.Aphid tissue and stage samples obtaining
The tissue samples ofM.persicaewere dissected from about 70 adults using insect dissection pins.The different stage samples ofM.persicaewere selected from the aphid population,which was raised on wild-type tobacco plants under controlled growth conditions (25°C,75-80%relative humidity,and a photoperiod of 16 h light/8 h dark),starting from the first-instar aphid nymph to adult.
2.6.Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from insect or plant samples using Trizol reagent (Invitrogen,Waltham,MA,USA) according to the manufacturer’s instructions.In addition,1 μg RNA was used for the synthesis of first-strand cDNA.The qRTPCR was performed using a CFX Connected Real-time System (BioRad,Hercules,CA,USA) with TB GreenTMPremixExTaqTM II (TaKaRa,Dalian,China) following the manufacturer’s instructions.The qPCR procedure consisted of an initial denaturation at 95°C for 2 min,followed by 39 cycles at 95°C for 5 s,60°C for 30 s,and 72°C for 30 s.Melt curve analysis was performed to assess the specificity of amplification.The relative expression levels of the genes were quantified using the aphid18SrRNAgene and rapeβ-actingene as internal reference genes.All qRT-PCR assays were performed in triplicate.Relative gene expression levels were calculated by using the 2-ΔΔCTmethod (Livak and Schmittgen 2001).The primer sequences for qRT-PCR are listed in Appendix A.
2.7.Aphid bioassays on whole plants
The aphid population was raised on wild-type tobacco plants under controlled growth conditions (25°C,75-80%relative humidity,and a photoperiod of 16 h light/8 h dark).To synchronize the developmental stage of aphids,adult aphids of 2-3 days were placed on 10-week-old rape plants to produce nymphs and then removed from these plants after 24 h.
When the wild type (Bn-wt) and transgenic type (BnnuPar6#1 andBn-nuPar6#7) were grown for 10 weeks,35 first-instar nymphs were allowed to feed on the indicated rape plants.Bioassays were performed in plastic cages closed with mesh to prevent aphids from escaping (Dongetal.2022).The number of progenies on each plant was counted,and survival rates were recorded daily.Six aphid samples were randomly selected as a sampling group for RNA isolation and qRT-PCR analysis on day 9.The bioassay was performed in triplicate.
2.8.Statistical analysis
Survival curves were analyzed using the Kaplan-Meier method.The log-rank test was used to evaluate significant differences between the two groups.The differences between multiple groups were analyzed using one-way ANOVA with Duncan’s multiple range test,and the differences between the two groups were analyzed using Student'st-test.
3.Results
3.1.Screening of potential lethal target genes of RNAi for aphid control
Based on the survival curves,we found that among the seven RNAi targets,dsAdar,dsPar6,or dsLamwere significantly higher than dsGFPcontrol (dsGFPvs.dsAdar:P<0.001;dsGFPvs.dsPar6:P<0.001;dsGFP vs.dsLam:P=0.001) (Fig.1-A and B).The mortality ofM.persicaefed on dsPar6was about 70%,significantly higher than that ofM.persicaefed on dsAdaror dsLam(dsAdarvs.dsPar6:P=0.05;dsLamvs.dsPar6:P=0.002).Meanwhile,cumulative mortality in the dsAdarand dsLamgroups was about 50%,significantly higher than that in the control group (about 10%).
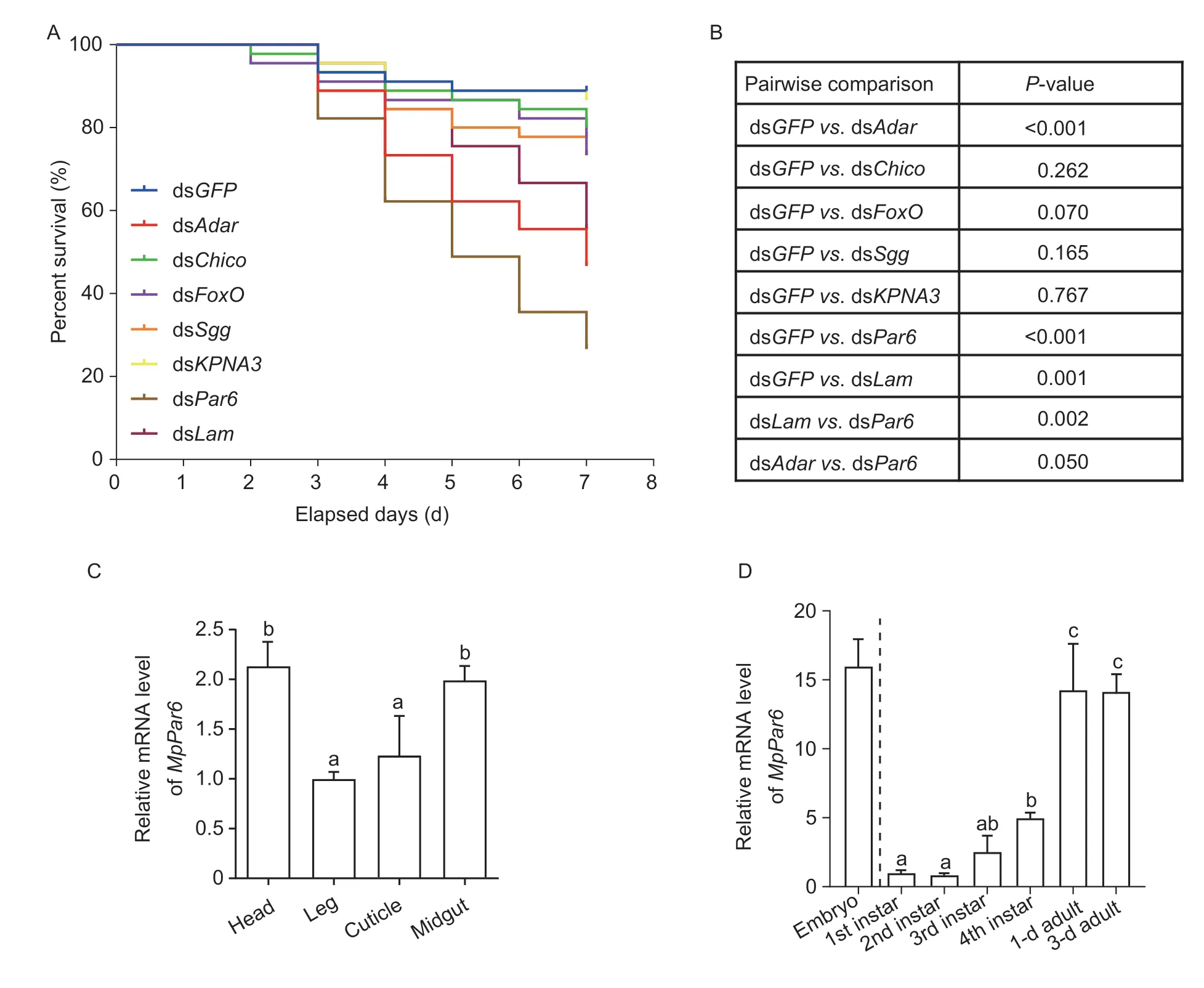
Fig.1 Feeding assays of Myzus persicae with artificial diets containing synthetic dsRNAs.A,survival rates of M. persicae fed with artificial diets containing synthetic dsRNAs.B,the significance analysis of survival curves.C,analysis of the expression levels of MpPar6 in the head,leg,cuticle,and midgut of the aphid adults by qRT-PCR.D,relative expression levels of the MpPar6 in the different developmental stages of M. persicae.The data represent mean±SD (n=3) and different lowercase letters above the columns represent significant differences (P<0.05,one-way ANOVA with Duncan’s multiple range test).
To confirm whether the increased mortality is due to the silencing of the target gene,we determined the expression level of thePar6gene inM.persicaeupon feeding with artificial diets by qRT-PCR (Appendix B).In agreement with the mortality data,the expression level ofPar6inM.persicaefed on dsPar6was significantly downregulated on the third and seventh days compared with the control group (Fig.1).These results suggest thatPar6is the most effective target for inducing RNAi inM.persicae.
We next dissectedM.persicaeadults and analyzed the expression pattern ofPar6in different tissues by qRTPCR.We found thatPar6is constitutively expressed in all the tissues,displaying a relatively higher expression in the head and midgut than that in the leg and cuticle(Fig.1-C).By detecting the expression profile ofMpPar6in the different developmental stages ofM.persicae,we revealed that the expression ofMpPar6is significantly higher in the embryonic or adult stage than in the nymph stage;additionally,its expression level is low at the first and second instar stages,and gradually increased at third and fourth instar stages (Fig.1-D).
3.2.Generation and molecular characterization of transgenic rape plants expressing dsPar6
Having confirmedPar6as a potent target for RNAi inM.persicae,we performed rape transformation for expression of dsPar6againstMpPar6byAgrobacteriummediated transformation (Fig.2-A).Twenty kanamycinresistant lines were obtained,and nine of them were found to be positive transgenic lines verified by PCR analysis (Fig.2-B).However,no PCR amplification signals were detected in the negative control (NC) or wild-type rape plants (Bn-wt).Subsequently,the relative expression of dsPar6in nine transgenic rape lines was measured by qRT-PCR (Fig.2-C).PCR analysis of the T2 transgenic rape plants were also measured (Appendix C).Two transgenic rape lines (Bn-nuPar6#1 andBnnuPar6#7) with relatively higher expression levels were selected for the next aphid bioassays.

Fig.2 Generation of the transgenic rape plants expressing RNAi constructs targeted against Myzus persicae.A,map of the T-DNA locus in nuclear transgenic rape lines (Bn-nuPar6 lines) transformed with a hairpin construct for expression of dsPar6.B,PCR analysis of the transgenic rape plants(Bn-nuPar6 lines).The Bn-nuPar6 lines were generated after selection for kanamycin resistance.The nuclear transformation vector pnuPar6 served as a positive control (PC),and PCR amplification with H2O as a template served as negative control (NC).C,analysis of the relative expression of dsPar6 in the transgenic lines by qRT-PCR.The rape β-actin mRNA was used as an internal standard.The data represent mean±SD (n=3)and different lowercase letters above the columns represent significant differences (P<0.05,one-way ANOVA with Duncan’s multiple range test).
3.3.Assessment of the resistance of transgenic rape plants to M.persicae
Seeds from the T2 generation of two independent transgenic lines (Bn-nuPar6#1 andBn-nuPar6#7) were soaked in a kanamycin-containing aqueous solution.After germination,the seedling of wild-type plants (Bn-wt)presented a lemon-yellow phenotype probably,while two transgenic lines showed normal green (Fig.3-A).When grown in soil,transgenic rape plants were grown normally and indistinguishable from wild-type plants (Fig.3-B).After 10 weeks of growth,35 newborn nymphs were selected and inoculated on the indicated rape plants.
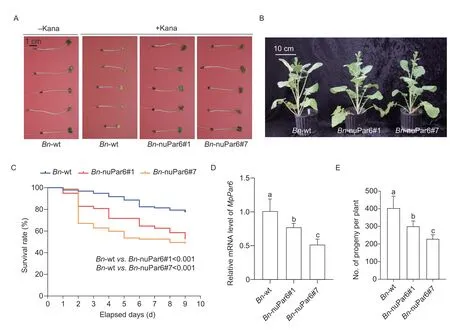
Fig.3 Feeding assays of Myzus persicae on the transgenic rape plants.A,seed assays to confirm the homozygous status of the transgenic rape plants.B,phenotypes of ten-week-old wild-type and the transgenic rape plants.C,survival rates of M. persicae fed on wild-type and transgenic rape plants.D,analysis of Par6 expression in M. persicae fed on wild-type and transgenic rape plants after nine days.E,the number of progenies of M. persicae fed on wild-type and transgenic rape plants.The data are represented as mean±SD (n=3) and different lowercase letters above the columns represent significant differences (P<0.05,oneway ANOVA with Duncan’s multiple range test).
The survival rates ofM.persicaeon wild-type (Bn-wt)and transgenic rape lines (Bn-nuPar6#1 andBnnuPar6#7) were monitored daily.The result showed that feedingM.persicaewith transgenic rape plants induced significantly higher mortality compared with wildtype plants (Bn-wtvs.Bn-nuPar6#1:P<0.001;Bn-wtvs.Bn-nuPar6#7:P<0.001).The mortality ofM.persicaereached around 50% on the ninth day of feeding on nuclear transgenic lines (Bn-nuPar6#1 andBn-nuPar6#7),while there was less than 20% mortality ofM.persicaeupon feeding with wild-type plants (Bn-wt).However,there was no significant difference in the mortality ofM.persicaefed on two nuclear transgenic lines (BnnuPar6#1vs.Bn-nuPar6#7:P=0.364) (Fig.3-C).
qRT-PCR analysis was next conducted to determine the expression of theMpPar6.We found thatMpPar6was significantly suppressed inM.persicaefed on the transgenic plants compared to that fed on wild-type plants.The suppressed expression level ofPar6inM.persicaefed onBn-nuPar6#7 was more pronounced than that onBn-nuPar6#1 (Fig.3-D).
Similarly,the number ofM.persicaeprogeny on the transgenic plants was obviously decreased compared to that on wild-type plants,and the number ofM.persicaeprogeny onBn-nuPar6#7 was much less than that onBn-nuPar6#1 (Fig.3-E).These results demonstrate that transgenic rape plants expressing dsPar6are resistanttoM.persicae,and the levels of resistance are positively correlated with the expression levels of dsPar6.
4.Discussion
It has been shown that plant-mediated RNAi can be successfully employed for the control of sap-sucking pests (Sunetal.2019;Dongetal.2020).It is important to screen ideal RNAi targets to increase the RNAi effects in the insect pests.In agreement with previous studies,our data showed thatPar6is highly expressed in the embryonic development stage ofM.persicae.We demonstrated that the transgenic rape plants expressing dsPar6are of significant resistance toM.persicae,indicating thatPar6is an effective target of RNAi for aphid control.
Par6is a conserved gene presenting inCaenorhabditis elegans,Drosophilamelanogaster,mice,and humans.It is involved in several important physiological processes in the living cells,including the establishment of mitotic spindle localization,gonad development,and specification of anterior/posterior axis polarity (Macara 2004;Buckley and St Johnston 2022).Feeding dsPar6-expressing bacteria toC.elegansled to over 90% of the mortality of their offspring (Fraseretal.2000).InD.melanogaster,interference withPar6expression in the neuroblasts or the intermediate neural progenitors resulted in a lethal phenotype (Neumülleretal.2011).Consistent with these studies,our results showed that mortality ofM.persicaesignificantly increased after feeding on an artificial diet containing dsPar6(Fig.1-A) or transgenic rape plants expressing dsPar6(Fig.3-C),indicating thatPar6is an effective target of RNAi for control ofM.persicae.Par6is a pivotal gene in the process of embryonic development and is necessary for the formation of apicobasal and anterior-posterior asymmetries associated with cell adhesion and gastrulation during the first few cell cycles of embryogenesis (Wattsetal.1996).These may explain the phenomenon that the expression ofMpPar6was the highest in the embryos (Fig.1-C and D),and a similar observation was also described inD.melanogaster(Huttereretal.2004).Moreover,M.persicaeused in this study is reproduced in a manner of parthenogenesis;in this way,the embryonic development stage is conducted in the body of the female parent.At the third to fourth instar stages ofM.persicae,the expression ofMpPar6began to increase and was highly expressed at the adult stage (Fig.1-D).This observation is probably related to the beginning and the gradual acceleration of the development of embryos in the body of nymphs or adults(Blackman 1978).Since the parthenogenetic viviparity of aphids is universal,it would be an ideal choice to interfere with the genes highly expressed in the embryonic stage for aphid control.
5.Conclusion
The study demonstrates that the expression of dsPar6is an effective strategy for RNAi-based protection of rape plants from aphids.The insect genes involved in embryo development could be the potential RNAi targets for efficient control of insect pests.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32102297 and 32272634).
Declaration of competing interests
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.05.027
杂志排行
Journal of Integrative Agriculture的其它文章
- Advances in DNA methylation and its role in cytoplasmic male sterility in higher plants
- Assessment of molecular markers and marker-assisted selection for drought tolerance in barley (Hordeum vulgare L.)
- 3D genome organization and its study in livestock breeding
- SUPER WOMAN 2 (SPW2) maintains organ identity in spikelets by inhibiting the expression of floral homeotic genes OsMADS3,OsMADS58,OsMADS13,and DROOPING LEAF
- Genetic dissection and validation of a major QTL for grain weight on chromosome 3B in bread wheat (Triticum aestivum L.)
- Coupling of reduced inorganic fertilizer with plant-based organic fertilizer as a promising fertilizer management strategy for colored rice in tropical regions
