High-throughput screening system of citrus bacterial cankerassociated transcription factors and its application to the regulation of citrus canker resistance
2024-01-17JiaFuJieFanChenxiZhangYongyaoFuBaohangXianQiyuanYuXinHuangWenYangShanchunChenYongruiHeQiangLi
Jia Fu ,Jie Fan ,Chenxi Zhang ,Yongyao Fu ,Baohang Xian ,Qiyuan Yu ,Xin Huang,Wen Yang,Shanchun Chen,,Yongrui He,#,Qiang Li,#
1 Integrative Science Center of Germplasm Creation in Western China (Chongqing) Science City,Citrus Research Institute,Southwest University,Chongqing 400712,China
2 School of Advanced Agriculture and Bioengineering,Yangtze Normal University,Chongqing 408100,China
3 National Citrus Engineering Research Center,Chongqing 400712,China
Abstract One of the main diseases that adversely impacts the global citrus industry is citrus bacterial canker (CBC),caused by the bacteria Xanthomonas citri subsp.citri (Xcc).Response to CBC is a complex process,with both protein-DNA as well as protein-protein interactions for the regulatory network.To detect such interactions in CBC resistant regulation,a citrus high-throughput screening system with 203 CBC-inducible transcription factors (TFs),were developed.Screening the upstream regulators of target by yeast-one hybrid (Y1H) methods was also performed.A regulatory module of CBC resistance was identified based on this system.One TF (CsDOF5.8) was explored due to its interactions with the 1-kb promoter fragment of CsPrx25,a resistant gene of CBC involved in reactive oxygen species (ROS) homeostasis regulation.Electrophoretic mobility shift assay (EMSA),dual-LUC assays,as well as transient overexpression of CsDOF5.8,further validated the interactions and transcriptional regulation.The CsDOF5.8-CsPrx25 promoter interaction revealed a complex pathway that governs the regulation of CBC resistance via H2O2 homeostasis.The high-throughput Y1H/Y2H screening system could be an efficient tool for studying regulatory pathways or network of CBC resistance regulation.In addition,it could highlight the potential of these candidate genes as targets for efforts to breed CBC-resistant citrus varieties.
Keywords: citrus bacterial canker (CBC),high-throughput screening system,transcription factor (TF),yeast-one hybrid (Y1H),CsPrx25
1.Introduction
One of the main diseases that threaten the global citrus industry is citrus bacterial canker (CBC),caused by the bacteriaXanthomonascitrisubsp.citri(Xcc) (Pengetal.2017;Lietal.2020a).Therefore,there is an urgent need to strengthen research on the prevention and control of CBC to sustain the citrus industry.In this context,molecular breeding has been rapidly developed and used for the efficient cultivation of disease-resistant germplasms (Ma and Mo 2017).
At the molecular level,transcription factors (TFs)represent a class of proteins that can specifically bind tocis-acting elements in the promoter regions for regulating the expression of target genes (Liuetal.1999).As far as citrus plants are concerned,TFs are important to control their growth,development,and resistance.For example,CiMADS43,a MADS-box TF,interacts with CiAGL9 to influence citrus flowering as well as the development of leaves (Yeetal.2021).Similarly,CitNAC62 in citrus acts alongside CitWRKY1 to upregulate CitAco3,resulting in the degradation of citric acid (Lietal.2017).Finally,in trifoliate orange (Poncirustrifoliata(L.) Raf.),ERF109 confers tolerance to cold conditions by regulating Prx1 expression,a gene involved in antioxidative process(Wangetal.2019).In a similar way to the above features,CBC resistance is also finely controlled by a complex network of TF.For instance,CsAP2-09,a AP2/ERF TF,provides resistance against CBC by regulating reactive oxygen species (ROS) homeostasis (Heetal.2019),while CsBZIP40,a BZIP TF,positively induces tolerance and response to CBC by regulating the salicylic acid (SA)signaling pathway (Lietal.2019).CsWRKY22,a WRKY TF,also influences susceptibility to CBC by enhancing cell enlargement as well as CsLOB1 expression (Longetal.2021).In addition to TFs,a number of other proteins are also important against citrus canker disease,with one example being CsPrx25,a class III peroxidase,that provides resistance to CBC by maintaining ROS homoeostasis as well as through cell wall lignification (Lietal.2020c).Similarly,citrus plants can display greater resistance to CBC through jasmonic acid (JA) signaling and ROS control,mediated by CsWAKL08,a pathogeninduced wall-associated receptor-like kinase (Lietal.2020b).However,the relationships between TFs as well as between TFs and other key regulators are poorly understood.Hence,exploring the upstream regulators and interaction of proteins of targets is of significance to clarify their regulatory mechanisms and construct their regulatory networks in view of identifying new potential candidate genes for breeding CBC-resistant varieties.
The yeast two-hybrid (Y2H) system,developed in 1989,can be used to study protein-protein interactions(Fields and Song 1989),and this can be followed by the yeast one-hybrid (Y1H) system that verifies protein-DNA interactions (Li and Herskowitz 1993).Together,these approaches allow the study of a large number of interacting factors to resolve complex transcriptional regulatory networks.However,even though cDNA libraries have been widely used for Y1H and Y2H,the low proportion of TFs and their low expression abundance made it difficult to screen interactive TFs with Y1H and Y2H (Mitsudaetal.2010).Indeed,TFs ofArabidopsisthalianaand rice respectively account for only 5 and 2.6% of all genes(Riechmannetal.2000;Xiongetal.2005).Therefore,a Y1H/Y2H library dedicated to TFs screening can greatly improve the efficiency and accuracy of the process.In this context,A.thaliana’s library of TFs on circadian rhythm regulation was created in 2009 (upgraded in 2014),and it allowed the geneCHE-centered regulatory pathway to be resolved (Pruneda-Pazetal.2009,2014).More recently,the upstream regulators and interacting proteins (PaPAL2 and PaMYB4) that influence wood formation were assessed though a high-throughput screening system forPopuluswood-associated TFs (Zhuangetal.2021).
Currently,there are no reports regarding the use of Y1H/Y2H-based screening systems specifically meant for the study of CBC.In this work,the TFs induced by CBC were explored before constructing a yeast library.To understand the regulatory mechanism ofCsPrx25involved in CBC resistance,one upstream regulator from the CBC-associated TF library,was screened to construct the regulatory pathway ofCsPrx25.The proposed high-throughput Y1H/Y2H screening system can be a powerful tool for efficiently and accurately constructing the regulatory pathways of CBC-associated genes.The findings,in turn,are expected to be of value for the study of CBC regulation and for the breeding of CBC-resistant varieties.
2.Materials and methods
2.1.Plants and bacteria
Citrus varieties Wanjincheng (Citrussinensis) and kumquat (Fortunellajaponica) were obtained from the National Citrus Germplasm Repository,Chongqing,China (19°51´N,106°37´E).Both species were used forXccassays,with Wanjincheng also used for transient transformation.The plants were kept at 28°C in greenhouses for growth,while theXccvariantXccYN1,obtained from infection-susceptible citrus leaves,was cultured at 28°C in peptone-yeast extract/malt extract containing D-glucose at a concentration of 1.5% (w/v) (Lietal.2021).
2.2.In silico analyses
Transcription factor binding sites were first predicted with JASPAR (http://jaspar.genereg.net) (Fornesetal.2020) before visualizing gene structures using the Gene Structure Display Server (GSDS V2.0,http://gsds.gao-lab.org) (Huetal.2015).In order to confirm the genes that were included in this study,BLAST was performed against the citrus genomic variations database (CitGVD,http://citgvd.cric.cn) (Lietal.2020b),and eventually,functional domains were predicted using the Simple Modular Architecture Research Tool (SMART,https://smart.embl.de)(Letunicetal.2021).
2.3.Xcc treatment
Wanjincheng and kumquat were processed for infection byXccas previously described (Lietal.2020a).Briefly,fresh leaves were placed in a petri dish and rinsed with 75% ethanol,followed by sterile distilled water (ddH2O).The leaves were then kept at 28°C with a 16-h-light/8-hdark cycle.Xccwas cultured overnight at 28°C until an OD600of 0.5 was reached.This culture was subsequently used for inoculating the leaves,and at 6,12,and 24 hours post-inoculation (hpi),samples were taken for analysis.
2.4.RNA-seq and identification of differentially expressed TFs
Three Wanjincheng and kumquat lines served as three biological replicates,infected withXccor ddH2O(mock) were used for RNA-seq assay.Total RNA was extracted from the leaves with an RNA miniprep kits(AidLab,China) as required by the manufacturer,and was shipped to Majorbio Inc.for RNA-seq on an Illumina Novaseq 6000 platform.The clean data were then mapped onto theC.sinensisgenome V3 before being assembled and annotated using the Citrus Pan-genome to Breeding Database (CPDB,http://citrus.hzau.edu.cn)(Liuetal.2022).Raw RNA-Seq data are archived as Sequence Read Archive (SRA) in National Center for Biotechnology Information (NCBI) with accession numbers PRJNA913776 and PRJNA913992.Differentially expressed genes (DEGs) were subsequently filtered using a fold change (FC)>3 and FC<0.3333 as thresholds.FC,in this case,was defined as the ratio ofXcc-induced gene expression to ddH2O-induced one.The differentially expressed TFs (DETFs) were finally obtained based on annotation from the NR (https://www.ncbi.nlm.nih.gov),Swissprot (https://www.sib.swiss/swiss-prot) (Bairoch and Apweiler 1999) and Pfam (http://pfam-legacy.xfam.org)(Mistryetal.2021) databases.
2.5.Creating a CBC-associated AD-TF library
Full-length CDS sequences of DETFs,retrieved from the CPBD,were amplified using primers designed by Primer 3.In this case,the lengths of the primers were 12-15 bp,and at their end was a 20-bp sequence that was complementary to the AD vector.cDNA,reversetranscribed from the mixed RNA of Wanjincheng and kumquat sampled at 0,6,12 and 24 hours postXccinfection by using a reverse transcription kit (TaKaRa,Japan),were selected as the template to clone the DETFs before mixing the resulting PCR products of 20 DETFs with AD vectors that had been linearized usingEcoRI andBamHI.EscherichiacoliDH5α competent cells were eventually transformed using the final ligated products.The mixture ofE.colicontaining all AD-TF plasmids formed the CBC-inducing TF library.
2.6.Evaluation of library capacity and quality
After coating plates with 10 mL of a 1,000-fold diluted library and incubating them overnight at 37°C,the clones were counted to calculate the capacity of the library according to the following formula:
Library capacity=(Number of clones on the plates/10 μL×1,000×103μL)×Total volume of library solution (mL)
Thirty clones grown were randomly selected and amplified by PCR using the primer pair FpGADT7(GTACCCATACGACGTACCAGA)/RpGADT7(TGCGGGGTTTTTCAGTATCTACGATTCAT) to determine the positivity rate of the library.In addition,30 pairs of primers for the DETFs were randomly selected for PCR amplification of the library plasmids in order to verify the coverage of the library.
2.7.Y1H screening
The bait was constructed by cloning the 1-kb promoter ofCsPrx25and inserting it into the pAbAi vector.Y1HGold yeast,with the bait,was subsequently added to SD/-Ura plates as well as SD/-Ura plates containing different concentrations of aureobasidin A (AbA).This was followed by a 3-day incubation at 30°C to determine the lowest AbA concentration that inhibits the growth of the bait strain.After transforming the Y1HGold yeast containing the bait with the library plasmid,the resulting Y1HGold yeast (with the library plasmid and the bait) were cultured on SD/-Leu+100 ng mL-1AbA plates prior to a 5-day incubation at 30°C.Positive clones were then identified and sequenced by PCR using universal primers of pGADT7.The CDS ofCsDOF5.8was also cloned into the pGADT7 vector to construct the prey.Finally,Y1HGold yeast competent cells were transformed with the prey and the bait vectors before being cultured on SD/-Leu+100 ng mL-1AbA plates for 5 days at 30°C in view of re-confirming the hybridization between the prey and the bait.
2.8.Dual-luciferase (dual-LUC) reporter assays
CsPrx25promoter regions (1 kb) were individually cloned into the pGreenII0800 vector for generating promoter-LUC reporter construct.Similarly,in a modified pBI121-M vector,the full-length CDS ofCsDOF5.8was individually inserted downstream of the CaMV35S promoter to generate an effector construct.These constructs,along with empty vectors that acted as controls,were used to transformAgrobacteriumtumefaciensGV3101 prior to agroinfiltration into tobacco leaves.A Dual-Glo®Luciferase Assay System(Promega,USA),was then used based on provided instructions to assess Firefly and Renilla luminescent signals.
2.9.Electrophoretic mobility shift assay (EMSA)
Cloning of the full-length CDS ofCsDOF5.8was achieved by ligating individual sequences into the pGEX-4T-1 prokaryotic expression vector containing a GST tag,and this construct was then introduced intoE.coliBL21.CsDOF5.8-GST protein expression and purification were then performed as reported before (Duanetal.2018).A single-stranded oligonucleotide,corresponding to either the wild-type probe WPCsDOF5.8(5´-AATTTAGTATTATT CTTTTTTTTTTACTTTACACTTGATGAAACTGATA-3´) or mutated probe MPCsDOF5.8(5´-AATTTAGTATTCTGCATCTAT CTGTCGTATTCTCATACAGAAACTGATA-3´),was synthesized before being labeled with biotin (Sangon Biotechnology,China).EMSA analyses were then conducted with the LightShift Chemiluminescent EMSA kit based on the provided instructions (Thermo Scientific,USA).
2.10.Citrus transient transformation
The overexpression plasmid pGLNe-CsDOF5.8was generated after amplifying the full-length CDS ofCsDOF5.8using the primer pair FOEC-CsDOF5.8(ggtaccATGTTTCAGATTCTCTC,KpnI site included)/ROEC-CsDOF5.8(gtcgacTCACTTGAGACCATTTCC,SalI site included).It was then added into the pLGNe vector with the CaMV35S promoter,with the resulting expression vectors transformed intoA.tumefaciensEHA105.The latter,containing overexpressing plasmids,was inoculated into Wanjincheng leaves before being incubated at 28°C for 5 days.Eventually,expression ofCsDOF5.8andCsPrx25was assessed by qRT-PCR.
2.11.Xcc growth rate assay
To measure the CBC resistance of citrus leaves transiently overexpressedCsDOF5.8,during inoculation,Xccgrowth assays were performed.Xccsuspension (OD600=0.5) was infiltrated in the position,that had been inoculated withA.tumefaciens(containing plasmid pGLNe-CsDOF5.8)24 hours before.A total of three 5-mm-diameter leaf discs were sampled from the inoculated leaves at 2,4 and 6 days,which were then ground and diluted in 1,000 μL ddH2O.The mixture was plated on LB media and incubated at 28°C for 2,4 and 6 days.At each time point,the bacterial colonies were counted,and the number of bacterial cells per square centimeter of leaf tissue was calculated (Pengetal.2015).The test was repeated three times.
2.12.qRT-PCR
qRT-PCR was performed on a Quantagene Real-Time System q225 (Novogene,China) using the SYBR premix kit (Bio-Rad,USA).In this case,each 12 μL reaction mixture contained 6 μL of SYBR Green PCR mix,0.5 μmol L-1of each primer and 100 ng of cDNA.In addition,the amplification conditions were as follows: 95°C for 5 min,followed by 40 cycles each at 95°C for 10 s and 56°C for 30 s.For the PCR,the following primer pairs were selected: FRTPCR-CsDOF5.8(CTCTCAAGTCCAATC)/RRTPCR-CsDOF5.8(GTCGGCATCTTTTGC),FRTPCR-CsPrx25(GCTTCAG CTTCTTCT)/RRTPCR-CsPrx25(CTTTTTTCAGGGCAT),withGAPDHbeing used as the reference gene (Mafraetal.2012)(GenBank ID: CX289161.1,CPBD ID: Cs_ont_1g004160)FRTPCR-CsGAPDH(GCTTTCCGTGTACCCACTGT)/RRTPCR-GAPDH(CTCTGACTCCGCCTTGATGG).These primers were constructedusing NCBI Primer-BLAST using theC.sinensismRNA database as a reference.Relative expression was then assessedviathe 2-ΔΔCTmethod (Livak and Schmittgen 2001).
2.13.Biochemical analyses
PRX activity and H2O2content were measured using SinoBestBio Kit (Shanghai,China).
2.14.Statistical analysis
The data were already analyzed using GraphPad Prism 8.0 (GraphPad,USA),with results reported as means±standard deviations (SDs).Results were subsequently compared by ANOVA along with Duncan’s multiple range test or two-tailedt-tests.
3.Results
3.1.DETFs of Wangjincheng and kumquat were induced by Xcc
To obtain CBC-associated TFs,DETFs between the CBC-susceptible Wangjincheng and the CBC-resistant kumquat were studied afterXccinfection.As a result,253Xcc-induced DETFs were identified as candidate genes for the construction of a CBC-associated TFs library (Fig.1-A;Appendix A).Of these,154 were found in Wanjincheng,204 were identified in kumquat,and 105 were common to both varieties (Appendix A).Hence,followingXccinfection,there were more DETFs in kumquat compared with Wanjincheng (Fig.1-B).More specifically,in Wanjincheng,88 DETFs were detected at 6 hpi,55 were detected at 12 hpi,and 90 were detected at 24 hpi,while 22 were detected at all three time points(Fig.1-C).Conversely,in kumquat,54 DETFs were detected at 6 hpi,109 were detected at 12 hpi and 137 were detected at 24 hpi (Fig.1-D).Unlike Wanjincheng,more DETFs were detected at 12 and 24 hpi in kumquat(Fig.1-C and D).The DETFs of Wangjincheng and kumquat were classified into 45 TF families according to the gene function annotations (Fig.1-E).
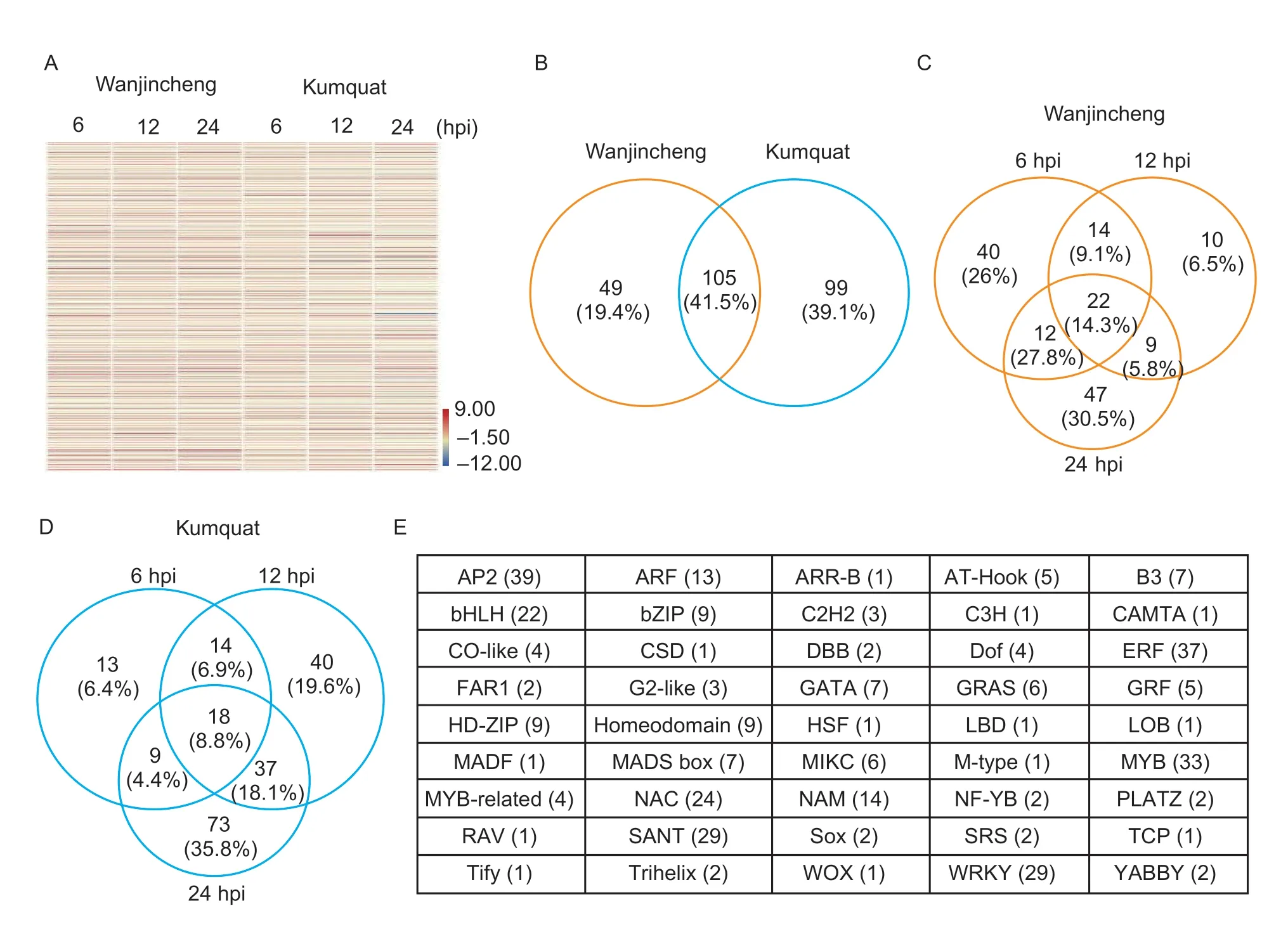
Fig.1 Differentially expressed transcription factors (DETFs) of Wangjincheng and kumquat induced by Xcc infection.A,heatmap showing expression of 253 Xcc-induced DETFs as determined by RNA-seq.Blue and red correspond to low and high expression as determined by log2FC value.B,number of DETFs in Wangjincheng and kumquat.C and D,number of DETFs in Wangjincheng(C) and kumquat (D) at different time points.In B-D,the percentages in brackets represent the ratio of the gene number of the group to the total.E,the TF families to which the 253 DETFs belong.The numbers in brackets represent the number of differential genes contained in the family.
3.2.Construction and evaluation of the CBCassociated AD-TF library
To clone the DETFs and build a CBC-associated AD-TF library,their full-length CDS sequences were obtained from the CPDB database before designing primers for the cloning process (Appendix B).A CBC-associated AD-TF library was then set up through three steps namely gene amplification,ligation to the vector and plasmid transformation (Fig.2-A).Finally,203 DETFs were successfully cloned and used in the AD-TF library with a capacity of 1×107CFU (Appendix C).In addition,30 clones were randomly selected,and clear insertion sequences could be detected,indicating that the positive rate of library cloning was 100% (Fig.2-B).The library plasmids were further detected using primer pairs for the 30 randomly selected DETFs.These primers were able to detect the correct size of the target fragments,indicating that the library of 203 TFs had a coverage of 100% (Fig.2-C;Appendix D).Thus,it could be said that the library was of high quality.
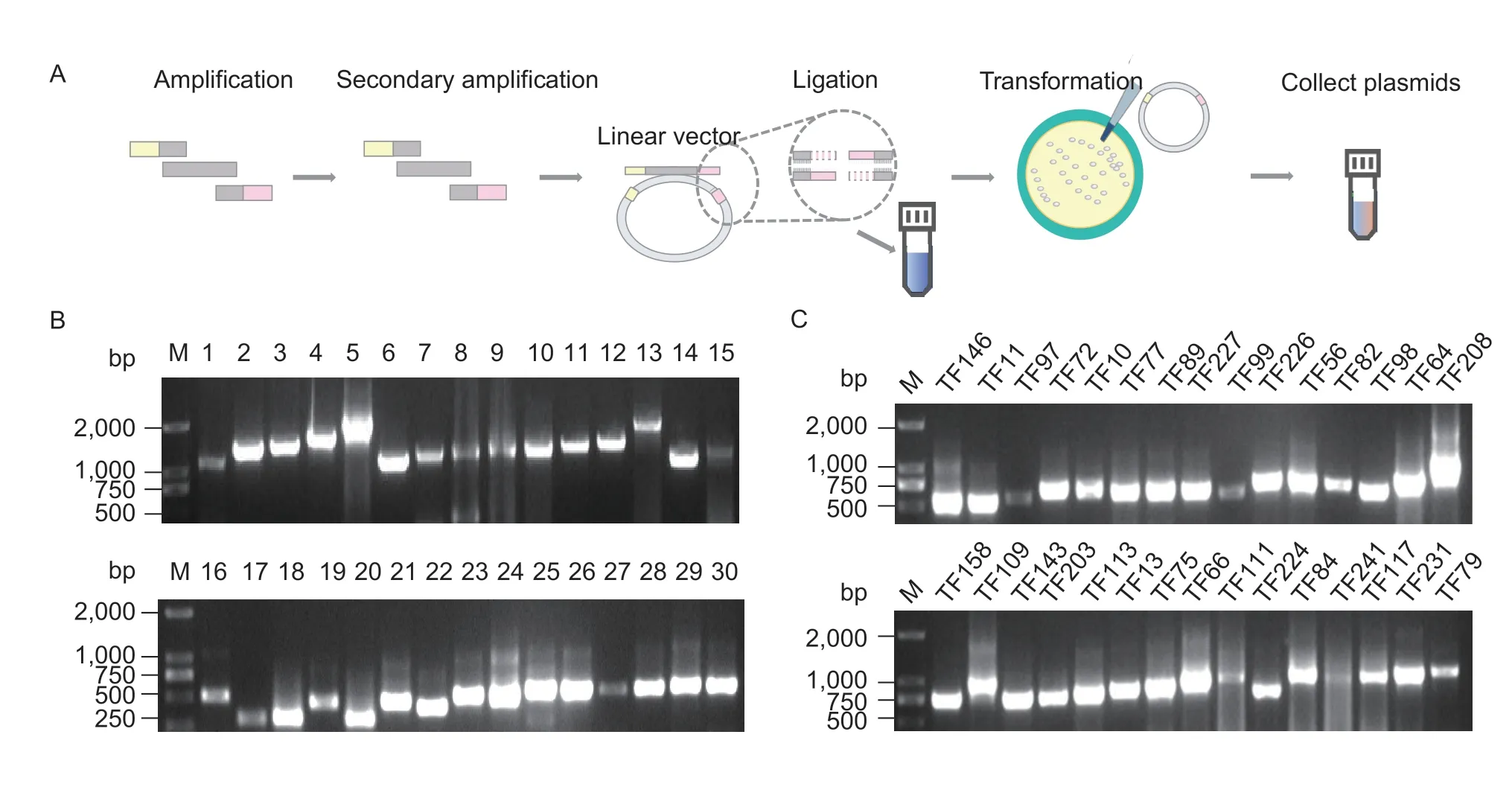
Fig.2 Construction and evaluation of a CBC-associated AD-TF library.A,the scheme of construction of a CBC-associated AD-TF library.B,positivity rate of the library.1-30,products of PCR amplification using 30 randomly selected library clones with vector primers.C,coverage determination of the library.TF#,products of PCR amplification using cloning primers for 30 randomly selected DETF genes.M,marker.
3.3.ldentification of TF that binds to the promoter of CsPrx25
To verify the usability of the CBC-associated ADTF library and explore the upstream regulators and regulatory mechanisms of the CBC resistance geneCsPrx25,the promoter ofCsPrx25was used to screen the library (Appendix E).This promoter was cloned and inserted into vector pAbAi to construct the bait (Fig.3-A).Determined by self-activation tests,the promoter ofCsPrx25(Pro25) has self-activating activity and needs to be inhibited with 100 ng mL-1of AbA (Fig.3-B).After library screening,positive library clones (Fig.3-C),eight of which had inserted genes,were obtained (Fig.3-D).These eight clones were subsequently sequenced,aligned,and finally a common DOF (DNA binding with one finger) family TF was obtained.The DOF family TF located on chromosome 1 ofC.sinensis,contains a 990-bp exon,no introns and possesses a typical DOF domain (Fig.3-E;Appendix E).After BLAST analysis,this DOF TF had the highest homology with AtDOF5.8 ofArabidopsis,and as such,it was termed as CsDOF5.8.CsDOF5.8expression in the Wanjincheng(CBC sensitive) and kumquat (CBC resistant) varieties was evaluated followingXccinfection to assess the relationships betweenCsDOF5.8and CBC.In both citrus varieties,CsDOF5.8displayed unique expression patterns in response to the pathogen.CsDOF5.8was significantly upregulated in kumquat during 0-24 hours,while overall was not induced in Wanjincheng (Fig.3-F).Furthermore,the prey vector ofCsDOF5.8was constructed and used to perform a Y1H verification of the interaction betweenCsDOF5.8andPro25(Fig.3-G).The results showed that only positive control and yeast containing the pGADT7-CsDOF5.8and pAbAi-Pro25plasmids could grow (Fig.3-H),indicating thatCsDOF5.8binds directly to theCsPrx25promoter.The successful screening of regulatory factors onCsPrx25indicates that the CBC-associated AD-TF library has good usability and practicability.
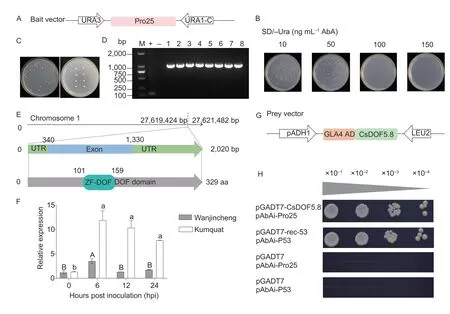
Fig.3 Identification of TF that binds to the promoter of CsPrx25.A,schematic overview of the bait vector utilized to conduct a library screening.URA3,URA3 gene;URA1-C,AbA resistance gene.B,self-activation of Pro25 and its inhibition in the presence of AbA.C,positive library clones on SD/-Ura with AbA (100 ng mL-1).D,PCR validation of positive library clones with primer pair FpGADT7/RpGADT7.M,DNA ladder;+,positive control with pGADT7 vector as the template;-,negative control with ddH2O2 as the template.E,bioinformatics characterization of CsDOF5.8.The chromosomal position of the gene was obtained from the CPDB.The gene structure was visualized with GSDS.The protein functional domains were analyzed with Simple Modular Architecture Research Tool (SMART).UTR,untranslated region;DOF,DNA binding with one finger;aa,amino acid.F,relative expression of CsDOF5.8 after Xcc infection.Relative expression in Wanjincheng and kumquat was determined by qRT-PCR and normalized to CsGAPDH.Duncan’s multiple range test for ANOVA was used to examine differences (P=0.05).Different letters on the bars indicate significant differences.To determine repeatability,all experiments were performed in triplicate (n=3).Data are mean±SD.G,schematic overview of the prey vector used to conduct a Y1H assay.pADH1,ADH1 promoter;GLA4 AD,GAL4 transcriptional activator;LEU2,LEU2 gene.H,Y1H assays conducted for the CsDOF5.8 and CsPrx25 promoter.In A and H,Pro25,1-kb promoter of CsPrx25.
3.4.CsDOF5.8 directly binds the CsPrx25 promoter to induce transcriptional activity
To verify the accuracy of the screening results of the CBC-associated AD-TF library,EMSA assay was used to confirm theinvitrointeractions betweenCsDOF5.8andCsPrx25promoter.JASPAR analysis revolved 1,740 binding sites in theCsPrx25promoter (1 kb),including a DOF5.8 binding site that exhibited relative scores of 88.6% (Fig.4-A;Appendix F).At the same time,probes were constructed using normal or mutant versions of thisCsPrx25promoter binding site (Fig.4-A).After purifying CsDOF5.8-GST and incubating it with the normal probe,a slow but consistent band migration was detected for the binding between the protein and the probe DNA sequence.This slowed band migration was reduced in a dose-dependent manner upon unlabeled competitor added.In contrast,CsDOF5.8-GST was unable to bind the mutated version of this probe (Fig.4-B).To examine whetherCsDOF5.8had a transcriptional effect onCsPrx25,a dual-LUC assay was then performed by inserting this promoter region into the pGreenII0800 vector upstream of the luciferase gene.This process generated a reporter construct (Pro25),with CaMV35S-drivenCsDOF5.8acting as an effector construct (Fig.4-C).When transiently expressed in tobacco leaves,a significantly stronger firefly luciferase signal was observed after being induced in both constructs as compared to individual leaves that carry thePro25orCsDOF5.8constructs,while no significant difference was observed between the promoter mutant group and control group (Fig.4-D).These results supported the ability ofCsDOF5.8to activate theCsPrx25promoter.

Fig.4 CsDOF5.8 directly binds the CsPrx25 promoter to induce transcriptional activity.A,wild type (WT) and mutant probes used for the EMSA assay.Mutated binding sites are shown in red.B,EMSA results corresponding to the specific binding of CsDOF5.8 to CsPrx25 promoter binding sites.The CsDOF5.8-GST protein was incubated with biotinylated probe constructs.+,presence;-,absence.C,the reporter and effector vectors used to conduct dual-LUC assays.REN,Renilla luciferase;LUC,firefly luciferase;P35S,CaMV35S promoter;T35S,CaMV35S terminator.D,the results of an assay in which promoter activity was analyzed upon the transient expression of the prepared constructs in tobacco leaves.Data represent ratios of LUC to REN signal.Results were compared via two-tailed t-tests.***,P<0.0001.In A,C and D,Pro25,1-kb promoter of CsPrx25;Pro25M,1-kb promoter of CsPrx25 with the mutated predicted binding site shown in (A).
3.5.Transient overexpression of CsDOF5.8 restores CsPrx25 expression and H2O2 homeostasis
To explore the functions ofCsDOF5.8in the regulation ofCsPrx25-mediated H2O2homeostasis,vectors were constructed for the transient overexpression ofCsDOF5.8in the Wanjincheng variety (Fig.5-A).Successful overexpression ofCsDOF5.8at 5 dpi was verified by qRT-PCR (Fig.5-B),which confirmed thatCsPrx25was significantly upregulated by over 28 folds in materials overexpressingCsDOF5.8(Fig.5-C).Both PRX activities and the H2O2level of materials overexpressingCsDOF5.8increased (Fig.5-D and E),and this reflected the effects of overexpressedCsPrx25as previously reported (Lietal.2020c).These findings further indicate thatCsDOF5.8positively regulates H2O2homeostasis by directly targeting the promoter ofCsPrx25and activating its expression (Fig.5-F).The subsequently enhanced H2O2content could improve the CBC resistance by activating hypersensitive responses and cell lignification(Lietal.2020c).To explore the functions ofCsDOF5.8in CBC resistance,Xccgrowth rate in Wanjincheng leaves with transiently overexpressedCsDOF5.8was assayed.Transient overexpression ofCsDOF5.8exhibited fewer colony-forming units (CFUs) relative to plants with empty vector at 2,4 and 6 dpi.After 4 dpi,CsDOF5.8overexpression reduced the significant proliferation ofXcc(Fig.5-F),suggesting thatCsDOF5.8does enhance CBC resistance.In summary,we proposed a mechanism by whichCsDOF5-CsPrx25positively regulates CBC resistance (Fig.5-G).
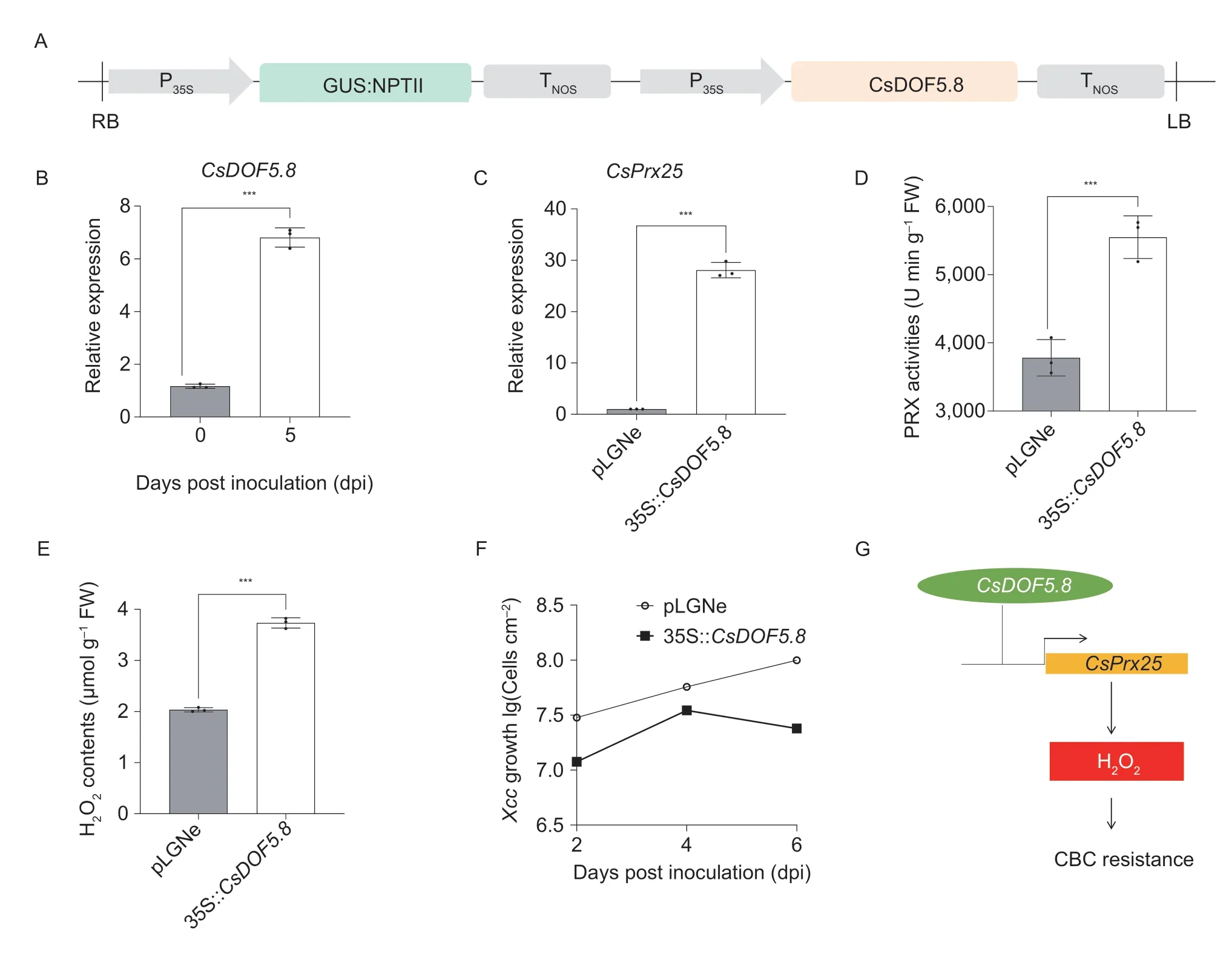
Fig.5 Transient overexpression of CsDOF5.8 restores CsPrx25 expression and H2O2 homeostasis.A,plasmid used for CsDOF5.8 overexpression.NPTII,NPTII gene;NOS,NOS terminator;P35S,cauliflower mosaic virus 35S promoter;GUS,β-glucuronidase coding gene;RB,right border;LB,left border.B and C,the relative expressions of CsDOF5.8 (B) and CsPrx25 (C) as determined by qRT-PCR prior to normalization against CsGAPDH.D and E,peroxidase (PRX) activities (D) and H2O2 concentrations (E) after transient overexpression of CsDOF5.8 in Wanjincheng.In B-E,differences were assessed with two-tailed t-tests (***,P<0.0001).In C-F,pLGNe,transient transformation of empty vector;35::CsDOF5.8,transient transformation of vector for CsDOF5.8 overexpression.F,growth of Xcc in leaves of materials overexpressing CsDOF5.8.G,hypothetical molecular mechanism of CsDOF5.8-CsPrx25-CBC resistance.
4.Discussion
To efficiently and accurately screen TFs which are closely associated with CBC,a library of 203 CBC-associated TFs was constructed in this study,and interestingly,all of them were induced byXccinfection (Appendix C).Consequently,they can be used to systematically study the regulatory network of CBC-associated genes and provide an effective method for the breeding of CBC resistant varieties.During the process of library construction,multiple measures were taken to ensure that the library was more representative and of better quality.Firstly,the transcriptome data of susceptible and disease-resistant varieties,induced byXcc,were analyzed at multiple time points to explore CBC-related TFs more comprehensively (Fig.1).Secondly,during the process of TF cloning,debugging of PCR programs,re-amplification of low amounts of PCR products as well as homogenization of PCR products according to their DNA concentration,were undertaken to ensure that the abundance of each TF in the library was almost consistent.Compared with the process of using restriction enzyme digestion and ligation of one gene at a time,this approach also increased efficiency and reduced costs by ligating the PCR products of 20 TFs to the AD vectors in one ligation system with homologous recombination.
As part of a plant-specific multigene family,CIII Prxs promotes suberization,the flexibility of cell walls,lignification (Fernández-Pérezetal.2015;Pandey and Dwivedi 2015) as well as disease resistance (Liszkayetal.2003;Lietal.2015).Previous research from the current authors found changes in the expression pattern ofCsPrx25in CBC-susceptible and CBC-resistant varieties,hence suggesting its role in CBC development(Lietal.2020b).In addition,overexpression strategies could be used to exploreCsPrx25’s functional role and it was noted that,in transgenic plants,CBC resistance was largely conferred by the gene (Lietal.2020c).By applying this CBC-associated AD-TF library,one upstream regulator ofCsPrx25was identified (Fig.3),with the interactions further validated by EMSA and dual-LUC assays (Fig.4).CsDOF5.8was investigated during the early stages ofXccinfection,finding thatCsDOF5.8was upregulated afterXccinfection in the CBC-resistant variety (Fig.3).This expression pattern is highly consistent with theXcc-inducible expression pattern ofCsPrx25reported previously (Lietal.2020c).CsDOF5.8may be associated with resistance toXccinfection.Overall,we speculated thatCsDOF5.8andCsPrx25are located in the same pathway to positively regulate CBC resistance.Thus,it was proposed thatCsDOF5.8positively regulates the H2O2homeostasis by directly targeting the promoter ofCsPrx25and activating its expression,thereby enhancing H2O2content and improving the CBC resistance (Fig.5).These data offer new insight into the mechanisms and regulatory pathways through whichCsPrx25regulates CBC resistance,thereby highlighting its potential as a target to breed CBC-resistant citrus varieties.
5.Conclusion
In this study,the library prepared was a specific one for CBC-associate TFs.It was of high quality,efficient and involved accurate screening,high construction efficiency and low construction costs.This powerful system can assist in understanding the regulatory mechanism of CBC resistance,generate comprehensive interaction networks of TFs as well as accelerate the breeding of CBCresistant citrus varieties.This library can further provide a reference for the construction of specific AD-TF libraries for a wide range of crop varieties or gene types.
Acknowledgements
This study was funded by the National Key Research and Development Program of China (2022YFD1201600),the earmarked fund for the China Agriculture Research System (CARS-26),the Fundamental Research Funds for the Central Universities,China (SWU-XDJH202308),and the Science and Technology Research Program of Chongqing Municipal Education Commission,China(KJQN202001418).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.11.011
杂志排行
Journal of Integrative Agriculture的其它文章
- Advances in DNA methylation and its role in cytoplasmic male sterility in higher plants
- Assessment of molecular markers and marker-assisted selection for drought tolerance in barley (Hordeum vulgare L.)
- 3D genome organization and its study in livestock breeding
- SUPER WOMAN 2 (SPW2) maintains organ identity in spikelets by inhibiting the expression of floral homeotic genes OsMADS3,OsMADS58,OsMADS13,and DROOPING LEAF
- Genetic dissection and validation of a major QTL for grain weight on chromosome 3B in bread wheat (Triticum aestivum L.)
- Coupling of reduced inorganic fertilizer with plant-based organic fertilizer as a promising fertilizer management strategy for colored rice in tropical regions
