A Pair of New Spirocyclic Alkaloid Enantiomers with TrxR Inhibitory Activities Were Isolated from Marine-Derived Aspergillus ruber TX-M4-1
2023-12-21WANGCongWANGYufeiSUNJianWANGShiyiDUWeishengZHOULimanandKONGFandong
WANGCong, WANG Yufei, SUN Jian, WANG Shiyi, DU Weisheng,ZHOU Liman, andKONG Fandong,
A Pair of New Spirocyclic Alkaloid Enantiomers with TrxR Inhibitory Activities Were Isolated from Marine-DerivedTX-M4-1
WANGCong1), 2), WANG Yufei1), 2), SUN Jian1), 2), WANG Shiyi1), 2), DU Weisheng1), 2),ZHOU Liman1), 2), andKONG Fandong1), 2),*
1),,,,530006,2),,,530006,
One new spirocyclic alkaloid, 5-isopentenyl-cryptoechinuline D (1), along with 11 known compounds (2–12), were iso- lated from a marine fungusTX-M4-1. The structures of compounds 1–12 were elucidated by spectroscopic evi- dences. Compound 1 was initially isolated as an enantiomer, and further separation of 1 by chiral HPLC afforded a pair of enantio- mers, including (-)-5-isopentenyl-cryptoechinuline D (1a) and (+)-5-isopentenyl-cryptoechinuline D (1b). Their absolute configura- tions were elucidated by ECD spectroscopic data. Compounds 1a, 5 and 10 could inhibit thioredoxin reductase (TrxR) activity with IC50values of 6.2, 36.3 and 18.6μmolL−1, respectively. Surface plasmon resonance (SPR) study also demonsrated the interactions between compounds 6, 8 and Niemann-Pick C1 Like 1 (NPC1L1) respectively, which indicate that compounds 6 and 8 are potential NPC1L1 inhibitors.
TX-M4-1; spirocyclic alkaloid; TrxR inhibitory activity
1 Introduction
Thioredoxin reductase (TrxR) plays important roles inmany biologicalprocesses, including cell growth regulation,DNA synthesis, and angiogenesis (Mahmood, 2013). TrxR inhibitors could lead to the inhibition of proliferation, and induction of apoptosis of cells. In recent years, more and more evidences suggested that TrxR is a potential anti-cancer target (Lei, 2018; Jovanović, 2020).
Marine fungi have adapted to the oceanic environment by special defense systems and physiological metabolic pathways. These genetic adaptations resulted in producingbioactive substances with novel chemical structures and uni- que biological functions (Fan., 2018). Marinesp. is an important microbial source of novel active metabolites (Carrol., 2022), showing rich chemical di-versity, including diketopiperazine alkaloids, sterols, terpe-noids, peptides, anthraquinones and so on. These chemicalconstituents show a variety of pharmacological activities,such as neuroprotective, cytotoxic, antiparasitic and anti-in-flammatory activities, and also show great biological po-tential. The natural products ofsp. have the cha- racteristics of rich halogen and nitrogen active compoundsand high proportion of new active compounds (Zhang., 2022), showing a good development prospect in drug de- sign and screening. Thus, it is important to find compoundswith novel structural scaffolds and excellent biological ac- tivity from chemometrics ofsp
In our previous study, one new benzaldehyde compound and 10 known compounds were isolated from petroleum ether layer ofTX-M4-1 (Wang, 2022).The methanol extracts of the culture of this fungus were further investigated in order to find new bioactive com-pounds. As a result, twelve compounds, including a pair of enantiomer 5-isopentenyl-cryptoechinuline D (1a and 1b) and eleven known ones (2–12), neoechinulin F (2) (Lin., 2021), tardioxopiperazine A [-L-alany-5-iso- pentenyl-2-(1’,1’-dimethylallyl)-L-tryptophan] (3) (Fujimo- to., 1999), alkaloid E-7 (4) (Liu., 2022), echinu- lin (5) (Liang., 2018), isoechinulin B (6) (Luo., 2015), isoechinulin A (7) (Yan., 2022), 2--Methyl-9- dehydroxyeurotinone (8) (Li., 2009), eurotinone (9) (Saad., 2021), epihevadride (10) (Williams., 2022), asperflavin (11) (Zhao., 2018) and variecolorquinone B (12) (Wang., 2007) were isolated (Fig.1). Herein, the isolation process, chemical structures, and bioactivities of these compounds were reported.
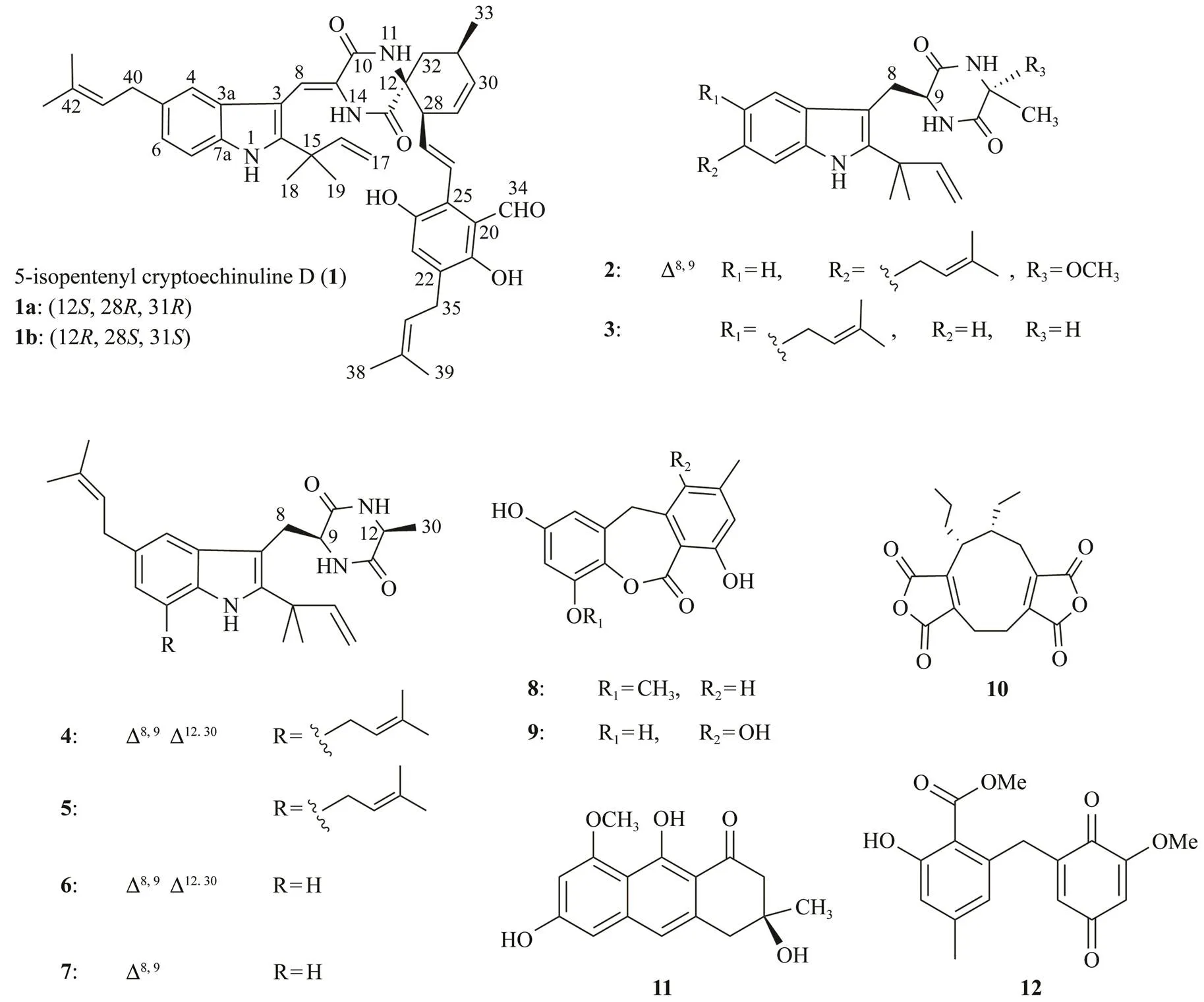
Fig.1 Structures of compounds 1−12. ‘△8,9’ means double bond between C-8 and C-9.
2 Materials and Methods
2.1 General Experimental Procedures
UV spectra were recorded on an Agilent Cary 60. Opti- cal rotations were measured on a JASCO P-1020 digital po- larimeter. IR spectra were obtained on a MAGNA-1R550spectrophotometer in KBr discs. NMR spectra were record-ed on a Bruker Avance 400 spectrometer (Bruker, Fallan- den, Switzerland), HRESIMS spectra were obtained using a Mariner API-TOF. Semi-preparative HPLC was perform- ed on a Waters 1525 system using a semi-preparative C18 (Cosmosil, 5μm, 10 ID×250mm) column coupled with a Varian 330 detector. Chiral HPLC of 1 was run on a Chi- ral CD-Ph (Shiseido, 5μm, 4.6 ID×250mm). Column chro-matography was performed over silica gel. Thin layer chro- matography was performed on plates precoated with sili- ca gel GF254. Experimental electronic circular dichroism (ECD)spectra in MeOH were recorded on a Chirascan cir- cular dichroism spectrometer.
2.2 Fungal Material
TX-M4-1was isolated from a mosssample collected from the WeizhouIsland (21˚1΄42.97΄΄N,109˚7΄46.29΄΄E) and identified by the 18S rDNA gene se-quence (GenBank accession no. OL989330) (Wang, 2018).
2.3 Fermentation and Isolation
TX-M4-1 was cultured on potato dex-trose agar medium at 28℃ for 3 days. Spores were cul- tured in 100mL×1000mL Erlenmeyer flasks (80g rice in120mL seawater). The flasks were cultued at room tem-perature under static conditions for 32 days. The solid cul- ture was extracted three times with EtOAc to yield 83.47gproduct.The extracts were extracted by 90%MeOH-H2O and petroleum ether to produce 25.1g of petroleum ether extract and 56.18g of MeOH-H2O extract. The MeOH- H2O extract was separated into 33 fractions (Fr. 1−33) on a silica gel VLC column,eluting with a stepwise gradient with EtOAc-petroleum ether (0−100%). Fr.11 (80mg) waspurified by semipreparative HPLC on an ODS column, elut- ed with 60% MeCN-H2O to yield 10 (18.9mg,R28.0min). Fr. 20 (302.3mg) was purified over Sephadex LH-20 with MeOH-CH2Cl2(v/v 1:1) into eleven subfractions. Fr. 20-3 (40mg) was further purified by HPLC over a ODS column (80% MeCN-H2O) to yield compound 1 (2.0mg,R17.5min), and 1was resolved to 1a(1.0mg) and 1b(1.0mg) by HPLC on a Chiral CD-Ph (75% MeOH-H2O). Fr. 20.4 (51mg) was subjected to HPLC over ODS (70% MeCN-H2O, v/v) to produce compound 4 (3mg,R24.2min).Fr. 24 (1.5g) was further separated into nine sub- fractions (Fr. 24-1−Fr. 24-9) eluted with MeOH-CH2Cl2(v/v 1:1) through Sephadex LH-20. Fr. 24-8 (180mg) was fur- ther purified by semipreparative HPLC on an ODS column,eluted with 50% MeCN-H2O to yield compounds 8 (5.0mg,R6.7min) and 12(5mg,R11.62min). Fraction 26 (2.0g)was applied to octadecylsilane silica gel with gradient elu- tion of MeOH-H2O to yield eight subfractions (Fr. 26-1− Fr. 26-8). Fraction 26.2 (59mg) was subjected to HPLC over ODS (75% MeOH-H2O, v/v) to yield compound 2(8.2mg,R11.1min).Fraction 31(3.19g) was applied to ODS silica gel with gradient elution of MeOH-H2O to yield fif- teen subfractions (Fr. 31-1−Fr. 31-15). Fr. 31-12 (54mg) was purified by semipreparative HPLC (75% MeOH-H2O, v/v) over a C18 column to afford compound 3 (5.7mg,R9.6min). Fr. 31-15 (380mg) was purified by semiprepara- tive HPLC (65% MeOH-H2O, v/v) over a C18 column to afford compound 5 (7mg,R16.8min). Fr. 31-13 (45mg) was subjected to semipreparative HPLC (YMC-pack ODS- A 45% MeCN-H2O) to produce compound 7 (6.7mg,R9.38min). Fr.31-1 (23mg) was subjected to semiprepara- tive HPLC (YMC-pack ODS-A, 40% MeOH-H2O to give compound 9 (14.6mg,R8.5min). Fr. 31-6 (509mg) was purified by semipreparative HPLC (53% MeOH-H2O, v/v) over a C18 column to afford compound 11 (13.0mg,R7.7min). Fr. 49 (171mg) from petroleum ether extract was further purified by semipreparative HPLC on an ODS co- lumn, eluted with 78% MeCN-H2O to yield compound 6 (3.5mg,R9.0min).
2.4 Physical-Chemical Property
5-isopentenyl-cryptoechinuline D (1): yellow solid; UV (MeOH)max(logε) 228 (4.1), 277 (3.7), 342 (3.3)nm; IR (KBr)max3447, 1647cm−1;1H NMR (400MHz, CDCl3) and13C NMR (100MHz, CDCl3) see Table 1; HRESIMS (positive)/688.3774 [M+H]+(calcd. for C43H49N3O5, 688.3745).
(-)-5-isopentenyl-cryptoechinuline D (1a): [α]25=−55.0 (0.1, CH3CN);R18.9min (Chiral CD-Ph (75% MeOH- H2O); ECD (0.15mmolL−1, MeOH)max215 (+14.49), 269 (−4.15), 341 (+1.80)nm.
(+)-5-isopentenyl-cryptoechinuline D (1b):[α]25=+58.1(0.1, CH3CN);R21.8min (Chiral CD-Ph (75% MeOH- H2O); ECD (0.15mmolL−1, MeOH)max215 (−10.71), 269(+4.94), 348 (−2.59)nm.
2.5 Biological Assays
The TrxR inhibition assay was performed as described by Zhu(2015).The NPC1L1 protein inhibition assay was performed as described by Zhang. (2021, 2022).
3 Results and Discussion
3.1 Structure Determination
5-isopentenyl-cryptoechinuline D(1) was obtained as a yellow solid and its molecular formula was determined as C43H49N3O5by the positive HRESIMS (/688.3774 [M+H]+, calcd. for 688.3745), requiring 20 degrees of unsatu- ration. The1HNMR spectrum (Table 1) of 1 showed the presence of four aromatic protons (4× Ar−H), eight olefi- nic protons (8× CH), two sp3methine protons (2× CH), one sp2methylene protons (1× CH2), three methylene protons (3× CH2), seven methyl groups (7× CH3) and one aldehyde(1× CHO) (Table 1). The13CNMR, DEPT, and HSQC spec-tra of 1 demonstrated the presence of 43 carbons includ- ing seventeen non-protonated carbons (17× −C), one car- bonyl (1× −CHO), two sp3methines (2× CH), twelve sp2methines (12× =CH), one sp2methylene (1× CH2), three methylenes (3× CH2), and seven methyls (7× CH3). The above data were quite similar to those of cryptoechinulin D(Gao, 2013). The main difference between them werethe absence of an aromatic proton in the indole ring and thepresence of additional NMR signals for an isopentenyl group (C/H34.5/3.34, 124.3/5.31, 132.1, 17.9/1.69, 25.9/1.71) in 1. The latter was confirmed by COSY correlations betweenthe olefinic proton H-41 with H2-40 as well as HMBC cor- relations from the two singlet methyl protons H3-43 and H3- 44 to the two olefinic carbons C-41 and C-42. The location of this isopentenyl group at C-5 of the indole moiety was demonstrated by HMBC correlations from H2-40 to C-4, C-5, and C-6, H-7 to C-5 and C-3a, as well as from H-6 to C-4, and C-7a. The remaining substructure of 1 was de-monstrated to be identical to that of cryptoechinulin D by analysis of HMBC and COSY data(Fig.2). The geometry of the ∆8double bond was assigned to beconfiguration by the downfield shift of H-8 (H7.18, s)because of the de- shielding effect of the carbonyl group on the β-vinyl proton (Gao., 2012; Zhong, 2019), which was also con-firmed by the lack of NOE effectbetween H-8 and H-14. The relative configuration of 1 was assigned the same ascryptoechinulin D by NOESY experiments (Fig.3), from which diagnostic cross peaks of Ha-32 (H1.83) with NH-11 and H3-33, NH-11 with H3-33, and H-28 with Hb-32 (H2.50) were observed. Based on the above results, 1was named as 5-isopentenyl-cryptoechinuline D.

Table 1 1H (400MHz) and 13C (100MHz) NMR data for compound 1 in CDCl3

Fig.2 Key 1H-1H COSY (bolds, blue), HMBC (solid arrows, red)key correlations of compound 1.
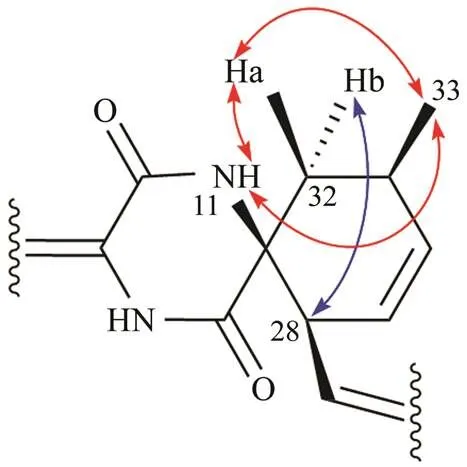
Fig.3 Key NOESY correlations of 1.
Due to no optical rotation and baseline ECD curve, 1 was deduced to be enantiomeric mixture, which was con- firmed by further separation of 1 to 1aand 1b by HPLC with chiral column. The ECD curves (Fig.4) of 1a and 1b were mirror symmetrical, which was similar to those of en- antiomers (-) and (+)-cryptoechinulin D (Gao, 2013). The ECD curve of 1a showed obvious positive CEs (cot- ton effects) around 215 and 341nm as well as negative CEsaround 269nm, and was nearly identical to that of (-)-(12,28,31)-cryptoechinulin D (Gao,2013), thus assign-ing the (12,28,31) configuration for 1a. As the conse- quence, the absolute configuration of 1b was assigned to be (12,28,31).
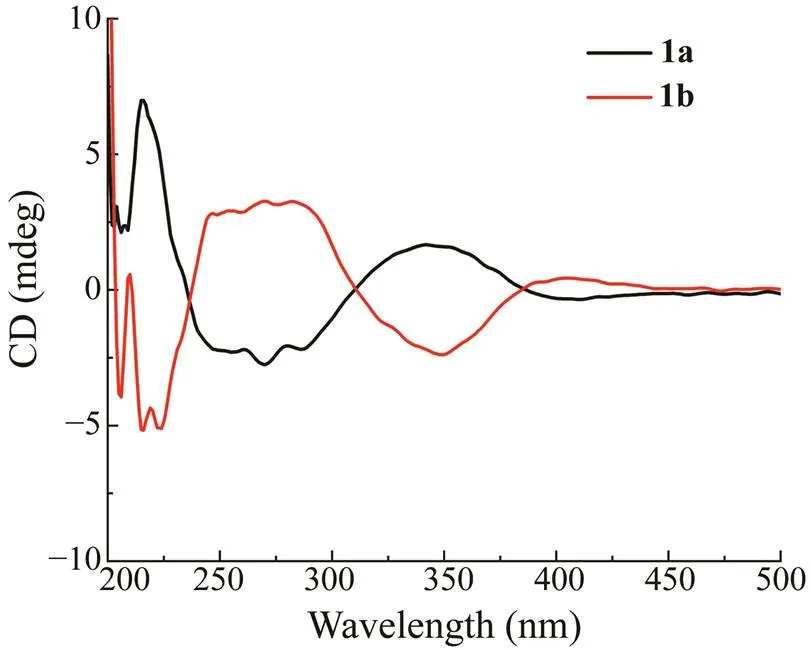
Fig.4 Experimental ECD curves of 1a and 1b.
3.2 Bioactivity
All of the isolated compounds were evaluated for enzyme inhibitory activity against thioredoxin reductase (TrxR) (Ta- ble 2). Compound 1a showed strong TrxR inhibitory activi-ty with IC50value of 6.2μmolL−1, about three times strong- er than the positive drug curcumin (IC50, 20.4μmolL−1). Compounds 5 and 10 could alsoinhibited TrxR with IC50values of 36.3 and 18.6μmolL−1, respectively. However, compound 1b was inactive against TrxR, although it bears the same planar structure as 1a. This once again proves the importance of the configurations of compounds for bioac- tivities. NPC1L1, as the specific transporters for cholesterol, has attracted more and more attentions. SPR study (Fig.5) demonstrated the interaction between compounds 6, 8 and NPC1L1 with comparable effects to that of the positive con-trol ezetimibe, indicating that compounds 6, 8 might be NPC1L1 inhibitors.
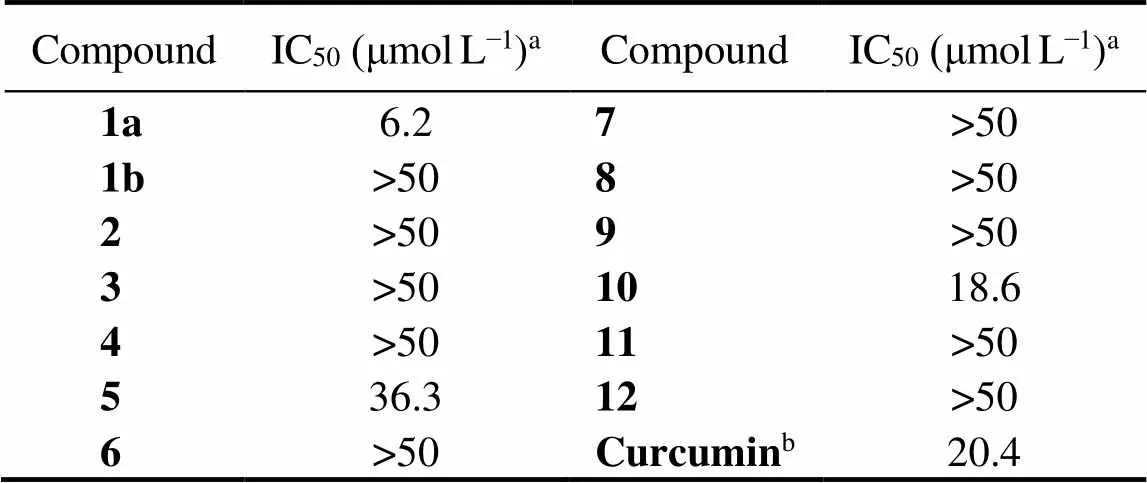
Table2 TrxR inhibitory activity of compounds (IC50)
Notes:aValues are represented as means.bPositive control.
4 Conclusions
One new spirocyclic alkaloid 5-isopentenyl-cryptoechi- nuline D (1) and 11 known compounds (2–12) were obtain- ed from a marine fungusTX-M4-1. Com- pound 1 is an enantiomer. In the bioassay, compounds 1a,5 and 10 could inhibit TrxR, with 1a being the most potent. Compounds 6 and 8 were active on NPC1L1 protein. In con- clusion, we find a new potent cryptoechinuline-type TrxR inhibitor from a marine fungus. These findings further de- monstrated the potential to obtain TrxR inhibitors with me- dicinal potential from marine natural products.
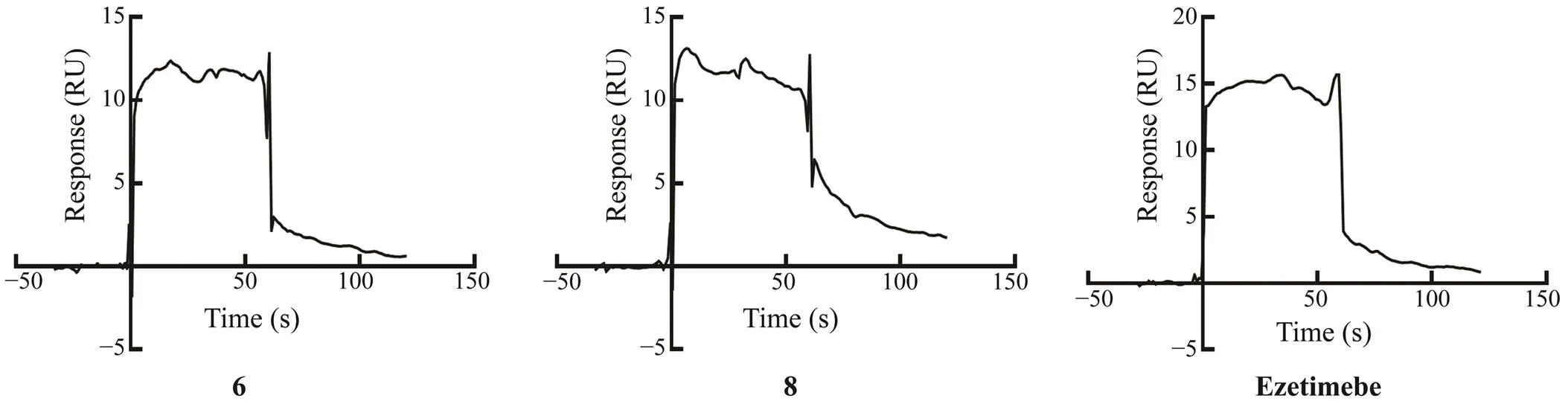
Fig.5 Binding sensorgram for compounds 6, 8, and the positive control ezetimibe interaction with immobilized NPC1L1.
Acknowledgements
Our project was supported by the National Natural Sci- ence Foundation of China (No. 82204276), the Guangxi Na-tural Science Foundation (No. 2021GXNSFBA075036), the Specific Research Project of Guangxi for Research Bases and Talents (Nos. AD22035018, AD202970 36), the 2021 University-Level Scientific Research Projects of Guangxi Minzu University (No. 2021MDKJ003), the Talent Scien- tific Research Initiation Project of Guangxi Minzu Univer- sity (No. 2021KJQD09), the Xiangsi Lake Youth Innova- tion Team Project of Guangxi Minzu University (No. 2021 RSCXSHQN01), and the Guangxi Scholarship Fund of Guangxi Education Department.
Carroll,A. R., Copp, B.R., Davis, R. A., Keyzers, R. A., and Prin- sep, M. R.,2022. Marine natural products., 39: 1122-1171, DOI: 10.1039/D1NP00076D.
Fan, Y. Q., Wang, C., Wang, L.,Chairoungdua, A., Piyachaturawat, P., Fu,P.,.,2018.New ansamycins from the deep-sea-de- rived bacteriumsp. OUCMDZ-2164., 16(8): 282, DOI: 10.3390/md16080282.
Fujimoto, H., Fujimaki,T.,Okuyama, E., and Yamazaki, M., 1999.Immunomodulatory constituents from an ascomycete,, 47(10): 1426-1432, DOI: 10.1248/cpb.47.1426.
Gao, H., Liu, W., and Li, D., 2012. Diketopiperazine alkaloids from a mangrove rhizosphere soil derived fungusH1-1., 10 (47): 9501- 9506, DOI:10.1039/c2ob26757h.
Gao, H.,Zhu, T.,Li,D., Liu, W.,and Liu, W., 2013. Prenylated indole diketopiperazine alkaloids from a mangrove rhizosphere soil derived fungusH1-1., 36 (8): 952-956, DOI: 10.1007/s12272-013-0107-5.
Haque,N.,Parveen, S., and Huang,Z., 2022. Marine natural pro- ducts in clinical use.,20 (8):528, DOI:10.3390/ MD20080528.
Jovanović, M., Zhukovsky, D.,Podolski-Renić, A., Žalubovskis,R., Dar’in, D., Sharoyko, V.,., 2020. Further exploration of DVD-445 as a lead thioredoxin reductase (TrxR) inhibitor for cancer therapy: Optimization of potency and evaluation of anticancer potential., 191: 112119, DOI: 10.1016/j.ejmech.2020.112119.
Lei, H., Wang, G., Zhang, J., and Han,Q., 2018. Inhibiting TrxR suppresses liver cancer by inducing apoptosis and eliciting po-tent antitumor immunity., 40 (6): 3447-3457, DOI: 10.3892/or.2018.6740.
Li, D. L.,Li, X. M., and Wang, B. G., 2009. Natural anthraqui- none derivatives from a marine mangrove plant-derived endo- phytic fungus: Structural elucidation andDPPH radical scavenging activity.,19 (7): 675-680, DOI: 10.4014/jmb.0805.342.
Liang,T. M., Fang,Y. W., Zheng,J. Y.,and Shao, C. L., 2018.Se- condary metabolites isolated from the gorgonian-derived fun- gusand their antiviral activity., 54(3): 559-561, DOI: 10.1007/s10600- 018-2406-z.
Lin,L. B., Gao, Y. Q.,Han, R., Xiao,J., Wang, Y. M.,Zhang, Q.,., 2021. Alkylated salicylaldehydes and prenylated indole alkaloids from the endolichenic fungusand their bioactivities.,69 (23): 6524-6534,DOI: 10.1021/ACS.JAFC.1C01148.
Liu, Z., Chen, Y., Li, S., Hu, C., Liu,H., and Zhang, W.,2022. Indole diketopiperazine alkaloids from the deep-sea-derived fungussp. FS445.,36(20): 5213-5221, DOI: 10.1080/14786419.2021.1925271.
Luo, M. H., Huang, H. B., Lu,L. C.,and Ju,J. H., 2015. Studies on the secondary metabolites of indole alkaloids from the ma- rine fungusSCSIO 151., 27(1):50-54, 147, DOI: 10.16333/j.1001-6880. 2015.01.010.
Mahmood, D. F.D.,Abderrazak, A., El Hadri,K., Simmet, T.,and Rouis, M., 2013. The thioredoxin system as a therapeutic tar- get in human health and disease.,19 (11): 1266-1303, DOI: 10.1089/ars.2012.4757.
Newman,D. J.,andCragg, G. M., 2022.Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019.,83 (3): 770-803, DOI: 10.1021/acs.jnatprod.9b01285.
Saad,M. M., Abdelgaleil, S.A., and Shiono, Y., 2021. Antibac- terial and herbicidal properties of secondary metabolites from fungi., 35 (23): 5446-5451, DOI: 10. 1080/14786419.2020.1779718.
Wang,C.,Liu,C. J.,and Wang, J. Z.,2018.A new diterpene gly- coside from marine fungussp. H1., 49 (24): 5746-5750, DOI: 10.7501/ j.issn.0253-2670.2018.24.003 (in Chinese with English abstract).
Wang, C., Wang, Y. F., Sun, J.,Wang, S. Y.,Lan, K.Y., Du, W. S.,., 2022.Studies on secondary metabolites of marineTX-M4-1.,53 (18): 5600-5606, DOI: 10.7501/j.issn.0253-2670.2022.18. 002 (in Chinese with English abstract).
Wang,W. L.,Zhu, T. J.,Tao, H., Lu, Z., Fang, Y.,Gu, Q.,.,2007. Two new cytotoxicquinone type compounds from the halotolerant fungus., 60 (10): 603-607, DOI: 10.1038/ja.2007.77.
Williams, K.,deMattos-Shipley, K.M., Willis,C. L., and Bai- ley,A. M., 2022. In silico analyses of maleidride biosynthetic gene clusters., 9 (1): 1-21, DOI: 10.1186/S40694-022-00132-Z.
Yan,W., and Liu, D., 2022.Studies on indole diketopiperazine alkaloids in marine fungusCTD-5000-2., 41(1): 31-37, DOI: 10.13400/ j.cnki.cjmd.2022.01.004 (in Chinese with English abstract).
Zhang, R., Liu,W.,Zeng,J.,Meng,J., Jiang, H.,Wang, J.,., 2022.Niemann-Pick C1-Like 1 inhibitorsfor reducing chole- sterol absorption.,230: 114111, DOI:10.1016/J.EJMECH.2022.114111.
Zhang, R.,Song,Z., Wang,X., Xue, J.,and Xing,D.,2021.One- step modification to identify dual-inhibitors targeting both pan- creatic triglyceride lipase and Niemann-Pick C1-like 1., 216: 113358, DOI: 10. 1016/j.ejmech.2021.113358.
Zhang, Y.,Li,Z., and Luo, X., 2022. Anti-osteoclastogenic and antibacterial effects of chlorinated polyketides from the Beibu Gulf coral-derived fungusGXIMD 02505., 20(3): 178, DOI:10.3390/MD20030178.
Zhao, D., Cao, F.,Guo, X. J., Zhang, Y. R., Kang, Z., and Zhu, H. J., 2018. Antibacteral indole alkaloids and anthraquinones from a sewage-derived fungussp.,54 (2): 399-401, DOI: 10.1007/s10600-018- 2361-8.
Zhong, W., Wang, J., and Wang, F., 2019. Three pairs of new spi- rocyclic alkaloid enantiomers from the marine-derived fungussp. SCSIO F452., 7: 350, DOI: 10.3389/fchem.2019.00350.eCollection2019.
Zhu, J.Y.,Lou, L. L., Guo, Y. Q., Li,W.,Guo,Y. H.,Bao,J. M.,.,2015.Natural thioredoxin reductase inhibitors from., 5(58): 47235-47243, DOI:10.1039/C5RA07274C.
(December 12, 2022;
February 1, 2023;
May 9, 2023)
© Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2023
. E-mail: kongfandong501@126.com
(Edited by Qiu Yantao)
杂志排行
Journal of Ocean University of China的其它文章
- Effects of 5-Azacytidine (AZA) on the Growth, Antioxidant Activities and Germination of Pellicle Cystsof Scrippsiella acuminata (Diophyceae)
- Using Rn-222 to Study Human-Regulated River Water-Sediment Input Event in the Estuary
- Parameterization Method of Wind Drift Factor Based on Deep Learning in the Oil Spill Model
- YOLOv5-Based Seabed Sediment Recognition Method for Side-Scan Sonar Imagery
- A Metadata Reconstruction Algorithm Based on Heterogeneous Sensor Data for Marine Observations
- Large Active Faults and the Wharton Basin Intraplate Earthquakes in the Eastern Indian Ocean
