PIWI相互作用RNA017724在肝细胞癌中的表达及其对细胞增殖、凋亡、侵袭和迁移的影响
2023-11-20吴依靖王婕王浩陈琳王慧璇居林玲袁柳霞李民王峰
吴依靖 王婕 王浩 陈琳 王慧璇 居林玲 袁柳霞 李民 王峰


[摘 要] 目的:分析PIWI相互作用RNA017724在肝細胞癌组织、人肝癌细胞系中的表达及其对细胞增殖、凋亡、侵袭及迁移的影响。方法:采用实时定量PCR(real-time quantitative PCR,RT-qPCR)检测肝细胞癌组织和肝癌细胞系中PIWI相互作用RNA017724的表达水平。将PIWI相互作用RNA017724模拟物和抑制剂转染SMMC-7721和PLC/PRF/5细胞,采用CCK-8法和细胞集落形成实验检测各组细胞增殖能力,流式细胞术检测细胞凋亡率,Transwell实验检测各组细胞迁移和侵袭能力。结果:肝癌组织中PIWI相互作用RNA017724表达水平显著低于匹配的相邻正常肝组织,差异有统计学意义(P<0.05);PIWI相互作用RNA017724低表达患者的生存期较短。PIWI相互作用RNA017724抑制剂促进SMMC-7721和PLC/PRF/5细胞的增殖、迁移和侵袭,而模拟物抑制SMMC-7721和PLC/PRF/5细胞的增殖、迁移和侵袭。PIWI相互作用RNA017724对细胞凋亡无明显作用。结论:PIWI相互作用RNA017724抑制HCC细胞的增殖、迁移和侵袭。
[关键词] 肝细胞癌;PIWI相互作用RNA017724;增殖;迁移;侵袭;凋亡
[中图分类号] R735.7 [文献标志码] A [DOI] 10.19767/j.cnki.32-1412.2023.03.003
The expression of piRNA-017724 inhepatocellular carcinoma andits effects
on cell proliferation, apoptosis, invasion and migration
WU Yijing1,2, WANG Jie1,2, WANG Hao1,2, CHEN Lin2, WANG Huixuan2, JU Linling2, YUAN Liuxia2, LI Min2, WANG Feng3
(1Medical School, Nantong University, Jiangsu 226001; 2Affiliated Nantong Hospital 3 of Nantong University/Institute of Liver Diseases, Nantong Third Peoples Hospital; 3Central Lab, Affiliated Hospital of Nantong University)
[Abstract] Objective:To explore the expression of piRNA-017724 in hepatocellular carcinoma tissue and human liver cancer cell line, and its effects on cell proliferation, apoptosis, invasion and migration. Methods:Relative piRNA-017724 expression in hepatocellular carcinoma tissue and liver cancer cell line was measured by real-time quantitative PCR (RT-qPCR). piRNA-017724 mimic and inhibitor were transfected into SMMC-7721 and PLC/PRF/5 cells. CCK8 method and cell colony formation assay were used to detect cell proliferation, flow cytometry was used to detect cell apoptosis, and Transwell assay was performed to detect cell migration and invasion. Results:piRNA-017724 in hepatocellular carcinoma tissue was significantly lower than that in adjacent normal liver tissue, the difference was statistically significant(P<0.05); and the patients with low expression of piRNA-017724 had shorter survival. The piRNA-017724 inhibitor promoted the proliferation, migration and invasion of SMMC-7721 and PLC/PRF/5 cells, while the piR-hsa-017724 mimic inhibited the proliferation, migration and invasion of SMMC-7721 and PLC/PRF/5 cells. However, piRNA-017724 had no affect on cell apoptosis. Conclusions:piRNA-017724 inhibits the proliferation, migration and invasion of HCC cells.
[Key words] hepatocellular carcinoma; piRNA-017724; proliferation; migration; invasion; apoptosis
肝癌是常见的恶性肿瘤之一,其中肝细胞癌(hepatocellular carcinoma,HCC)占绝大多数。HCC的危险因素包括自身免疫性肝炎、酗酒、糖尿病和慢性乙型肝炎病毒(HBV)感染[1-3]。虽然近年来涌现许多HCC免疫疗法和靶向治疗,但患者总体预后仍較差,术后复发和转移率很高[4-6]。HCC中存在多种基因表达变化,识别在发病机制中具有重要作用的基因有助于开发有效治疗靶点。近年来,PIWI与RNA相互作用成为新的研究热点。研究表明,PIWI相互作用RNA可能在癌症的发展中发挥重要作用,并可能作为诊断和预后的生物标志物[7-10]。RIZZO等[11]发现PIWI相互作用RNA 017724(piRNA-017724)在肝细胞肝癌中异常表达。本研究选取南通大学附属南通第三医院/南通市第三人民医院行肝癌根治术45例肝癌组织及其匹配的相邻正常肝组织,分析PIWI相互作用RNA 017724的表达水平及其与预后的相关性,探索其在HCC发病机制中的潜在作用。
1 资料与方法
1.1 一般资料 肝癌患者45例,经病理证实均为肝细胞癌,其中男性36例,女性9例,≤50岁13例,>50岁32例;血清AFP≤20 ng/mL 18例,>20 ng/mL 27例;肿瘤直径≤3 cm 1例,>3 cm 44例;无侵袭32例,发生侵袭13例;肿瘤TNM分期I/II期11例,III/IV期34例。本研究经南通市第三人民医院临床研究伦理委员会批准,患者均知情同意。
1.2 细胞培养 永生LO2人肝细胞、人肝癌细胞系(SMMC-7721、Li-7、PLC/PRF/5、SK-HEP-1、Huh-7、MHCC-97H、HCC-LM3)均购自上海细胞库。将LO2细胞、SMMC-7721、Li-7细胞置于RPMI-1640培养基(Gibco)中培养,PLC/PRF/5、SK-HEP-1细胞在添加碳酸氢钠和丙酮酸钠的MEM培养基中培养,Huh-7、MHCC-97H、HCC-LM3细胞在Dulbecco改良伊格尔培养基(Gibco)中培养,所有培养基均添加10%胎牛血清(Cell Sciences,Canton,MA),在37 °C、5%CO2培养箱中培养。
1.3 实时定量PCR(RT-qPCR)检测 使用Trizol试剂从细胞和手术切除肝癌组织及其匹配的相邻正常肝组织中提取总RNA,使用Bulge-LoopTM qPCR试剂盒(锐博生物,中国广州)进行RT-qPCR,检测PIWI相互作用RNA017724的表达水平。采用U6小核RNA作为内参对照,RNA相对表达量以幂值(2-△△Ct)计算。
1.4 细胞转染 PIWI相互作用RNA017724模拟物和抑制剂及相应的阴性对照物由锐博生物合成。将SMMC-7721和PLC/PRF/5细胞接种在6孔板中,培养过夜。分为过表达PIWI相互作用RNA017724组(mi-017724)及其空白对照组(mi-NC),敲低PIWI相互作用RNA017724组(in-017724)及其空白对照组(in-NC)。根据制造商说明书,两种溶液混合后,将混合物添加到6孔板中。转染48 h后收集细胞,进行后续实验。
1.5 细胞增殖试验 采用CCK-8法检测细胞活力,按照试剂盒(Sigma,USA)说明书操作。将转染48 h的肝癌细胞用胰蛋白酶消化,以每孔3×103个细胞密度接种到96孔板,培养过夜。每孔加入10 μL CCK-8试剂,37 °C孵育2.5 h。在酶标仪(Thermo Fisher,USA)450 nm处测量吸光度。
1.6 细胞集落形成试验 转染48 h的肝癌细胞经胰蛋白酶化,制备单细胞悬液,以每孔2 000个细胞密度接种到6孔板中。每3天更换1次培养基,培养2周以获得细胞集落。磷酸盐缓冲液清洗,4%多聚甲醛固定液固定,结晶紫染色。洗涤并在空气中干燥后,对细胞集落进行拍照和计数。
1.7 细胞凋亡检测 应用Annexin V/7-AAD Apoptosis Detection Kit(BD公司,美国)对细胞进行染色,使用FlowJo软件(BD公司)通过流式细胞术评估细胞凋亡。
1.8 Transwell试验 采用transwell室(Corning公司,NY,USA)测试细胞侵袭和迁移能力。转染48 h后肝癌细胞经胰蛋白酶消化,收集后离心,基培重悬。以每孔2×105个细胞接种到小室中,将600 μL含20%FBS培养基加至下室中。37 °C、5%CO2培养箱中培养24 h,用棉签去除transwell膜上表面的残留细胞。transwell膜下表面迁移的细胞用4%多聚甲醛固定,PBS洗涤,结晶紫染色8 min。侵袭实验需在transwell膜预涂基质胶(BD Biosciences,USA)。用光学显微镜(Olympus,Japan)对transwell膜下表面的细胞进行拍照,计算3个随机视野的细胞数。
1.9 统计学处理 应用SPSS 25.0统计学软件对数据进行分析处理。计量资料以■±s表示,组间比较采用t检验。P<0.05为差异有统计学意义。
2 结 果
2.1 肝癌组织中PIWI相互作用RNA017724表达及其与患者预后的关系 RT-qPCR检测显示,与正常肝组织比较,肝癌组织中PIWI相互作用RNA017724低表达,差异有统计学意义(P<0.01)(图1A)。根据PIWI相互作用RNA017724临界值,将45例患者分为PIWI相互作用RNA017724-high组和low组,结果显示PIWI相互作用RNA017724水平与患者的预后密切相关,与高表达组患者比较,低表达组患者生存期较短,差异有统计学意义(P<0.05)(图1B)。
2.2 肝癌细胞PIWI相互作用RNA017724表达及转染效果 RT-qPCR检测显示,与正常LO2肝细胞比较,各肝癌细胞系PIWI相互作用RNA017724表达显著减少,差异均有统计学意义(P<0.05)(图2A)。SMMC-7721和PLC/PRF/5细胞转染PIWI相互作用RNA017724抑制剂后,PIWI相互作用RNA017724表达降低,转染PIWI相互作用RNA017724模拟物后,PIWI相互作用RNA017724表达增加(图2B、C),说明转染成功。
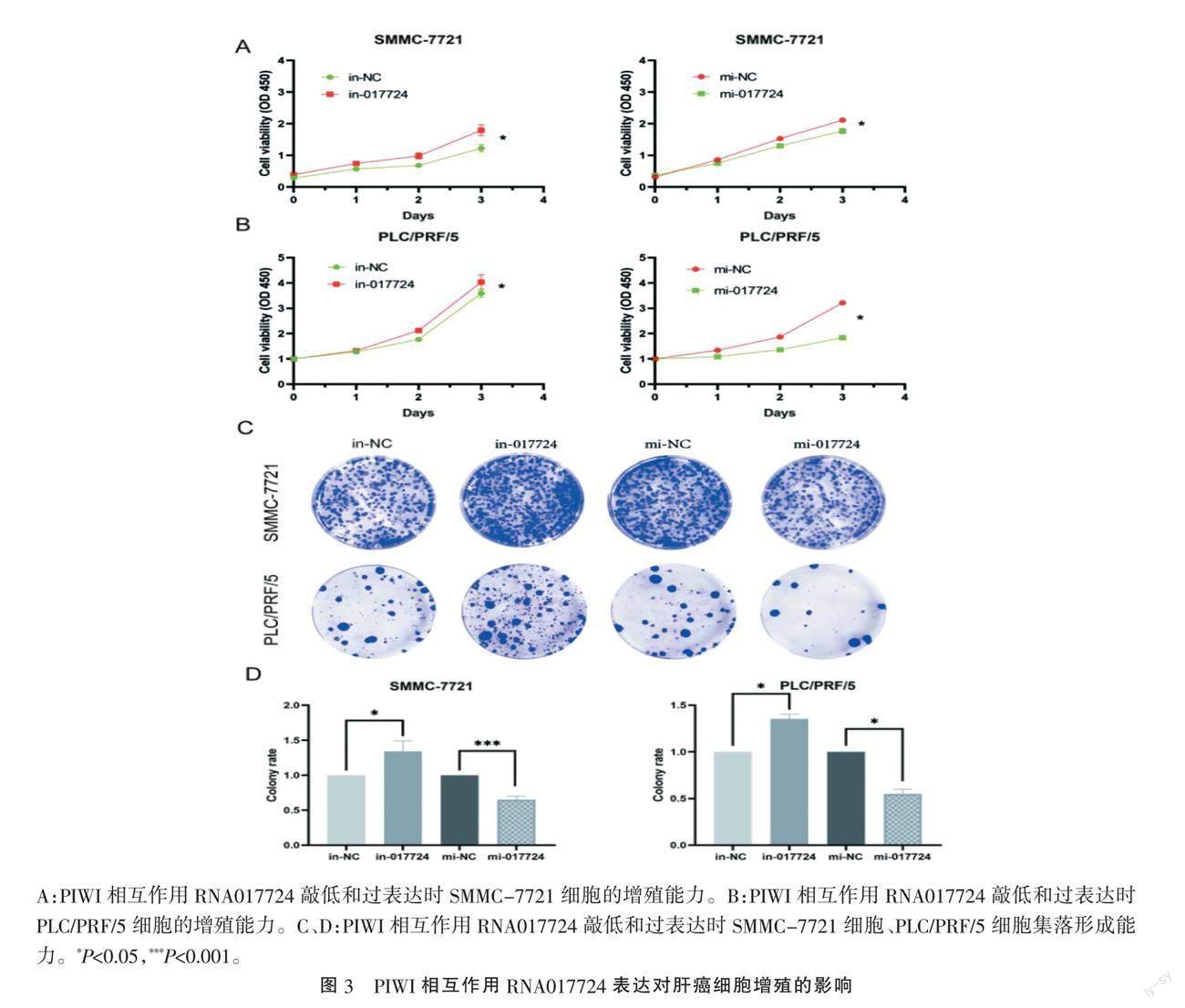
2.3 PIWI相互作用RNA017724抑制肝癌细胞增殖能力 CCK-8实验显示,抑制PIWI相互作用RNA017724表达后SMMC-7721和PLC/PRF/5细胞活力显著增加,而增加PIWI相互作用RNA017724表达后两种细胞活力降低(图3A、B)。细胞集落形成实验显示,PIWI相互作用RNA017724降低SMMC-7721和PLC/PRF/5细胞集落数量(图3C、D)。
2.4 PIWI相互作用RNA017724表达对肝癌细胞凋亡的影响 流式细胞仪检测发现,过表达PIWI相互作用RNA017724和敲除PIWI相互作用RNA017724对SMMC-7721和PLC/PRF/5细胞凋亡无显著影响(图4)。
2.5 PIWI相互作用RNA017724抑制肝癌细胞迁移和侵袭能力 transwell实验显示,与in-NC组比较,敲低PIWI相互作用RNA017724表达增加SMMC-7721和PLC/PRF/5细胞的迁移能力,而与mi-NC组比较,PIWI相互作用RNA017724过表达抑制SMMC-7721和PLC/PRF/5细胞的迁移能力(图5A、B)。与mi-NC组比较,PIWI相互作用RNA017724过表达抑制SMMC-7721和PLC/PRF/5细胞的侵袭能力,而与in-NC组比较,抑制PIWI相互作用RNA017724表达增强细胞侵袭能力(图5C、D)。表明PIWI相互作用RNA017724可以抑制肝癌细胞的侵袭和迁移能力。
3 讨 论
PIWI相互作用RNA是一类新发现的小非编码RNA(ncRNA)[12-13],迄今为止已鉴定出20 000多个PIWI相互作用RNA[14-15]。一些研究表明,PIWI相互作用RNA在生殖細胞中广泛表达。高通量测序表明,PIWI相互作用RNA信号通路在人类癌细胞中也很活跃[16-20],可能在癌症发展过程中发挥作用。PIWI相互作用RNA通过调节转录和转录后水平的基因表达影响癌细胞多种过程,包括细胞凋亡、增殖、迁移和侵袭[17,21]。已证明,PIWI相互作用RNA通过直接结合PIWI蛋白表现出促癌或抗癌作用[22]。PIWI相互作用RNA的异常表达与多种癌症有关,可能参与癌症发生、发展和转移。PIWI相互作用RNA与胃癌的恶性程度密切相关,胃癌组织中PIWI相互作用RNA651和PIWI相互作用RNA823异常表达与患者预后密切相关,可作为胃癌的诊断工具或治疗靶点[23-25]。PIWI相互作用RNA在乳腺癌的发病中具有重要作用[26-28]。PIWI相互作用RNA与肾癌的转移和预后密切相关[29-31]。PIWI相互作用RNA作为结直肠癌诊断工具和治疗靶点具有重要临床意义[32-37]。PIWI相互作用RNA55490在肺癌组织中的表达降低,抑制PIWI相互作用RNA55490表达可以通过抑制肺癌细胞中mTOR通路的激活来促进肺癌细胞的增殖。这表明PIWI相互作用RNA55490在肺癌的发生中可能具有抗癌作用[19]。PIWI相互作用RNA651参与非小细胞肺癌的发生、侵袭和转移,可能是一种潜在的癌症诊断工具[38-40]。
既往研究表明,PIWI相互作用RNA017724在肝细胞癌中异常表达。本研究结果显示,PIWI相互作用RNA017724在肝细胞癌组织中显著下调,PIWI相互作用RNA017724低表达患者预后较差。当上调PIWI相互作用RNA017724时,肝癌细胞侵袭、迁移和增殖能力受到抑制,下调PIWI相互作用RNA017724则促进肿瘤细胞侵袭、迁移和增殖。有研究表明,上皮间充质转化(EMT)[41]、WNT/β-连环蛋白通路[42-43]影响细胞增殖、侵袭和迁移。推测PIWI相互作用RNA017724可能通过这些途径影响肝细胞癌的发生发展,有待后续进一步实验研究。PIWI相互作用RNA017724在HCC的发生和发展中发挥重要作用,并可能作为诊断和预后的生物标志物。
[参考文献]
[1] ?魣LVAREZ E G,DEMEULEMEESTER J,OTERO P,et al. Aberrant integration of Hepatitis B virus DNA promotes major restructuring of human hepatocellular carcinoma genome architecture[J]. Nat Commun,2021,12(1):6910.
[2] LOCKART I,YEO M G H,HAJARIZADEH B,et al. HCC incidence after hepatitis C cure among patients with advanced fibrosis or cirrhosis: a meta-analysis[J]. Hepatology,2022,76(1):139-154.
[3] SEO W,GAO Y,HE Y,et al. ALDH2 deficiency promotes alcohol-associated liver cancer by activating oncogenic pathways via oxidized DNA-enriched extracellular vesicles[J]. J Hepatol,2019,71(5):1000-1011.
[4] EUROPEAN ASSOCIATION FOR THE STUDY OF THE LIVER. EASL clinical practice guidelines: management of hepatocellular carcinoma[J]. J Hepatol,2018,69(1):182-236.
[5] FORNER A,REIG M,BRUIX J. Hepatocellular carcinoma[J]. Lancet,2018,391(10127):1301-1314.
[6] SIM H W,KNOX J. Hepatocellular carcinoma in the era of immunotherapy[J]. Curr Probl Cancer,2018,42(1):40-48.
[7] ASSUMP?覶?魨O C B,CALCAGNO D Q,ARAJO T M,et al. The role of PiRNA and its potential clinical implications in cancer[J]. Epigenomics,2015,7(6):975-984.
[8] MEI Y,WANG Y,KUMARI P,et al. A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells[J]. Nat Commun,2015,6:7316.
[9] NG K W,ANDERSON C,MARSHALL E A,et al. Piwi-interacting RNAs in cancer: emerging functions and clinical utility[J]. Mol Cancer,2016,15:5.
[10] ZHONG F D,ZHOU N,WU K,et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes[J]. Nucleic Acids Res,2015,43(21):10474-10491.
[11] RIZZO F,RINALDI A,MARCHESE G,et al. Specific patterns of PIWI-interacting small noncoding RNA expression in dysplastic liver nodules and hepatocellular carcinoma[J]. Oncotarget,2016,7(34):54650-54661.
[12] GIRARD A,SACHIDANANDAM R,HANNON G J,et al. A germline-specific class of small RNAs binds mammalian Piwi proteins[J]. Nature,2006,442(7099):199-202.
[13] GRIVNA S T,BEYRET E,WANG Z,et al. A novel class of small RNAs in mouse spermatogenic cells[J]. Genes Dev,2006,20(13):1709-1714.
[14] SAI LAKSHMI S,AGRAWAL S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs[J]. Nucleic Acids Res,2008,36(suppl_1):D173-D177.
[15] ZHANG P,SI X,SKOGERB?覫 G,et al. piRBase: a web resource assisting piRNA functional study[J]. Database (Oxford),2014,2014:bau110.
[16] FATHIZADEH H,ASEMI Z. Epigenetic roles of PIWI proteins and piRNAs in lung cancer[J]. Cell Biosci,2019,9:102.
[17] GUO B,LI D,DU L,et al. piRNAs: biogenesis and their potential roles in cancer[J]. Cancer Metastasis Rev,2020,
39(2):567-575.
[18] HYUN S. Small RNA pathways that protect the somatic genome[J]. Int J Mol Sci,2017,18(5):E912.
[19] ROSS R J,WEINER M M,LIN H. PIWI proteins and PIWI-interacting RNAs in the soma[J]. Nature,2014,505(7483):353-359.
[20] PENG L P,SONG L,LIU C Y,et al. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling[J]. Tumour Biol,2016,37(2):2749-2756.
[21] LIU Y,DOU M,SONG X,et al. The emerging role of the PiRNA/piwi complex in cancer[J]. Mol Cancer,2019,18 (1):123.
[22] ZENG Q,WAN H,ZHAO S,et al. Role of PIWI-interacting RNAs on cell survival: proliferation,apoptosis,and cycle[J]. IUBMB Life,2020,72(9):1870-1878.
[23] LIN X,XIA Y,HU D,et al. Transcriptome-wide PiRNA profiling in human gastric cancer[J]. Oncol Rep,2019,41 (5):3089-3099.
[24] MARTINEZ V D,ENFIELD K S S,ROWBOTHAM D A,et al. An atlas of gastric PIWI-interacting RNA transcriptomes and their utility for identifying signatures of gastric cancer recurrence[J]. Gastric Cancer,2016,19(2):660-665.
[25] CHENG J,DENG H,XIAO B,et al. piR-823,a novel non-coding small RNA,demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells[J]. Cancer Lett,2012,315(1):12-17.
[26] HASHIM A,RIZZO F,MARCHESE G,et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer[J]. Oncotarget,2014,5(20):9901-9910.
[27] KRISHNAN P,GHOSH S,GRAHAM K,et al. Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer[J]. Oncotarget,2016,7(25):37944-37956.
[28] HUANG G,HU H,XUE X,et al. Altered expression of PiRNAs and their relation with clinicopathologic features of breast cancer[J]. Clin Transl Oncol,2013,15(7):563-568.
[29] LI Y,WU X,GAO H,et al. Piwi-Interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival[J]. Mol Med,2015,21(1):381-388.
[30] BUSCH J,RALLA B,JUNG M,et al. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas[J]. J Exp Clin Cancer Res,2015,34:61.
[31] ZHAO C,TOLKACH Y,SCHMIDT D,et al. Mitochondrial PIWI-interacting RNAs are novel biomarkers for clear cell renal cell carcinoma[J]. World J Urol,2019,37(8):1639-1647.
[32] MAI D,DING P,TAN L,et al. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma[J]. Theranostics,2018,8(19):5213-5230.
[33] YIN J,JIANG X Y,QI W,et al. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1[J]. Cancer Sci,2017,108(9):1746-1756.
[34] YIN J,QI W,JI C G,et al. Small RNA sequencing revealed aberrant PiRNA expression profiles in colorectal cancer[J]. Oncol Rep,2019,42(1):263-272.
[35] WENG W,LIU N,TOIYAMA Y,et al. Novel evidence for a PIWI-interacting RNA (PiRNA) as an oncogenic mediator of disease progression,and a potential prognostic biomarker in colorectal cancer[J]. Mol Cancer,2018,17(1):16.
[36] SUN R,GAO C L,LI D H,et al. Expression status of PIWIL1 as a prognostic marker of colorectal cancer[J]. Dis Markers,2017,2017:1204937.
[37] PETRA V F,KAROLINA S,LENKA R,et al. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer[J]. Cancer Epidem Biomar,2018,27(9):1019-1028.
[38] YAO J,WANG Y W,FANG B B,et al. piR-651 and its function in 95-D lung cancer cells[J]. Biomed Rep,2016,
4(5):546-550.
[39] ZHANG S J,YAO J,SHEN B Z,et al. Role of piwi-interacting RNA-651 in the carcinogenesis of non-small cell lung cancer[J]. Oncol Lett,2018,15(1):940-946.
[40] LI D,LUO Y,GAO Y,et al. piR-651 promotes tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4[J]. Int J Mol Med,2016,38(3):927-936.
[41] HUANG K,LIN Y,WANG K,et al. ARFIP2 regulates EMT and autophagy in hepatocellular carcinoma in part through the PI3K/Akt signalling pathway[J]. J Hepatocell Carcinoma,2022,9:1323-1339.
[42] YUAN W,ZHAO H,ZHOU A,et al. Interference of EFNA4 suppresses cell proliferation,invasion and angiogenesis in hepatocellular carcinoma by downregulating PYGO2[J]. Cancer Biol Ther,2022,23(1):1-12.
[43] TONG Q,YI M,KONG P,et al. TRIM36 inhibits tumorigenesis through the Wnt/β-catenin pathway and promotes caspase-dependent apoptosis in hepatocellular carcinoma[J]. Cancer Cell Int,2022,22(1):278.
[收稿日期] 2022-12-13
(本文編辑 赵喜)
