Mechanism of FXR alleviating the liver fibrosis by regulating perilipin 5
2023-11-20HUANGXiaoxiaZHENGZhiminPANGBiyingHUANGNanaLIXinXIONGWentingKONGBoLIUJisheng
HUANG Xiao‑xia, ZHENG Zhi‑min, PANG Bi‑ying, HUANG Na‑na, LI Xin, XIONG Wen‑ting, KONG Bo, LIU Ji‑sheng
School of Life Sciences, Guangzhou University, Guangzhou 510006, China
Keywords:
ABSTRACT Objective: To investigate the regulatory mechanism in liver fibrosis progression by nuclear receptor of farnesoid X receptor (FXR) and the lipid droplet‑associated protein of perilipin 5(PLIN5).Methods: FXR response element (FXRE) upstream of PLIN5 gene was found by bioinformatics, and confirmed by a dual luciferase reporter gene system; a hepatic fibrosis model based on human hepatic stellate cell LX‑2 was established by induction of transforming growth factor‑β1 (TGF‑β1); mRNA and protein levels of α‑smooth muscle actin (α‑SMA)and collagen Ⅰ were measured by qPCR and Western blot after transient overexpression of FXR or PLIN5; Oil red O staining was used to study the formation of lipid droplets.Results:The promoter region of the PLIN5 gene contained a known reverse repeats‑1 (IR‑1); the gene expression of PLIN5 in LX‑2 cells was up‑regulated after FXR activation (P<0.01);overexpression of PLIN5 promoted the formation of lipid droplets and significantly reduced the TGF‑β1 induced fibrosis gene expression (P<0.05); FXR activation showed no effects on the inhibition of LX‑2 cells activation.Conclusion: Overexpression of PLIN5 promotes the formation of lipid droplets and inhibits activation of LX‑2 cells.FXR might bind to the FXRE site upstream of PLIN5 gene and regulate its gene expression.In summary, FXR may prevent liver fibrosis progression partially by regulating lipid droplet‑associated protein of PLIN5.
1.Introduction
Non‑alcoholic fatty liver disease (NAFLD) is the most prevalent liver disease around the world at present, which is characterized by liver damage caused by excessive accumulation of fat in hepatocytes[1].and is closely related to type 2 diabetes and obesity.About 25% of people in the world are suffering from NAFLD.Liver fibrosis is a self‑repair process after liver injury, and its pathological features include a large amount of extracellular matrix deposition and fibrous tissue proliferation in the portal area and liver lobules.Risks factors such as alcohol, lipid metabolism imbalance,viruses, and so on[2], cause liver damage and eventually lead to liver fibrosis.During the progression of liver fibrosis, hepatocytes,sinusoidal endothelial cells, macrophages, as well as hepatic stellate cells (HSCs) in the liver interact each other, and produce various cytokines and oxidative active products, which further stimulate the liver inflammation and fibrosis[3].Without intervention, fibrosis can further develop to cirrhosis and even hepatocellular carcinoma in the late stage[4].
The key character of liver fibrosis is the activation of hepatic stellate cells (HSCs)[5].HSC is located in the disse gap between hepatocytes and sinusoidal endothelial cells, accounting for approximately 10% of all liver cells.In a healthy physiological state, HSCs are under a quiescent state, storing a large amount of lipid droplets containing vitamin A.After external stimuli, HSCs differentiate into myofibroblasts, transit to an activated state of proliferation, and subsequently lose lipid droplets.After activation,the gene and protein expression of α‑SMA increase, which produce a large amount of extracellular matrix and collagen[6].Due to the irreversible nature of liver fibrosis, preventing and reversing HSC activation are key approaches for preventing liver fibrosis[7].
PLIN5 belongs to the lipid droplet protein family, and is highly expressed in myocardial, skeletal muscle, and HSCs, and has the function of maintaining lipid droplet homeostasis[8].HSCs lose their lipid droplets during the activation, indicating that exogenous PLIN5 might be involved in the HSC activation and the formation of lipid droplets[9,10].
FXR belongs to the nuclear receptor superfamily and is highly expressed in the liver, intestine, and kidneys.As a nuclear factor,bile acids are the physiological ligands for FXR[11].Moreover, FXR regulates multiple physiological functions such as regulating bile acid homeostasis and lipid metabolism, and reducing fibrosis[12].FXR is a popular drug target for liver diseases[13].Loss of FXR function in mouse leads to increased liver cholesterol and triglycerides, and promotes the development of atherosclerosis,which indicated the roles of FXR in regulating lipid and lipoprotein metabolism[14].However, there is no researches showing FXR prevent or decrease the fibrosis progress in NAFLD patience.In this study, human hepatic stellate cell line LX‑2 was chosen to verify whether FXR in the stellate cells could prevent HSCs activation through increasing the PLIN5 expression and decreasing the loss of lipid drops, which could provide scientific theoretical evidences for FXR regulating lipid metabolism and inhibiting liver fibrosis.
2.Materials and methods
2.1 Cells and reagents
Human embryonic kidney cells line HEK293T was presented by Qiao Yunbo group of Guangzhou university, and kept by laboratory;The human hepatic stellate cells line LX‑2 was purchased from Wuhan Procell; the reporter gene plasmid pGL4‑Luc (101788) was purchased from Addgene; The overexpression plasmid pCI‑GFP was presented by the Wanggang group of Guangzhou university;2×Taq Master Mix PCR, T4 DNA ligase, Exfect transfection reagent,M‑MLV (H ‑) reverse transcriptase, RNA‑easy total RNA extraction kits, ChamQ real-time quantitative PCR kits, DH5α competent cells and BCA kits were purchased from Nanjing Vazyme; TGF‑β1 was purchased from PeproTech of the United States; Opti‑MEM medium, fetal bovine serum, DMEM medium, and trypsin were purchased from Invitrogen; ECL kits, protease inhibitor, phosphatase inhibitor, rabbit secondary antibodies, mouse secondary antibodies,β‑actin antibodies, 4% paraformaldehyde, and Oil Red O kits were purchased from Shanghai Beyotime; Triglyceride assay kits(tissue, cell) were purchased from Beijing Applygen; α‑SMA primary antibodies, Collagen I primary antibodies were purchased from Abcam, and PLIN5 primary antibodies were purchased from Proteintech.
2.2 Instruments
Nanodrop, Thermofisher; ABI mini AMP PCR amplifier,Thermofisher; ABI QuantStudio 3 quantitative PCR system,Thermofisher; CO2 cell incubator, Thermofisher; SW‑CJ‑1FD clean bench, Suzhou Airtech; Infinite M Plex multimode reader, Tecan;Biospectrum Imaging System, UVP; SCILOGEX SLK‑O3000‑S horizontal shaker, Scilogex.
2.3 Cells Culture
LX‑2 cells were cultured in DMEM medium containing 10%FBS and grew in an environment of 37 ℃ and 5% CO2.Induction conditions of cellular liver fibrosis in vitro model: using TGF‑β1(10 ng/mL) to induce LX‑2 cells for measuring mRNA and protein level at 24 and 48 h, respectively.
2.4 qPCR
Referred to the protocol of the total RNA extraction kit for total RNA extraction, and quantified the RNA by Nanodrop for reverse transcription.Referred to the protocol of the reverse transcription kit for reverse transcription to obtain cDNA.Referred to the protocol of ChamQ Universal SYBR qPCR Master Mix for the reaction system and used ABI QuantStudio 3 for qPCR.The primers were synthesized by Tsingke, and the primers used for qPCR are shown in Table 1.

Tab 1 Primer sequences for qPCR
2.5 Prediction of PLIN5 FXRE
Determined the DNA sequence information of the regulatory region of the human and mouse PLIN5 gene by UCSC browser (https://genome.ucsc.edu/).Further predicted the possible cis element FXRE in these DNA fragments by NubiScan (https://www.nubiscan.unibas.ch/), and the results was shown in Fig.1A.
2.6 Plasmids construction
Referred to the protocol of 2×The Taq Master Mix PCR reagent for PCR cloning to obtain the target fragments, and the primers used are shown in Table 2.Referred to the protocols of endonuclease, T4 DNA ligase and DH5α for digestion, ligation and transformation, and FXRE fragments were inserted to upstream region of the luciferase gene of pGL4‑Luc to construct pGL4‑PLIN5 FXRE wt‑Luc , pGL4‑PLIN5 FXRE mut-Luc and pGL4-3×PLIN5 IR-1-Luc plasmids (Fig 1B).Removed the GFP gene in the downstream region of the CMV promoter of the over‑expression plasmid pCI‑GFP, and inserted the coding sequence (CDS) of target gene to construct pCI‑PLIN5 and pCI‑FXR plasmids (Fig 1C).

Tab 2 Primer sequences for DNA cloning

Fig 1 Prediction of FXRE sites(A) and constructed reporter gene plasmid(B‑C)
2.7 Cells transfection
Seeded LX‑2 cells into a 6‑wells culture plate, and when the cell confluency is 70%‑80%, referred to the protocol of the transfection reagent, transiently transfected with 1.5 μg plasmids and 3 μL transfection reagent per well, diluted the plasmids and transfection reagent with 200 μL opti-MEM medium respectively before the transfection and mix gently, stood for 15 min, added dropwise to a 6‑wells culture plate and mix gently, transfected for 6 hours, and then replaced the medium.The transfection conditions for HEK293T are described above.
2.8 Oil red O staining
After treatment, cells were fixed and stained according to the protocol of Beyotime, and images were taken with an inverted microscope.
2.9 Dual luciferase reporter gene assay
GW4064 is a well‑known FXR agonist, and the concentration used in our laboratory to activate FXR is 2 μM.Transfected HEK293T cells for 6 h according to the method 1.6, the cells were digested with trypsin and centrifuged, and uniformly seeded into a 96‑wells culture plate to ensure consistent transfection efficiency.Set the same volume of DMSO as vehicle and 2 μM GW4064 as treatment, there were three replicates in each group, and the dual luciferase reporter gene was detected at 18 h and 42 h, respectively.After inducing cells, removed the culture medium, washed twice with PBS and then discarded the solution.Add 50 μL reporter gene lysis solution to each well on a 96‑wells culture plate, stood at room temperature for 5 min, blew and pipetted the cell lysate into a 1.5 mL microcentrifuge tube, centrifuged the samples at 4 ℃, 12 000×g for 3 min and pipetted the supernatant for the assay.Pipetted 10 μL supernatant into a 96-wells white plate and added 50 μL luciferase substrate incubated to 37 ℃ to the well, quickly mixed and immediately detected the fluorescence value of firefly luciferin with a multimode reader.Pipetted another 10 μL supernatant,added 50 μL Renilla working solution incubated to 37 ℃, quickly mixed, and immediately detected the fluorescence value of renilla luciferin with a multimode reader.The ratio of activity between the firefly luciferase and the renilla luciferase (Fluc/Luc) indicates the activity of luciferase, thus indicating the activity of transcription.And indicated the relative luciferase activity by the ratio of GW4064 treatment to vehicle (GW/DMSO).
2.10 Western blot
Lysed the cells with RIPA lysate, centrifuged at 4 ℃ and pipetted the supernatant into a 1.5 mL microcentrifuge tube.Determined the protein concentration of the sample by BCA assay.After adding protein loading buffer to the sample, heated it in a 100 ℃ metal bath for 10 min for denaturation.According to the molecular weight of the samples, electrophoresed at 80 V/30 min and 120 V/60 min in 8%, 10%, or 12% SDS‑PAGE resolving gels.The proteins were transferred to PVDF activated by methanol at 230 mA/90 min,blocked by blocking buffer with 5% nonfat powdered milk at room temperature for 1 hour, a primary antibody was added and incubated at 4 ℃ for 14‑18 h.The PVDF was washed three times with TBST buffer (10 minutes each time); the secondary antibody (1:10 000)labeled with horseradish peroxidase was added and incubated at room temperature for 1 hour, the PVDF was washed 3 times with TBST buffer (10 min each time); added ECL chemiluminescence solution and incubated for 3 min for imaging.
2.11 Triglyceride measurement
After cells treatment, referred to the protocol to measure TG content in cells; quantified the total protein of cells by BCA assay,and corrected TG content by the concentration of total protein.
2.12 Statistical processing
Performed statistical analysis and plotting with GraphPad Prism 9.0.The results were showed as mean ± SD.The significance between the two groups was determined by t‑test, and the significance between the multiple groups was determined by ANOVA.P<0.05 indicates a statistically significance.
3.Results
3.1 FXR binds to PLIN5 IR-1
FXR is a transcription factor which can recognize and bind to FXR response elements in downstream target genes.IR‑1 is a typical FXRE fragment, which is a reverse repetitive sequence separated by one nucleotide[15].The IR‑1 fragments in the upstream regulatory sequences of the mouse and human PLIN5 gene were confirmed by NUBIScan, and the sequences containing IR‑1 fragments and their upstream and downstream sequences were compared (Fig 1A).A luciferase reporter gene plasmid pGL4‑PLIN5 FXRE wt‑Luc(FXRE wt group) was constructed by inserting the FXRE of IR‑1(‑176/+179) to the upstream region of the luciferase gene for the ability of FXR binding to the PLIN5 promoter., while pGL4‑PLIN5 FXRE mut‑Luc (FXRE mut group) was constructed by inserting the mutant sequence of FXRE (Fig 1B).The HEK293T cells were transiently co‑transfected with dual luciferase reporter gene system,and the Vec group was transfected with pGL4‑Luc.There was no significance in the relative luciferase activity (GW/DMSO) between FXRE wt group and FXRE mut group (Fig 2B).The luciferase activity of both DMSO treatment and GW4064 treatment of FXRE wt group and FXRE mut group is more than 150 times higher than that of Vec group (Fig 2A), which may be due to the presence of sites regulated by other regulatory factors in LX‑2 cells, which prevents GW4064 from further inducing the reporter gene after activating FXR.A reporter gene plasmid with three PLIN5 IR‑1 repeatedly connected was constructed (3×IR‑1 group) to confirm the binding effect to the FXR.The relative luciferase activity (GW/DMSO) of 3×IR-1 group was 10 times higher than that of Vec group (P<0.000 1) (Fig 2B).A FXR response element of IR‑1 in the upstream region of the PLIN5 promoter was confirmed by the above results, which binds to FXR and regulates downstream target genes.
3.2 Low expression of FXR in LX‑2 cells
There was no significance in the transcription level of the known FXR target gene SHP between the GW4064 treatment and DMSO treatment (Fig 3A).The low expression of FXR in LX‑2 cells was revealed by the qPCR assay (not shown in the results), and the target genes of FXR was not activated by the GW4064 treatment.The activation effect of FXR was measured by co‑transfecting FXR over‑expression plasmid and dual luciferase reporter gene system into LX‑2 cells, while the pCI‑GFP transfection was the negative control(Vec group).The relative luciferase activity (GW/DMSO) of the FXR over‑expression group in LX‑2 cells was 12 times higher (P<0.001)(Figure 3B) and 6 times higher (P<0.01) (Fig 3C) than that of the Vec group respectively after GW4064 treatment for 24 h and 48 h.The activation of over‑expression of FXR in LX‑2 cells by the FXR agonist GW4064 was confirmed by the above results.

Fig 2 Binding effect of PLIN5 FXRE fragments with FXR

Fig 3 Detection of FXR activation by qPCR and dual luciferase reporter gene system in LX‑2 cells
3.3 PLIN5 was upregulated by FXR
The expression level of PLIN5 in LX‑2 cells was measured after FXR over‑expression and GW4064 treatment.The control group was not transfected with any plasmids, and the pCI‑GFP transfection group was negative control (Vec group).The mRNA transcription and protein expression levels of PLIN5 compared to the Control and Vec groups were significantly increased by the over‑expression of FXR (P<0.01) (Fig 4).The upregulation of expression of PLIN5 by the FXR activation was confirmed by the above results.
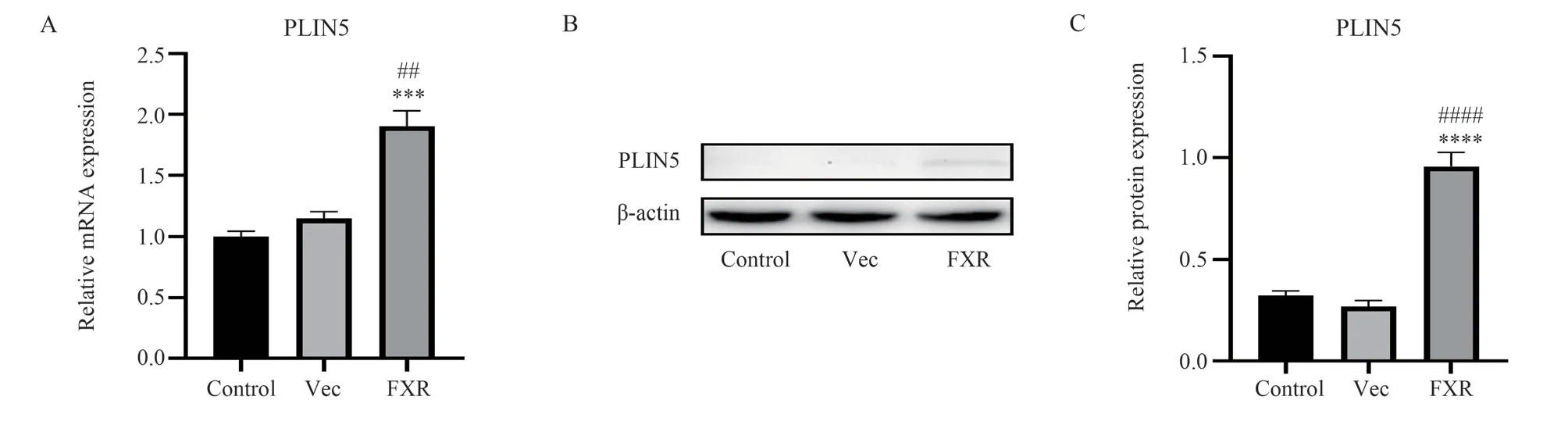
Fig 4 Activation of FXR up‑regulates PLIN5 expression
3.4 FXR activation cannot promote the formation of lipid droplets and inhibit the activation of LX‑2
In vitro models, oleic acid is normally used to provide the source of free fatty acids (FFA)[16].The formation of lipid droplets in LX‑2 cells can be promoted by Oleic acid, and may be related to the activation of HSC.The lipid droplets that can be stained with Oil red O were insufficient to be formed by the 30 μM oleic acid (Fig 5A), and the formation of lipid droplets cannot be promoted by over‑expression of FXR and activation of GW4064 in LX‑2 cells.TGF‑β1 is a strong cytokine that induces HSC activation, which was used to stimulate LX‑2 to established a hepatic stellate cell activation model.There was no significant inhibitory effect on the mRNA levels of fibrotic genes (α-SMA and Collagen I) induced by TGF-β1 when the FXR was activated (Fig 5C).
3.5 Over‑expression of PLIN5 promotes lipid droplet formation and inhibits LX‑2 activation
Previous studies have shown that the gene expression of PLIN5 is significantly reduced or even disappeared in activated HSC cells[10].LX‑2 cells are hyperactivated in vitro.Whether over‑expression of PLIN5 can promote the formation of lipid droplets and inhibit the activation of LX‑2 needs to be verified base on the results of 2.4.The formation of lipid droplets treated with 30 μM oleic acid can be promoted by over‑expression of PLIN5 (Fig 6A‑B).The fibrotic gene mRNA levels (P<0.001) and protein levels (P<0.05) of α-SMA and Collagen I induced by TGF‑β1 can be significantly inhibited by over‑expression of PLIN5 (Fig 6C‑D).

Fig 5 Effect of FXR over‑expression on lipid droplet and genes of fibrosis
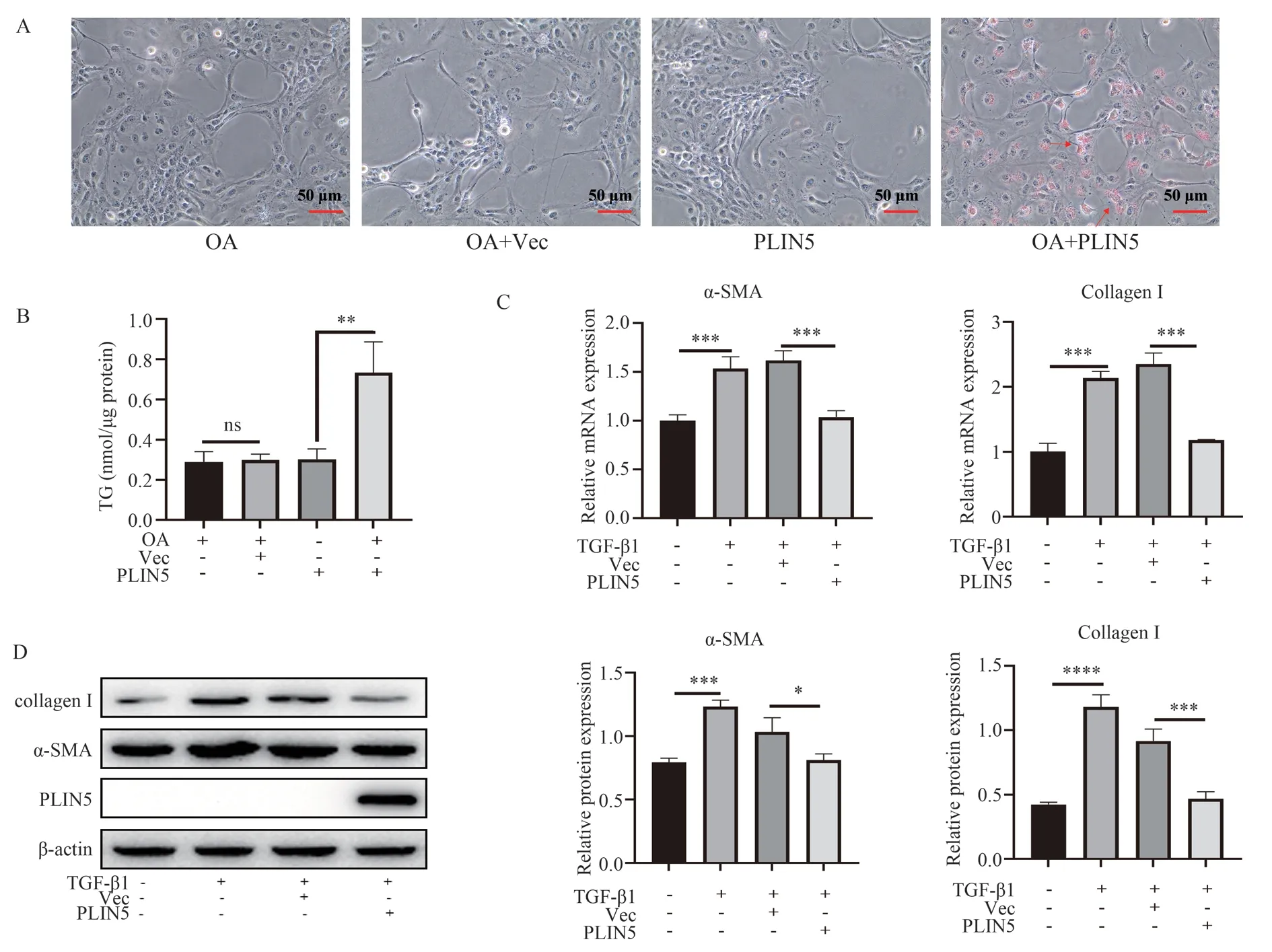
Fig 6 Effect of PLIN5 over‑expression on lipid droplet and genes of fibrosis
4.Discussion
As a nuclear receptor, FXR is activated after binding to ligands, and binds to FXRE on its target genes as a heterodimer with the retinoid X receptor α (RXRα), thus regulates the transcription of target genes.IR‑1 is a typical FXRE binding site.The possible FXRE sites in the human PLIN5 promoter region were predicted by the bioinformatics,and determined by dual luciferase reporter gene system.Moreover,the expression level of PLIN5 can be increased by FXR activation,which may be a partial mechanism by which FXR activation inhibits liver fibrosis.
There are sites of other regulatory factors in the upstream or downstream regions of FXRE in LX‑2 cells base on the results of FXRE wt group and FXRE mut group in Fig 2A, so that high luciferase activity even in DMSO treatment, and GW4064 cannot further activate the reporter gene after activating FXR.The relative luciferase activity (GW/DMSO) of 3×PLIN5 IR-1 group was 10 times higher than that of Vec group, which indicated that PLIN5 IR‑1 could bind to FXR.That FXR can bind to PLIN5 IR‑1 and upregulate the expression of luciferase was determined by the dual luciferase reporter gene system, a highly sensitive detection system,but the regulation of this binding activity on the transcription level of PLIN5 is still unknown.
It was found that the expression level of PLIN5 were increased in LX‑2 after over‑expressing and activating FXR (Fig 4), therefore the formation of lipid droplets and the activation of LX‑2 may be regulated by the up‑expression of PLIN5, but over‑expression of FXR and activation with GW4064 did not restore lipid droplets and inhibit mRNA levels of fibrosis genes α-SMA and Collagen I (Fig 5).Previous studies have shown that FXR can be expressed in rat HSC, HSC‑T6 and LX‑2.FXR/SHP could promote the quiescence and apoptosis of HSC by reducing collagen I, inhibiting fibrogenic signals which are in the downstream signals of AP‑1, inhibiting TIMP‑1, and increasing MMP‑2 activity, thereby preventing the development of liver fibrosis, therefore FXR ligands may be beneficial for the treatment of liver fibrosis[17,18].However, the above researches are not supported by this study, possibly due to the following reasons: on the one hand, it was found that expression level of FXR in LX‑2 cells was very low, which results in no significance in the mRNA level of the FXR target gene SHP after GW4064 treatment (Fig 3A).There may be too high activation level of the in vitro model LX‑2 to get some functions of quiescent state, while get some functions of myofibroblasts, FXR may also be downregulated during this process and only exerts limited biological functions.On the other hand, protein level of PLIN5 induced by exogenous FXR was at a low level (Fig 4), which shows a significant difference compared to the protein level of exogenous PLIN5 in Fig 6.Therefore, the PLIN5 induced by FXR is not sufficient to exert the function in restoring lipid droplets and inhibiting LX‑2 activation.
That FXR exists in HSC and plays an anti‑fibrotic role was demonstrated by previous studies[19,20].Plin5 is also expressed in freshly isolated mouse primary HSCs.With activation of HSCs,Plin5 rapidly loses along with lipid droplets.Over‑expression of Plin5 can restore lipid droplets and delay HSC activation[9], and the results of Fig 6 also supported that PLIN5 can inhibit the activation of LX‑2.Although FXR showed limited effects in liver fibrosis in vitro model, there were differences between the LX‑2 cell line in vitro and the HSC in vivo.The role of FXR binding to PLIN5 FXRE in HSC activation is unknown in vivo and further research is necessary.
In summary, this study found that FXR activation can bind to PLIN5 IR‑1 and upregulate the mRNA and protein levels of PLIN5; meanwhile, over‑expression of PLIN5 can downregulate the expression of liver fibrosis genes.Although the effect of FXR in liver fibrosis after upregulating PLIN5 is limited in the model of this study, the effect of FXR in liver fibrosis still cannot be ruled out base on the latest research.The gap in the regulation of lipid metabolism gene PLIN5 by FXR is supplemented by these data, which provide theoretical support for further in vivo research to reveal the mechanism of FXR’s effect in HSC activation and protection against liver fibrosis.
The authors declare that there are no conflict of interests.
杂志排行
Journal of Hainan Medical College的其它文章
- Advances in cellular and molecular mechanisms of oral submucosal fibrosis
- Research progress of natural products in treating polycystic ovary syndrome
- Research progress of related signal pathways in the prevention and treatment of heart failure with traditional Chinese medicine
- Network pharmacology and preliminary cell screening studies on the anti-liver cancer activity of Nauclea Officinalis
- Quality of life of hospitalized patients after lung cancer operation and analysis of influencing factors
- Meta-analysis of the efficacy and safety of extended right liver transplantation versus whole liver transplantation
