Automated Segmentation of the Brainstem, Cranial Nerves and Vessels for Trigeminal Neuralgia and Hemifacial Spasm
2023-11-14YuqingYangZhiwenLiu
Yuqing Yang, Zhiwen Liu
Abstract: Accurate localization of cranial nerves and responsible blood vessels is important for diagnosing trigeminal neuralgia (TN) and hemifacial spasm (HFS).Manual delineation of the nerves and vessels on medical images is time-consuming and labor-intensive.Due to the development of convolutional neural networks (CNNs), the performance of medical image segmentation has been improved.In this work, we investigate the plans for automated segmentation of cranial nerves and responsible vessels for TN and HFS, which has not been comprehensively studied before.Different inputs are given to the CNN to find the best training configuration of segmenting trigeminal nerves, facial nerves, responsible vessels and brainstem, including the image modality and the number of segmentation targets.According to multiple experiments with seven training plans, we suggest training with the combination of three-dimensional fast imaging employing steady-state acquisition (3D-FIESTA) and three-dimensional time-of-flight magnetic resonance angiography (3DTOF-MRA), and separate segmentation of cranial nerves and vessels.
Keywords: trigeminal nerves; facial nerves; responsible vessels; medical image segmentation
1 Introduction
Trigeminal neuralgia (TN) and hemifacial spasm(HFS) are the most common cranial nerve diseases, which may have a serious impact on patients’ life [1].Accurate localization of cranial nerves and responsible blood vessels is important for the diagnosis of TN and HFS.TN and HFS are both divided into two categories: primary and secondary.The compression of blood vessels on the nerves in or near their root entry/exit zone(REZ) is the causing factor of primary TN(pTN) and primary HFS (pHFS) [2].The most common cause of pTN is focal compression of the trigeminal nerve root, near its entry point into the pons, called the root entry zone, through an abnormal loop of arteries or veins [3], which is now thought to account for 80%–90% of cases [4, 5].PHFS is often thought to be the result of hyperexcitation of the facial nerve and its nucleus,which is caused by vascular compression in the root exit zone of the facial nerve [6–8].These vessels are called responsible vessels.To reduce the pain made by responsible vessels, patients are treated by a surgical method called microvascular decompression (MVD), which will remove responsible vessels from the compressed nerves by adding a spacer between them [9,10].
It is important to locate the cranial nerves and responsible vessels.Before surgery, patients are required to take different examinations, for instance, computed tomography (CT), magnetic resonance imaging (MRI), magnetic resonance angiography (MRA) images, etc.CT with contrast is the most common way to detect whether the pain of HFS is made by the tumor [11,12].Also, hospitals provided 2D MRI, but weaknesses such as low resolution and missing spatial structure make it inadequately for constructing neurovascular spatial relationships in REZ.To overcome these tackles, [13,14] used 3D MRI which provided high spatial resolution while delineating vessels and nerves to perform neurovascularity visualization [15].Standard T2-weighted MRI sequences only fully depict larger cranial nerves, whereas current steady-state free precession (SSFP) MRI sequences have the ability to display the cisternal segments of all 12 cranial nerves.SSFP sequences provide submillimetric spatial resolution and high contrast resolution between cerebrospinal fluid and solid structures, allowing the reconstruction of elegant multiplanar images that highlight the course of each nerve [16].[17] found that with high resolution SSFP, it is simple to distinguish arteries from veins by determining whether the compressed vessels can be traced back to the maternal artery.
SSFP sequences are often referred to as three-dimensional fast imaging employing steadystate acquisition (3D-FIESTA).Conventional MRI sequences can provide the best soft tissue resolution, but may not be sufficient to define the spatial resolution required for cranial nerves [18].3D-FIESTA sequences produce a very high-resolution T2 weighted image which can generate high-resolution images with excellent image contrast and high signal-to-noise ratio among cerebrospinal fluid, blood vessels and cranial nerves,making small structures eye-catching [19].In addition, the 3D-TOF-MRA sequences increase the contrast between blood flow and static tissue,with thinner sequence layers, higher arterial resolution, and direct visualization of small vessels with a diameter of 0.8 mm, improving the vascular and nerve reconstruction.
Manual delineation of responsible vessels and cranial nerves on patient images by doctors is time-consuming and labor-intensive.As convolutional neural networks (CNNs) have made great achievement in the medical image segmentation tasks [20–23], researchers have started to train CNNs, such as U-Net like models [24–27], to automatically segment nerves and vessels.[28]proposed a basic CNN for the segmentation of vessels in coronary angiogram.[29] first applied CNN into cerebrovascular segmentation by utilizing a relatively shallow neural network in 3DTOF-MRA sequences.[30] segmented cerebral artery in 3D-TOF-MRA sequences only setting half channels in each layer of U-Net.[31] integrated 3D U-Net, multi-scale, and deep supervision methods to indentify small blood vessels in 3D-TOF-MRA sequences, guiding the middle layer of the network to better generate discriminative features, avoiding gradient explosion and vanishing, and improving model convergence [32].[33] proposed the scale-space approximated CNN,which retained the receptive field, increased the network depth, and showed excellent performance in blood vessels segmentation [34].[35]applied a standard U-Net for segmenting fascicles from micro CT sequences of human vagus nerves.[36] used diffusion MRI (dMRI) to segment cranial nerves, but dMRI acquisition is usually too long for clinical use.[15] segmented trigeminal nerves for TN diagnosis based on the 3D-TOF-MRA sequences with a CNN-based method.
During the diagnosis of TN and HFS, doctors need to identify nerves and vessels concurrently to find whether they are intersecting.However, either sequence alone is not sufficient to distinguish among the nerves and vessels.3DFIESTA sequences have excellent contrast between structures of trigeminal nerves and adjacent blood vessels [37], while it is difficult to distinguish arteries and veins.3D-TOF-MRA sequences selectively display fast-flowing blood and provide visualization of arteries and nerves,but an inherent flaw of the technique is insufficient visualization of veins [38].[39] registered and reconstructed 3D-FIESTA and 3D-TOFMRA sequences through an open source software 3D Slicer to generate a neurovascular model,which would display a better neurovascular relationship in three dimensional view.[40] generated fusion MRIs combined with 3D-TOF-MRA and three dimensional constructive interference in steady state (CISS, another name of 3DFIESTA) on Osirix software, showing a more accurate result in diagnosis through omitting subjective correlation between two modalities.
Inspired by taking multi modality images into software before surgery, we focus on automated segmentation approaches in multimodal sequences of TN and HFS.Due to the small structure of cranial nerves and vessels, we need the help of clear anatomical structures such as brainstem to locate them.Therefore, it is desired to investigate how to automatically and jointly segment the brainstem, cranial nerve and responsible vessel with multimodal images.
In this work, we explore the optimal configuration of automated segmentation of the brainstem, cranial nerve and responsible vessel with multimodal images for the clinical diagnosis of trigeminal neuralgia and hemifacial spasm, which has not been previously studied.We consider seven different configurations, including different combinations of image modalities with segmentation tasks.The state-of-the-art medical image segmentation method nnU-Net [41] is used for the segmentation.We found that the best plan for the segmentation task is to train two separate models, one for segmenting cranial nerves together with brainstem both 3D-FIESTA and 3D-TOF-MRA, and the other for segmenting the vessels as long as 3D-TOF-MRA is used.
The remaining of the paper is organized as follows.Section 2 presents the configurations for segmentation and the segmentation method.Section 3 describes the segmentation results.Section 4 discusses these results and concludes the paper.
2 Method
In this section, we first introduce the modalities used for segmenting cranial nerves and blood vessels.Then, the different segmentation configurations for consideration are presented.Finally, the implementation details are given.
2.1 Modalities for Cranial Nerve and Blood Vessel Segmentation
In the proposed method, we obtain two modalities both 3D-TOF-MRA and 3D-FIESTA for segmenting blood vessels and cranial nerves.In 3DFIESTA, the cranial nerves and blood vessels are both dark (see Fig.1(a)), whereas in 3D-TOFMRA, the vessels are bright (see Fig.1(b)).Due to these appearances, 3D-FIESTA is used for locating cranial nerves while 3D-TOF-MRA is used for segmenting vessels.However, it is unclear if the joint use of the two modalities for segmenting cranial nerves and blood vessels brings further benefits.Therefore, in this work we explore both modalities for segmentation and investigate what is the best practice of modality selection.

Fig.1 Visualization of the multimodal images used for segmentation: (a) 3D-FIESTA; (b) 3D-TOF-MRA; (c) 3DTOF-MRA after being registered to 3D-FIESTA
2.2 Implementation Details
In our problem, there are multiple anatomical structures to segment, including the brainstem,trigeminal nerves, facial nerves and responsible blood vessels.Note that here the brainstem is also considered because vessels and nerves are thin and spread throughout the brain while brainstem is relatively large and easy to localize,the branches of vessels and nerves can be determined by the position of blood vessels and nerves relative to the brainstem.To examine the best configuration for segmentation, seven different combinations of image modalities and structures to segment are selected as candidate configurations.The combinations are listed in Tab.1.

Tab.1 The candidate configurations for segmentation
First, all structures are jointly segmented,where 3D-TOF-MRA and 3D-FIESTA are used individually or together.In addition, the brainstem are jointly segmented with the cranial nerves on both two sequences or 3D-FIESTA only, as the nerves are not clearly visible on 3DTOF-MRA.Finally, the brainstem are jointly segmented with the vessels nerves on 3DFIESTA and 3D-TOF-MRA or 3D-TOF-MRA only, as the vessels are not clearly visible on 3DFIESTA.
2.3 Candidate Configurations
The 3D-FIESTA and 3D-TOF-MRA images are aligned (3D-TOF-MRA to 3D-FIESTA) with rigid registration so that they can be jointly used for segmentation, where the ANTs toolbox [42] is used.An example of the original 3D-FIESTA,3D-TOF-MRA and the aligned 3D-TOF-MRA image is shown in Fig.1.
Furthermore, as the cranial nerves and responsible vessels are located near the brainstem, the complete 3D-FIESTA and 3D-TOFMRA images are too large and their use may introduce confounding factors.Thus, the original images were cropped to preserve the center region of each slice.
We trained the segmentation models with the state-of-the-art nnU-Net methods [41], which has achieved consistently optimal performance across a variety of medical image segmentation tasks.This framework uses the U-Net architecture [43,44] and automatically determines data configuration, including intensity normalization,choice of 2D or 3D processing, patch size and batch size, etc.More details about nnU-net can be found in [41].
3 Results
3.1 Data Description and Experimental Settings
The investigation was performed on an in-house dataset for TN and HFS.It comprises 211 3DFIESTA and corresponding 3D-TOF-MRA images with TN or HFS.The scans were annotated by experienced radiologists with four labels,including the brainstem, trigeminal nerve, facial nerve, and responsible vessel.All 3D-FIESTA scans were acquired on a 1.5T GE scanner with a 0.35 mm×0.35 mm×0.40 mm resolution, whereas 3D-TOF-MRA images were acquired on 1.5T Siemens scanner with a 0.43 mm×0.43 mm×0.80 mm resolution.The images were aligned as described in Section 2.2.Then, the 512×512 images were cropped to 250×150 for each slice.Fig.2 shows the image and corresponding manual label after cropping.For each experimental configurations, we split the whole dataset into two sets.The same 168 cases were used for training, and the other 43 cases were used for testing.In addition, the same 20% of the training images were used as a validation set.
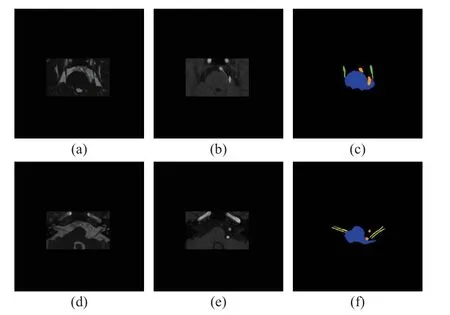
Fig.2 Visualization of the image and manual annotation.Two different slices are shown: (a) and (d) 3D-FIESTA; (b)and (e) 3D-TOF-MRA; (c) manual annotations of the brainstem, trigeminal nerve and responsible vessel; (f)manual annotations of the brainstem, facial nerve and responsible vessel (Blue represents the brainstem, green represents the trigeminal nerve, yellow represents the facial nerve, and orange represents the vessel)
3.2 Evaluation
Fig.3, Fig.4, Fig.5 and Fig.6 show the visualization of the segmentation results together with images for each configuration.Fig.3 and Fig.4 showing two different slices give the results for the first three configurations that segment all structures, Fig.5 gives the results for the middle configurations that segment the brainstem and cranial nerves, and Fig.6 gives the results for the last two configurations that segment the brainstem and vessels.
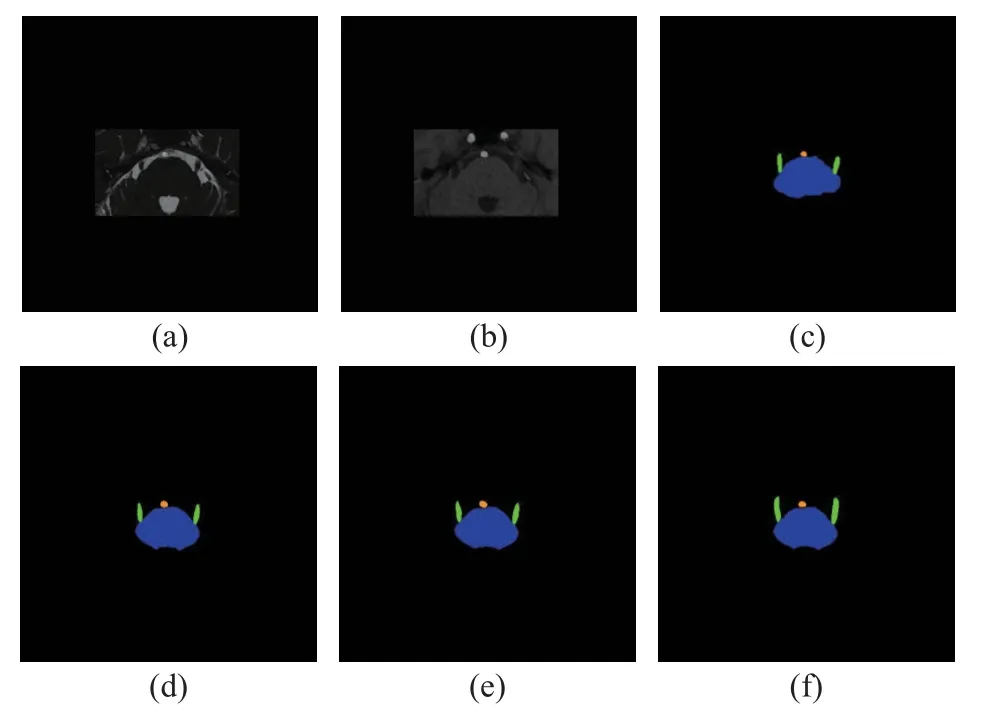
Fig.3 Visualization of results of joint segmentation of the brainstem, trigeminal nerve, facial nerve and responsible blood vessel for a slice with the trigeminal nerve,shown together with 3D-FIESTA and 3D-TOF-MRA:(a) 3D-FIESTA; (b) 3D-TOF-MRA; (c) manual annotations of the brainstem, trigeminal nerve and vessel;(d) the segmentation achieved with both 3D-FIESTA and 3D-TOF-MRA; (e) the segmentation achieved with 3D-FIESTA only; (f) the segmentation achieved with 3D-TOF-MRA only (Blue represents the brainstem,green represents the trigeminal nerve, and orange represents the vessel)
From Fig.3 and Fig.4, we can see that the configuration with 3D-FIESTA and 3D-TOFMRA together performed more stable than 3DTOF-MRA or 3D-FIESTA only, where both the segmented trigeminal nerve and facial nerve better resemble the manual annotation.Fig.5 shows that with both modalities the cranial nerves are better preserved than with one single modality.Fig.6 shows that the vessel is slightly better segmented with both modalities than one.

Fig.4 Visualization of results of joint segmentation of the brainstem, trigeminal nerve, facial nerve, and responsible blood vessel for another slice with the facial nerve,shown together with 3D-FIESTA and 3D-TOF-MRA:(a) 3D-FIESTA; (b) 3D-TOF-MRA; (c) manual annotations of the brainstem, facial nerve, and vessel; (d)the segmentation achieved with both 3D-FIESTA and 3D-TOF-MRA; (e) the segmentation achieved with 3DFIESTA only; (f) the segmentation achieved with 3DTOF-MRA only (Blue represents the brainstem, yellow represents the facial nerve, and orange represents the vessel)
Quantitatively, we computed the Dice coefficients between the segment-ation results and the manual labels to evaluate the segmentation quality.Tab.2 lists the results for the seven configurations for each structure.The best results for each structure are in bold.For the brainstem and cranial nerves, their segmentation accuracy is best when they are segmented on both 3DFIESTA and 3D-TOF-MRA without segmenting the vessels.For the vessels, the best segmentation accuracy is achieved with the joint segmentation of all structures on 3D-TOF-MRA.However, as long as 3D-TOF-MRA is included, the segmentation accuracy of the vessel is close,whether 3D-FIESTA is included or not and whether it is segmented together with the cranial nerves.Therefore, in practice, it is recommended that 1) we perform separate segmentation of the cranial nerves and vessels, where the cranial nerves are segmented together with the brainstem using 3D-FIESTA and 3D-TOF-MRA,and 2) the vessels should be segmented with at least with 3D-TOF-MRA, and it can be segmented together with the cranial nerves and the brainstem or the brainstem alone.

Fig.5 Visualization of results of joint segmentation of the brainstem, trigeminal nerve, and facial nerve, shown together with 3DFIESTA and 3D-TOF-MRA.Two different slices are shown: (a) and (f) 3D-FIESTA; (b) and (g) 3D-TOF-MRA; (c) and (h)manual annotations of the brainstem, facial nerve, and trigeminal nerve; (d) and (i) the segmentation achieved with both 3DFIESTA and 3D-TOF-MRA; (e) and (j) the segmentation achieved with 3D-FIESTA only (Blue represents the brainstem,green represents the trigeminal nerve, and orange represents the facial nerve)
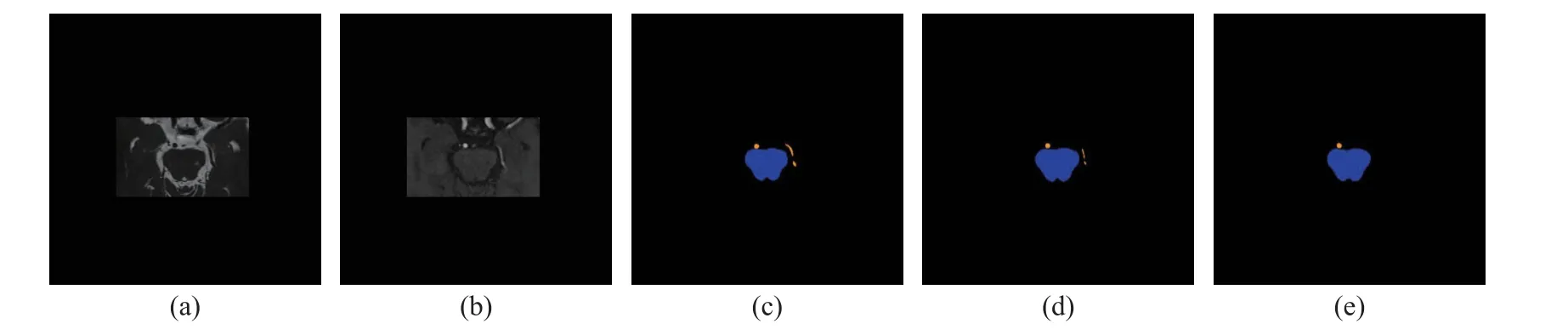
Fig.6 Visualization of segmentation results of joint segmentation of the brainstem and vessel, shown together with 3D-FIESTA and 3D-TOF-MRA: (a) 3D-FIESTA; (b) 3D-TOF-MRA; (c) manual annotations of the brainstem and vessel; (d) the segmentation achieved with both 3D-FIESTA and 3D-TOF-MRA; (e) the segmentation achieved with 3D-TOF-MRA only (Blue represents the brainstem, and orange represents the vessel)
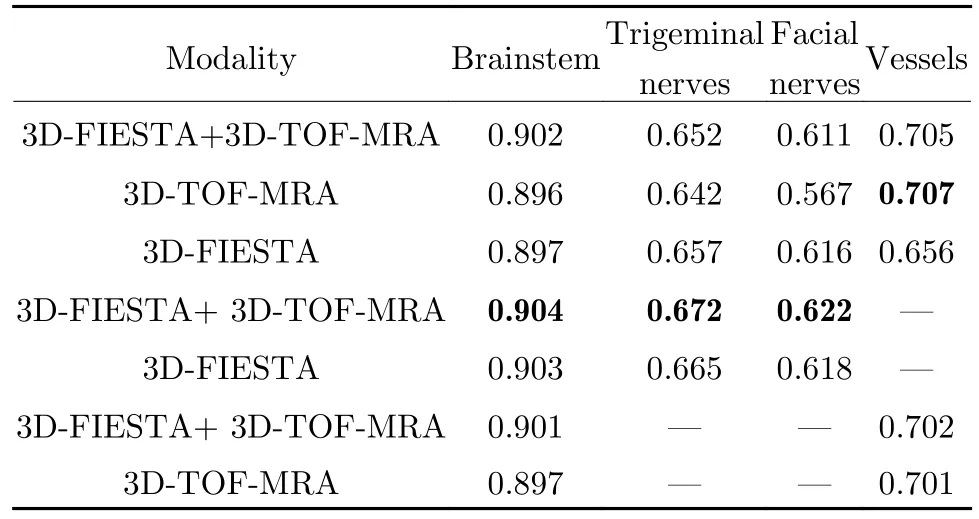
Tab.2 The averge Dice coefficients of the segmentation results(The best results are highlighted in bold)
4 Conclusion
We have investigated the configurations for segmenting the cranial nerves and vessels for TN and HFS.We have observed that for the cranial nerves both 3D-FIESTA and 3D-TOF-MRA should be used for segmentation, whereas for the vessels at least 3D-TOF-MRA should be included.Also, for the cranial nerves, it is best segmented separately instead of jointly with the vessels, whereas for the vessels there is little difference between the joint and separately segmentation.We expect that our empirical observation may provide guidance for future TN and HFS studies.
杂志排行
Journal of Beijing Institute of Technology的其它文章
- Topology and Semantic Information Fusion Classification Network Based on Hyperspectral Images of Chinese Herbs
- Structural Connectivity Enhanced Anisotropic 3D Network for Brain Midline Delineation
- Spatial-Spectral Joint Network for Cholangiocarcinoma Microscopic Hyperspectral Image Classification
- Tissue Microstructure Estimation of SANDI Based on Deep Network
- Application of Opening and Closing Morphology in Deep Learning-Based Brain Image Registration
- Nitrogen Content Inversion of Corn Leaf Data Based on Deep Neural Network Model
