BAM复合体及其靶向性抗革兰阴性菌药物研究进展
2023-08-25朱小红郑怡凡张晶李妍司书毅
朱小红?郑怡凡? 张晶?李妍?司书毅
摘要: 革兰阴性菌耐药问题是目前全球面对的最严重的公共卫生问题之一,耐药菌的不断出现与传播使得细菌感染的预防与治疗面临严峻挑战。外膜蛋白(outer membrane proteins, OMPs)是革兰阴性菌特有的外膜结构的重要蛋白,对于外膜的形成和稳定起到关键的作用,而OMPs在外膜上的正确折叠和组装是其功能正常发挥的基础。β-桶状蛋白折叠组装辅助因子(β-barrel assembly machine, BAM)即BAM复合体在外膜蛋白的折叠装配中起到至关重要的作用。本文对BAM复合体的结构与功能,以及靶向BAM复合体的抗革兰阴性菌先导药物研究进展等方面进行综合论述,以期为新型抗革兰阴性菌药物的发现提供参考。
关键词: 革兰阴性菌;耐药性;OMPs;BAM复合体
中图分类号:R978.1 文献标志码:A
Research progress of BAM complex and its targeted anti-Gram-negative
bacteria drugs
Zhu Xiao-hong, Zheng Yi-fan, Zhang Jing, Li Yan, and Si Shu-yi
(Beijing Key Laboratory of Antimicrobial Agents, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences
and Peking Union Medical College, Beijing 100050)
Abstract The problem of drug resistance of Gram-negative bacteria is one of the most serious public health problems facing the world. The continuous emergence and spread of drug-resistant bacteria has brought serious challenges to the prevention and treatment of bacterial infections. Outer membrane proteins (OMPs) are important components unique to the outer membrane of Gram-negative bacteria, which play a key role in the formation and stability of the outer membrane. The correct folding and assembly of OMPs on the outer membrane is the basis for their normal function. The β-barrel assembly machine (BAM complex) plays a crucial role in the folding and assembly of outer membrane proteins. This review comprehensively discusses the structure and function of BAM complexes, as well as the research progress of anti-Gram-negative bacteria leading drugs targeting BAM complexes, in order to provide references for the discovery of new anti-Gram-negative bacteria drugs.
Key words Gram-negative bacteria; Drug resistance; OMPs; BAM complex
多重耐藥(multidrug resistant, MDR)细菌是当前对公众健康最大的威胁之一,在过去的20年中,由这些病原体引起的感染变得日趋严重,是高发病率和死亡率以及长期住院的重要因素之一[1-3]。其中,革兰阴性菌耐药情况尤为严重,美国疾病预防控制中心(CDC)发布的报告中威胁公共卫生的14种病原体其中有9种是革兰阴性菌[2-3]。药物作用靶标发生改变而引起的耐药是主要耐药机制之一,因此针对新靶标,发现新型抗生素对抗菌药物的研发具有重要的意义。
革兰阴性菌外膜(outer membrane,OM)是革兰阴性菌特有的结构,它包围在菌体的最外层,由外层脂多糖和内层非对称脂质双分子层及外膜蛋白组成[4]。OM具有选择渗透性,保护菌体免受外部环境的有害影响,也是造成革兰阴性菌固有耐药的关键因素[5-8]。外膜蛋白(outer membrane proteins, OMPs)是存在于外膜上的重要蛋白,对于外膜的组装、稳定以及选择性渗透功能起到非常关键的作用[5,8-9]。OMPs在外膜中多数以β-折叠的形式存在,这种折叠结构是其稳定存在于外膜并正常发挥其功能的重要结构基础。BAM复合体在OMPs折叠、插入外膜及其在外膜的定位组装中起着至关重要的作用。靶向破坏BAM复合体可能会造成OMPs的异常,进而破坏外膜的形成,未折叠外膜蛋白在周质空间中的堆积也对菌体的生长产生毒性,导致菌体死亡。此外,营养物质获取和宿主细胞粘附等基本细胞过程使OMPs对感染的发病机制尤为重要,靶向破坏BAM的功能也将下调毒力因子的产生,从而抑制病原体对抗宿主免疫反应的能力。因此,以BAM复合体为靶标的抗革兰阴性菌药物的研究受到越来越多的关注。本文将对BAM复合体的结构与功能和作为抗菌药物靶标的潜在可能性以及目前以BAM复合体为靶标的抗革兰阴性菌药物研究进展等方面进行综合论述。
1 BAM复合体
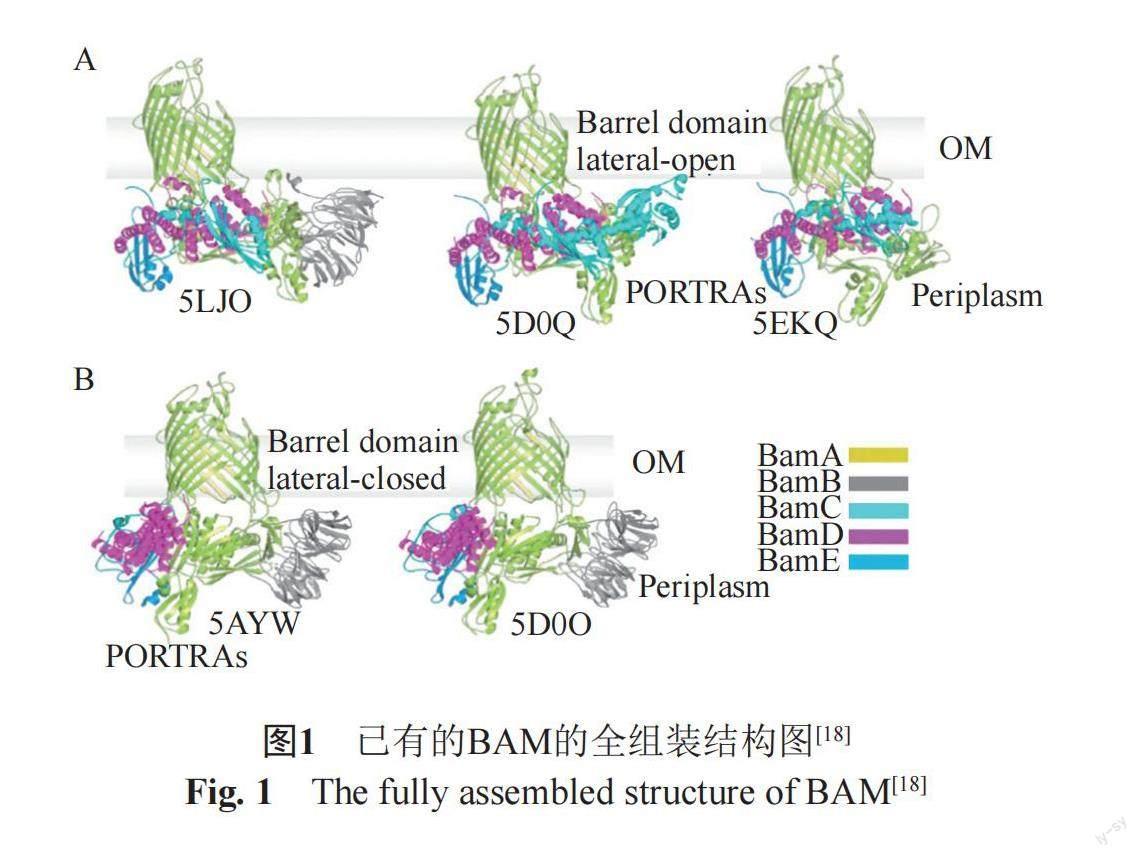
BAM复合体由多个组分组成,其构成因物种不同而有所差异,但是绝大多数革兰阴性菌中的BAM复合物包含BamA、B、C、D和E 5个蛋白(图1)[10-17]。BamA是BAM复合体的核心成分,是外膜蛋白在外膜上折叠和装配的主要场所。BamA在革兰阴性菌中高度保守,是菌体生存的必需蛋白[10-15]。除了BamA之外,多数革兰阴性菌中的BAM复合物还包括4种脂蛋白BamB、BamC、BamD和BamE,它们通过直接或间接与BamA相互作用锚定在周质空间中[18-21]。在这4个蛋白中,BamD在绝大多数革兰阴性菌中是必需且保守的,BamB和BamE主要存在于α-,β-和γ-变形杆菌中,而BamC仅存在于β-和γ-变形杆菌中[10-15]。在α-变形杆菌中发现了一个新的脂蛋白亚基BamF[12]。
BamA本身也是一个由16条β-折叠链组成的外膜蛋白,是Omp85超家族的成员。它的C-末端为整合到外膜的16链β-桶结构域,其中β1和β16接缝的横向分离是BAM功能所需的,未折叠的底物蛋白可沿BamA蛋白β-桶的管腔横向穿过而整合到OM中[22]。BamA的N-末端伸向周质空间,由多肽转运相关(polypeptide transport-Ass[11,22-24], POTRA)重复序列组成[23-24]。来自不同細菌物种的BamA蛋白的POTRA结构域数目不同,但是包括大肠埃希菌在内的大多数革兰阴性菌的BamA蛋白具有5个POTRA结构域,从N-末端到C-末端方向依次被命名为POTRA1-POTRA5[23-25]。POTRA结构有高/低柔韧性的不同区域,该结构被认为是BAM复合体辅助蛋白的支架[22,25-28],募集结合其他成分共同组成BAM复合体。
BamD包含5个四肽重复(tetratricopeptide repeat, TPR)结构域,其直接与外膜上的BamA的POTRA5结合[18,29-33]。BamD的N-末端结构域接收从周质空间伴侣分子转运的未折叠外膜蛋白(uOMPs),与uOMPs羧基端的β信号肽结合,以促进其递送至BamA的β-桶结构域后组装/整合到OM中[34-35]。在细胞质中,BamD也能够与未折叠的BamA蛋白C-末端的β信号肽结合,促进BamA蛋白定位到外膜进行折叠组装[36]。BamD的C-末端结构域对其与BamA,BamC和BamE蛋白的相互作用至关重要[26,37-40]。
单独的BamB、C和E对于细胞生存是非必需的,但它们的整体缺失会严重损害细胞生长和OMP的生成[41-42]。BamB具有八叶β-螺旋折叠,它与BamA的POTRA结构域主要沿着POTRA2和POTRA3之间的铰链区相互作用[43-44]。有研究认为BamB是一种支架蛋白,可以最佳定向BamA的柔性周质结构域,使其与其他BAM组分和/或底物相互作用,并有助于将底物从分子伴侣中转移至BAM[39,45-48]。BamA-BamB相互作用复合物对于在外膜形成有组织的组装区非常重要,但是具体机制有待深入研究[39,45-48]。
BamC的N-末端带有一个高度保守的区域,存在于周质中,其C-末端结构域暴露在菌体的表面,因此BamC的拓扑结构仍然是一个有争议的话题[43-44,49]。BamC的N-末端柔性结构域只有在与BamD结合时才能观察到,该结构直接与BamD相互作用,但尚未报道存在稳定相互作用,因此BamC以及BamC与BamD相互作用在BAM中的作用机制尚不明确[32-33,50-51]。BamE具有ααβββ折叠结构,与BamD直接相互作用,并通过桥接其他相互作用来实现增强BamD与BamA的结合[52-55]。BamB-E与BamA均沿POTRA域和桶状结构域的基部相互作用,其中BamCDE复合体主要与POTRA5和部分POTRA4相互作用[53-57]。与BamC相似,尽管BamE不是生存必需蛋白,但BAM中某些底物或其他未知功能可能需要BamE参与,缺失BamE会阻碍OMP/RcsF复合物的装配,然而BamE的作用机制仍然有待确证[42,52,54,56-59]。
2 BAM介导的OMP蛋白折叠机制
虽然已经明确BAM复合体在OMPs的折叠过程中起到重要的作用,但是BAM复合体在这个过程中扮演的角色目前并不是特别的清晰,其介导的OMP生物折叠组装机制目前主要有两种理论[60-61]。一种是BamA辅助机制,该机制认为BamA作用下会造成其周边外膜的不稳定性,BAM复合体在接收了分子伴侣稳定的OMP底物后,催化其插入外膜中[62-64]。一些OMPs体外折叠研究支持这一观点,这些研究发现单一的BamA能够有效地促进OMPs在外膜上的折叠,而且这种折叠催化作用在变薄或者不稳定的膜层中更加有效[25]。在此过程中,关于BamA的状态存在两种矛盾的观点,一种观点认为处于稳定状态的BamA对于OMPs插入外膜是必须的,但是另外一个观点认为BamA在这个过程中需要构象的改变,尤其是它的侧缝区域。一种更为激进的BamA辅助机制认为OMPs在插入外膜之前已经在周质空间中部分或者完全完成折叠,这种折叠需要分子伴侣和(或)BAM复合体的介导或协助[25]。
另外一种可以称作BamA萌芽机制,在这种机制中,分子伴侣稳定的未折叠OMP的β信号区域识别BAM复合体,BamA折叠桶状结构的第一条链的裸露边缘作为新OMP折叠的模板链与OMP结合形成BamA:OMP杂交体,然后每条添加的链以此为核心形成下一条链。当新产生的OMP的β信号区域从BamA桶的第一链解离,并且以更高的亲和力和特异性结合到它自己的第一条链时,OMP的折叠终止。为了防止在外膜上形成超核,新OMP将经历一个“萌芽”或气泡样冒出而从BamA的核心桶状结构域中脱离。新OMP的两亲链的极性残基朝向内,疏水残基朝向桶外,介导与膜的相互作用。这种理论来源于多项研究支持,研究发现BamA蛋白桶状结构的侧向会发生向外张开和向内张开的构象转换。通过交联阻止侧向接缝开口会阻碍BAM的功能,也表明BamA的作用应该不仅仅是局部破坏膜的稳定[56,65]。此外,所有OMP的β-桶状结构域均为反平行链的线性排列形式,这表明折叠过程是系统的而不是随机的[63,66-70]。
3 以BAM复合物为靶标的抗革兰阴性菌药物研究进展
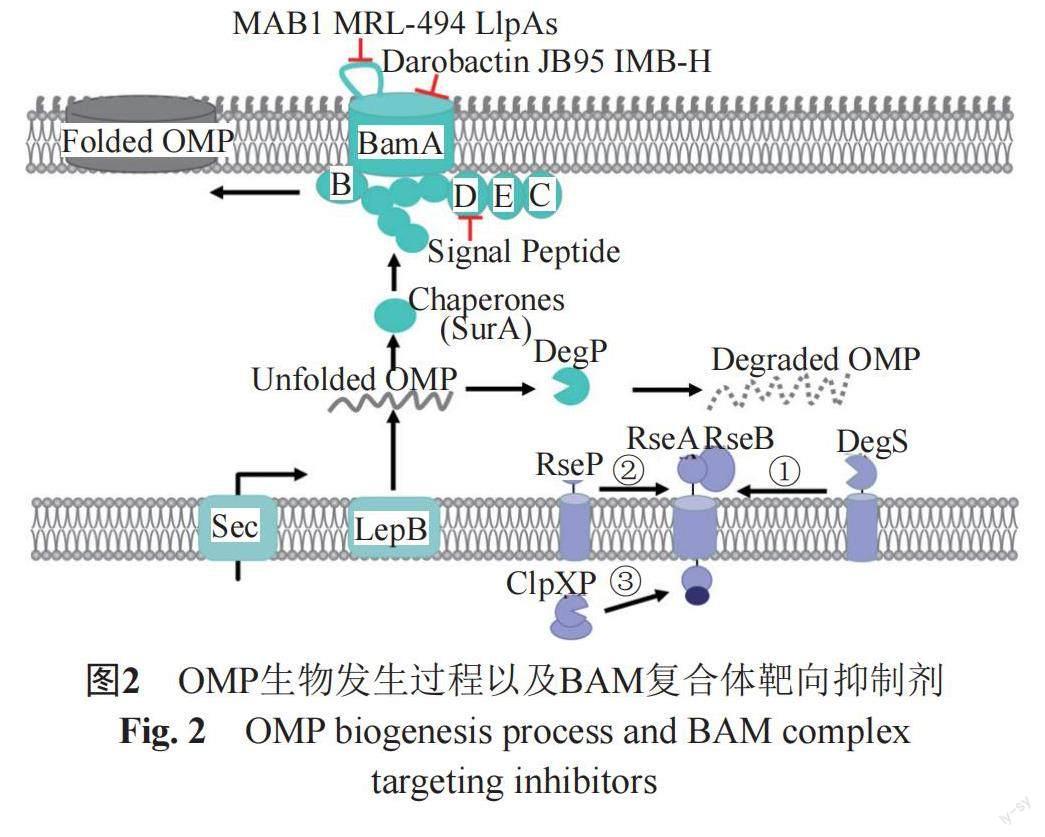
随着BAM功能和结构的解析,其作为抗革兰阴性菌药物靶标受到越来越多的关注,目前已发现多个化合物通过作用于BAM复合体发挥抗菌活性[66,69,71]。BamA和BamD是BAM复合体中最重要的核心组成部分,存在于所有的革兰阴性菌中,并具有高度的保守性,当其功能受到破坏时细菌无法存活[55,72],其他Bam蛋白的单独缺失对菌丝生长没有致死性,因此以BAM为靶标的抗菌药物研究目前集中在BamA和BamD这两个组分(图2~3)[73]。
3.1 以BamA为靶标的抗革兰阴性菌药物
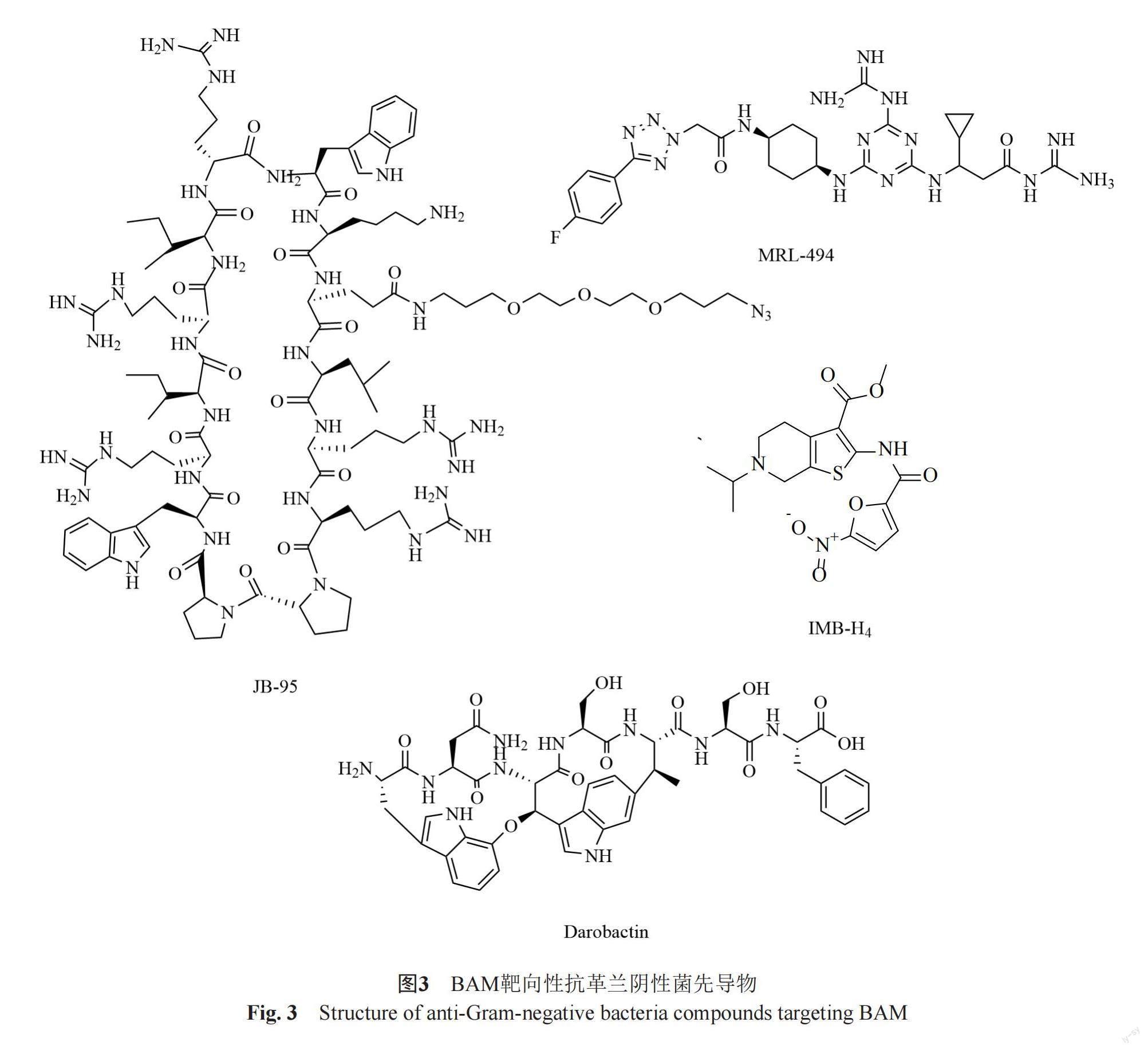
BamA是BAM复合体的核心成分,并且它的β-折叠结构域延伸到外膜外侧,因此靶向BamA的化合物有可能不需要跨膜发挥其抗菌作用,这克服了外膜带来的固有耐药性,因此BamA作为抗菌药物筛选靶标受到更多的关注。随着BamA结构和功能的深入解析,已发现了多个针对BamA的具有抗菌活性的化合物。Urfer等[69]研究发现,新型β-hairpin大环肽JB-95有较强的抗菌活性,尤其对大肠埃希菌的MIC为0.25 μg/mL。它能够选择性破坏大肠埃希菌OM,上调OM应激反应基因ClpX、DegP、PhoQ和RstB的表达。应用JB-95的光反应性衍生物检测发现其能够与BamA和LptD(LPS转运蛋白D)结合,同时下调外膜上OMPs的含量,这些结果提示其可能通过靶向BAM复合体发挥抗菌活性。Storek等[71]在BamA抗体的研究中筛选鉴定出单克隆抗体(monoclonal antibodies, mAbs)MAB1,该抗体在体外对大肠埃希菌表现出杀菌活性。MAB1能够识别结合BamA细胞外的loop4表位,并且能够抑制BamA的外膜蛋白折叠功能,由于MAB1的抗菌活性具有抗原表位的局限性,这限制了它在临床治疗的潜在应用性,但是该抗体的研究为新型的抗体类抗生素研发提供了很好的思路。Ghequire等[74]在一项对凝集素样细菌素 (lectin-like bacteriocins, LlpAs)的研究中发现,在铜绿假单胞菌中,BamA表面暴露的Loop6结构是细菌素靶标识别的关键,LlpAs可以靶向BamA引起BAM机制损伤,从而发挥其杀菌作用。Hart等[75]报道了BamA的一种小分子抑制剂MRL-494,该化合物作用下外膜上的多個OMPs含量下降,菌体通透性改变,而BamA的突变株对该化合物产生耐药。该化合物并未进入到菌体内,因此推测该化合物可能是通过直接结合BamA的β-berral结构域从而抑制OMPs的合成,或者通过影响脂多糖层的稳定性进而影响BAM复合体的功能。MRL-494作用于外膜发挥抗菌活性,避免了外膜的屏障作用和外排泵的影响,因此是一个极具潜力的抗革兰阴性菌先导化合物。Imai等[76]从Photorhabdus分离株中发现了新抗生素darobactin,它对多种临床上重要的革兰阴性致病菌具有良好的抑制活性,对重要的大肠埃希菌和肺炎克雷伯菌的临床耐药株也具有良好的杀伤活性,MIC在2~16 μg/mL。但是该化合物对革兰阳性细菌和哺乳动物细胞在128 μg/mL未见生长抑制活性。机制研究发现darobactin作用下菌体的外膜的完整性受到破坏,进一步深入研究发现这种外膜的损伤与BamA相关。Darobactin与BamA结合,可稳定BamA的侧向封闭构象阻止其打开,使得OMPs底物不能在外膜中正确折叠组装,进而抑制了外膜的形成。该化合物在动物模型上对大肠埃希菌、铜绿假单胞菌以及肺炎克雷伯菌感染均具有良好的抑菌活性,因此,Darobactin是目前非常具有抗革兰阴性菌药物开发前景的BamA靶向性先导化合物。
BamA本身也是外膜蛋白,它在菌体内合成后,跨膜转运到外膜进行β-折叠和组装,它的高效折叠和组装也需要BAM复合体的协助。未折叠的BamA蛋白其N-末端能够被BamD蛋白识别并与其结合,传递到BAM复合体进行高效折叠。Li等[77]成功构建了未折叠状态的BamA和BamD蛋白相互作用酵母双杂交模型,并且通过高通量筛选获得化合物IMB-H4。选该化合物能够与未折叠的BamA结合阻断其与BamD之间的相互作用,抑制OMPs的组装,破坏外膜完整性。IMB-H4在体外对大肠埃希菌具有较好的生长抑制活性,对铜绿假单胞菌和肺炎克雷伯菌等革兰阴性菌也具有一定的抑制作用。该工作也是首次针对BAM复合体进行的靶向性药物高通量筛选的尝试。
3.2 以BamD为靶标的抗革兰阴性菌药物
以另外一个保守的BAM复合体组分BamD蛋白为靶标的抗菌药物研究也多有报道。Mori等[78]在一项研究中基于铜绿假单胞菌的BamD上与BamA结合区域的保守氨基酸序列合成了多肽FIRL,该多肽序列在大肠埃希菌等革兰阴性菌上也具有一定的保守性。FIRL单独使用时抗菌活性较差,但是其对OM具有较强的亲和力,能够增加OM的通透性。体内外研究表明FIRL能够增强多个抗生素对铜绿假单胞菌的药敏活性,且在动物体内与左氟沙星(LVX)和美罗培南(MEM)联用,能够增强它们的抗菌活性。Hagan等[79-80]研究发现,来自未折叠BamA的C-末端β-barrel折叠区域的肽段能够在菌体内外抑制BamA与BamD的结合,并且抑制BamA本身的折叠和外膜蛋白的折叠,过表达该肽段的大肠埃希菌生长变慢,并且能够增加其他透膜困难的抗生素的抗菌活性。
4 展望
鉴于BAM复合体在外膜蛋白折叠装配中的重要作用,BAM复合体具备作为新型抗革兰阴性菌药物靶标的潜能。尽管迄今为止发现靶向BAM复合体化合物很少,且尚未有应用于临床,但这些探索为发现新型抗革兰阴性菌药物提供了新思路与新方向。BAM复合体尤其是BamA本身结构复杂,BAM复合体折叠机制尚未阐明,因此很难实现以BAM复合体尤其是外膜上折叠的BamA为靶标的抗革兰阴性菌先导化合物的高通量筛选。目前发现的多个靶向抑制剂中,除了IMB-H4是来自于靶向未折叠BamA蛋白的高通量筛选,其他抑制剂多来源于抗菌机制的研究。因此,深入解析BAM复合体的结构和功能,进而建立筛选模型有针对性地进行靶向BAM复合体功能的先导化合物的研究,将会更好地促进新型抗革兰阴性菌药物的研发。
參 考 文 献
van Duin D, Paterson D L. Multidrug-resistant bacteria in the community: Trends and lessons learned[J]. Infect Dis Clin North Am, 2016, 30(2): 377-390.
Solomon S L, Oliver K B. Antibiotic resistance threats in the United States: stepping back from the brink[J]. Am Fam Physician, 2014, 89(12): 938-941.
Frieden T. Antibiotic resistance threats.[EB/OL].(2013-4-23)[2020-04-15]. https://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf.
Ruiz N, Kahne D, Silhavy T J. Advances in understanding bacterial outer-membrane biogenesis[J]. Nature Rev Microbiol, 2006, 4(1): 57-66.
Tokuda H. Biogenesis of outer membranes in Gram-negative bacteria[J]. Biosci Biotechnol Biochem, 2009, 73(3): 465-473.
Delcour A H.Outer membrane permeability and antibiotic resistance[J]. Biochim Biophys Acta, 2009, 1794(5): 808-816.
Bos M P, Robert V, Tommassen J. Biogenesis of the Gram-negative bacterial outer membrane[J]. Annu Rev Microbiol, 2007, 61: 191-214.
Nikaido H. Molecular basis of bacterial outer membrane permeability revisited[J]. Microbiol Mol Biol Rev, 2003, 67(4): 593-656.
Henderson I R, Nataro J P. Virulence functions of autotransporter proteins[J]. Infect Immun, 2001, 69(3): 1231-1243.
Noinaj N, Kuszak A J, Gumbart J C, et al. Structural insight into the biogenesis of beta-barrel membrane proteins[J].Nature, 2013, 501(7467): 385-390.
Paramasivam N, Habeck M, Linke D. Is the C-terminal insertional signal in Gram-negative bacterial outer membrane proteins species-specific or not?[J]. BMC Genomics, 2012, 13(1): 510.
Anwari K, Webb C T, Poggio S, et al. The evolution of new lipoprotein subunits of the bacterial outer membrane BAM complex[J]. Mol Microbiol, 2012, 84(5): 832-844.
Webb C T, Heinz E, Lithgow T. Evolution of the beta-barrel assembly machinery[J]. Trends Microbiol, 2012, 20(12): 612-620.
Anwari K, Poggio S, Perry A, et al. A modular BAM complex in the outer membrane of the alpha-proteobacterium Caulobacter crescentus[J]. PLoS One, 2010, 5(1): e8619.
Volokhina E B, Beckers F, Tommassen J, et al. The beta-barrel outer membrane protein assembly complex of Neisseria meningitidis[J]. J Bacteriol, 2009, 191(22): 7074-7085.
Konovalova A, Kahne D E, Silhavy T J. Outer membrane biogenesis[J]. Ann Rev Microbiol, 2017, 71(1): 539-556.
Wu R, Stephenson R, Gichaba A, et al.The big BAM theory: An open and closed case?[J]. Biochim Biophys Acta Biomembr, 2020, 1862(1): 183062.
Wu T, Malinverni J, Ruiz N, et al.Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli[J]. Cell, 2005, 121(2): 235-245.
Gentle I E, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins[J]. Mol Microbiol, 2005, 58(5): 1216-1225.
Voulhoux R, Tommassen J. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly[J]. Res Microbiol, 2004, 155(3): 129-135.
Genevrois S, Steeghs L, Roholl P, et al. The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane[J]. Embo J, 2003, 22(8): 1780-1789.
Gatzeva-Topalova P Z, Walton T A, Sousa M C. Crystal structure of YaeT: Conformational flexibility and substrate recognition[J]. Structure, 2008, 16(12): 1873-1881.
Koenig P, Mirus O, Haarmann R, et al. Conserved properties of polypeptide transport-associated (POTRA) domains derived from cyanobacterial Omp85[J]. J Biol Chem, 2010, 285(23): 18016-18024.
Arnold T, Zeth K, Linke D. Omp85 from the thermophilic cyanobacterium Thermosynechococcus elongatus differs from proteobacterial Omp85 in structure and domain composition[J]. J Biol Chem, 2010, 285(23): 18003-18015.
Plummer A M, Fleming K G. BamA alone accelerates outer membrane protein folding in vitro through a catalytic mechanism[J]. Biochemistry, 2015, 54(39): 6009-6011.
Gatzeva-Topalova P Z, Warner L R, Pardi A, et al. Structure and flexibility of the complete periplasmic domain of BamA: The protein insertion machine of the outer membrane[J]. Structure, 2010, 18(11): 1492-1501.
Knowles T J, Jeeves M, Bobat S, et al. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes[J]. Mol Microbiol, 2008, 68(5): 1216-1227.
Kim S, Malinverni JC, Sliz P, et al. Structure and function of an essential component of the outer membrane protein assembly machine[J]. Science, 2007, 317(5840): 961-964.
Sikora A E, Wierzbicki I H, Zielke R A, et al. Structural and functional insights into the role of BamD and BamE within the beta-barrel assembly machinery in Neisseria gonorrhoeae[J]. J Biol Chem, 2018, 293(4): 1106-1119.
Dong C, Hou H F, Yang X, et al. Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly[J]. Acta Crystallogr D Biol Crystallogr, 2012, 68(Pt 2): 95-101.
Sandoval C M, Baker S L, Jansen K, et al. Crystal structure of BamD: An essential component of the beta-Barrel assembly machinery of gram-negative bacteria[J]. J Mol Biol, 2011, 409(3): 348-357.
Albrecht R, Zeth K. Structural basis of outer membrane protein biogenesis in bacteria[J]. J Biol Che, 2011, 286(31): 27792-27803.
Kim K H, Aulakh S, Paetzel M. Crystal structure of beta-barrel assembly machinery BamCD protein complex[J].J Biol Chem, 2011, 286(45): 39116-39121.
Lee J, Sutterlin H A, Wzorek J S, et al. Substrate binding to BamD triggers a conformational change in BamA to control membrane insertion[J]. Proc Natl Acad Sci U S A, 2018, 115(10): 2359-2364.
Lee J, Xue M, Wzorek J S, et al. Characterization of a stalled complex on the beta-barrel assembly machine[J]. Proc Natl Acad Sci U S A, 2016, 113(31): 8717-8722.
Hagan C L, Westwood D B, Kahne D. Bam lipoproteins assemble BamA in vitro[J]. Biochemistry, 2013, 52(35): 6108-6113.
Bergal H T, Hopkins A H, Metzner S I, et al. The structure of a BamA-BamD fusion illuminates the architecture of the beta-barrel assembly machine core[J]. Structure, 2016, 24(2): 243-251.
Fleming P J, Patel D S, Wu E L, et al. BamA POTRA domain interacts with a native lipid membrane surface[J].Bioph J, 2016, 110(12): 2698-2709.
Jansen K B, Baker S L, Sousa M C. Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the beta-barrel assembly machine[J]. J Biol Chem, 2015, 290(4): 2126-2136.
Voulhoux R, Bos M P, Geurtsen J, et al. Role of a highly conserved bacterial protein in outer membrane protein assembly[J]. Science, 2003, 299(5604): 262-265.
Tellez R J, Misra R. Substitutions in the BamA β-barrel domain overcome the conditional lethal phenotype of a ΔbamB ΔbamE strain of Escherichia coli[J]. J Bacteriol, 2012, 194(2): 317-324.
Sklar J G, Wu T, Gronenberg L S, et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli[J]. Proc Natl Acad Sci U S A, 2007, 104(15): 6400-6405.
Gunasinghe S D, Shiota T, Stubenrauch C J, et al. The WD40 Protein BamB mediates coupling of BAM complexes into assembly precincts in the bacterial outer membrane[J].Cell Reports, 2018, 23(9): 2782-2794.
Rassam P, Copeland N A, Birkholz O, et al. Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria[J]. Nature, 2015, 523(7560): 333-336.
Jansen K B, Baker S L, Sousa M C. Crystal structure of BamB from Pseudomonas aeruginosa and functional evaluation of its conserved structural features[J]. PLoS One, 2012, 7(11): e49749.
Noinaj N, Fairman J W, Buchanan S K. The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex[J]. J Mol Biol, 2011, 407(2): 248-260.
Vuong P, Bennion D, Mantei J, et al. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry[J]. J Bacteriol, 2008, 190(5): 1507-1517.
Ureta A R, Endres R G, Wingreen N S, et al. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment[J].J Bacteriol, 2007, 189(2): 446-454.
Webb C T, Selkrig J, Perry A J, et al. Dynamic association of BAM complex modules includes surface exposure of the lipoprotein BamC[J]. J Mol Biol, 2012, 422(4): 545-555.
Warner L R, Varga K, Lange O F, et al. Structure of the BamC two-domain protein obtained by Rosetta with a limited NMR data set[J]. J Mol Biol, 2011, 411(1): 83-95.
Kim K H, Aulakh S, Tan W, et al. Crystallographic analysis of the C-terminal domain of the Escherichia coli lipoprotein BamC[J]. Acta Crystallogr Sect F Struct Biol Cryst Commun, 2011, 67(Pt 11): 1350-1358.
Han L, Zheng J, Wang Y, et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins[J]. Nat Struct Mol Biol, 2016, 23(3): 192-196.
Gu Y, Li H, Dong H, et al.Structural basis of outer membrane protein insertion by the BAM complex[J]. Nature, 2016, 531(7592): 64-69.
Bakelar J, Buchanan S K, Noinaj N. The structure of the beta-barrel assembly machinery complex[J]. Science, 2016, 351(6269): 180-186.
Malinverni J C, Werner J, Kim S, et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli[J]. Mol Microbiol, 2006, 61(1): 151-164.
Iadanza M G, Higgins A J, Schiffrin B, et al. Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM[J]. Nature Commun, 2016, 7: 12865.
Hart E M, Gupta M, Wuhr M, et al. The synthetic phenotype of Δ bamB Δ bamE double mutants results from a lethal jamming of the bam complex by the lipoprotein RcsF[J]. MBio, 2019, 10(3): e00662-19.
Tata M, Konovalova A. Improper coordination of BamA and BamD results in Bam complex jamming by a lipoprotein substrate[J]. MBio, 2019, 10(3): e00660-19.
Gu Y, Li H, Dong H, et al. Structural basis of outer membrane protein insertion by the BAM complex[J]. Nature, 2016, 531(7592): 64-69.
Wu R, Stephenson R, Gichaba A, et al. The big BAM theory: An open and closed case?[J]. Biochim Biophys Acta Biomembr, 2020, 1862(1): 183062.
Noinaj N, Gumbart J C, Buchanan S K. The beta-barrel assembly machinery in motion[J]. Nature Rev Microbiol, 2017, 15(4): 197-204.
Danoff E J, Fleming K G. Novel kinetic intermediates populated along the folding pathway of the transmembrane beta-Barrel OmpA[J]. Biochemistry, 2017, 56(1): 47-60.
Gessmann D, Chung Y H, Danoff E J, et al. Outer membrane beta-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA[J]. Proc Natl Acad Sci U S A, 2014, 111(16): 5878-5883.
Burgess N K, Dao T P, Stanley A M, et al. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro[J]. J Biol Chem, 2008, 283(39): 26748-26758.
Noinaj N, Kuszak A J, Balusek C, et al. Lateral opening and exit pore formation are required for BamA function[J].Structure, 2014, 22(7): 1055-1062.
Choi U, Lee C R. Antimicrobial agents that inhibit the outer membrane assembly machines of Gram-negative bacteria[J]. J Microbiol Biotechnol, 2019, 29(1): 1-10.
Doyle M T, Bernstein H D. Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA beta-barrel[J]. Nature Comm, 2019, 10(1): 3358.
Hohr A, Lindau C, Wirth C, et al. Membrane protein insertion through a mitochondrial beta-barrel gate[J]. Science, 2018, 359(6373): eaah6834.
Urfer M, Bogdanovic J, Lo Monte F, et al. A peptidomimetic antibiotic targets outer membrane proteins and disrupts selectively the outer membrane in Escherichia coli[J]. J Biol Chem, 2016, 291(4): 1921-1932.
Plummer A M, Fleming K G. From chaperones to the membrane with a BAM![J]. Trends Biochem Sci, 2016, 41(10): 872-882.
Storek K M, Auerbach M R, Shi H, et al. Monoclonal antibody targeting the beta-barrel assembly machine of Escherichia coli is bactericidal[J]. Proc Natl Acad Sci U S A, 2018, 115(14): 3692-3697.
Werner J, Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli[J]. Mol Microbiol, 2005, 57(5): 1450-1459.
Lehman K M, Grabowicz M. Countering Gram-negative antibiotic resistance: Recent progress in disrupting the outer membrane with novel therapeutics[J]. Antibiotics (Basel), 2019, 8(4): 163.
Ghequire M, Swings T, Michiels J, et al. Hitting with a BAM: Selective killing by lectin-like bacteriocins[J]. MBio, 2018, 9(2): e02138-17.
Hart E M, Mitchell A M, Konovalova A, et al. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier[J]. Proc Natl Acad Sci U S A, 2019, 116(43): 21748-21757.
Imai Y, Meyer K J, Iinishi A, et al. A new antibiotic selectively kills Gram-negative pathogens[J]. Nature, 2019, 576(7787): 459-464.
Li Y, Zhu X, Zhang J, et al. Identification of a compound that inhibits the growth of gram-negative bacteria by blocking BamA-BamD interaction[J]. Front Microbiol, 2020, 11: 1252.
Mori N, Ishii Y, Tateda K, et al. A peptide based on homologous sequences of the beta-barrel assembly machinery component BamD potentiates antibiotic susceptibility of Pseudomonas aeruginosa[J]. J Antimicrob Chemother, 2012, 67(9): 2173-2181.
Hagan C L, Wzorek J S, Kahne D. Inhibition of the beta-barrel assembly machine by a peptide that binds BamD[J].Proc Natl Acad Sci U S A, 2015, 112(7): 2011-2016.
Kutik S, Stojanovski D, Becker L, et al. Dissecting membrane insertion of mitochondrial beta-barrel proteins[J]. Cell, 2008, 132(6): 1011-1024.
