水稻碱性亮氨酸拉链(bZIP)蛋白家族功能研究进展
2023-07-18韩聪何禹畅吴丽娟郏丽丽王磊鄂志国
韩聪 何禹畅 吴丽娟 郏丽丽 王磊 鄂志国
水稻碱性亮氨酸拉链(bZIP)蛋白家族功能研究进展
韩聪 何禹畅 吴丽娟 郏丽丽 王磊 鄂志国*
(中国水稻研究所, 杭州 310006;*通信联系人,email: ezhiguo@caas.cn)
碱性亮氨酸拉链(basic leucine zipper, bZIP)是一类重要的转录调控因子,广泛存在于真核生物中,因含有高度保守的bZIP结构域而得名。bZIP结构域由紧密相邻的碱性区域和亮氨酸拉链区域两部分组成。粳稻基因组中注释有89个bZIP基因,其中45个已得到功能验证,它们参与调节水稻生长发育、生物与非生物胁迫应答,包括种子休眠和萌发、成花转变、光形态建成,以及胁迫和激素信号通路等。
水稻;碱性亮氨酸拉链蛋白;bZIP转录因子;基因功能
种子的休眠、萌发,植株的生长发育及生物与非生物胁迫应答等生物学过程中,不同基因通常有着不同的时空表达特性和胁迫诱导反应,它们的表达受转录因子精准调控[1]。碱性亮氨酸拉链(basic leucine zipper, bZIP)蛋白是一类重要的转录因子,在植物中广泛分布,参与调控植物各种生物学过程。
水稻是我国最重要的粮食作物之一,种植面积约占粮食作物面积的25.4%,产量接近粮食总产量的31.2%[2]。然而,水稻在种植过程中极易遭受恶劣环境影响和病虫害袭击,造成减产和品质下降,严重威胁粮食安全。因此,水稻的生长发育和胁迫抗性机制,一直是农学与生物学研究的热点。本文对水稻bZIP转录因子的研究进展进行综述,旨在为水稻功能基因组学研究提供参考。
1 bZIP蛋白结构
bZIP蛋白因含有一个保守的bZIP结构域而得名。在bZIP蛋白分子结构上(如图1-A所示),bZIP域分为两个紧密相邻的部分,一个是碱性区域(蓝色),高度保守,约由20个氨基酸残基组成,包含一个核定位信号和一个能结合DNA的N-X7-R/K基序;第二个是亮氨酸拉链区(灰色),由7个氨基酸残基组成一个重复单位,亮氨酸或相关疏水氨基酸位于第7位,重复单元的数量则从3个到8个不等[3]。每个bZIP结构域形成一个连续的α螺旋,两个bZIP蛋白的α螺旋因侧链疏水性交错对插,形成拉链结构(图1-B)。bZIP蛋白形成同源或异源二聚体后,才能正常行使其功能。
通过全基因组相似性比对,Nijhawan等[4]和Ji等[5]分别从粳稻品种日本晴基因组中鉴定了89个bZIP位点,Corrêa等[6]报道了92个bZIP位点。这些基因的染色体分布和系统进化树等生物信息学分析,前人文献已有较透彻的阐述,本文不再赘述。本文聚焦于已报道的bZIP基因的功能,如表1所示。

A―bZIP结构域示意图, 由碱性DNA结合区(蓝色)和相邻的亮氨酸拉链区(灰色)组成; B―bZIP二聚体的结构模型。
2 bZIP蛋白调控水稻生长发育
2.1 bZIP蛋白调控水稻成花转变
成花素Hd3a和RFT1与14-3-3蛋白GF14c互作,形成复合物转至核内,与转录因子OsFD1(即OsbZIP77)结合形成三元成花素激活复合物(florigen activation complex, FAC),诱导表达,从而分别在短日照和长日照下启动水稻的成花转变[14, 66, 94-95]。蛋白激酶OsCIPK3能磷酸化修饰OsFD1第192位的丝氨酸位点,促进含RFT1的FAC形成[95]。研究表明,OsFD7(即OsbZIP62)[73]、OsFD4(即OsbZIP69)[83]也能与成花素和14-3-3蛋白形成FAC,促进开花,突变体和RNAi植株均表现为迟抽穗。转录因子Ehd1能诱导成花素基因和表达,形成FAC,而FAC又能进一步增强表达[14]。有意思的是,HBF1(即OsbZIP42)能与Hd3a直接互作,也能通过GF14c间接与RFT1互作,这两种互作形成的复合物在叶中能抑制表达,在顶端分生组织(SAM)中能抑制表达,从而推迟水稻的成花转变[14]。研究还发现,HBF2(即OsbZIP09)在叶中的功能与HBF2冗余,也能抑制表达[12]。此外,一类RCN蛋白(RICE CENTRORADIALIS)能与Hd3a竞争,与14-3-3蛋白和OsFD1互作,形成成花抑制复合物(florigen repression complex, FRC),阻止Hd3a正常行使功能,抑制水稻开花[101-102]。另一类CONSTANS转录因子DHD4,能与14-3-3竞争性结合OsFD1,干扰OsFD1与14-3-3的互作,从而影响FAC的形成,导致和的表达量降低,最终延迟开花[103]。
OsRE1(即OsbZIP01)[7]、OsABF1(即OsbZIP12)[21]、OsbZIP65[76]和OsbZIP71[84]均负调控水稻的成花转变。OsRE1、OsbZIP65和OsbZIP71各自能直接结合到的启动子区并抑制其表达;OsABF1则是通过激活表达,间接抑制表达。过表达、或的转基因植株均表现出晚花表型,而突变体和表现为早花表型。因OsABF1功能与OsbZIP40冗余,单突抽穗期无显著变化,但利用RNAi同时敲减和会导致明显的早花表型。OsRIP1能与OsRE1协作,通过精细调节的表达对抽穗期进行微调。
2.2 bZIP蛋白调控水稻种子休眠与萌发
脱落酸(abscisic acid, ABA)和赤霉素(gibberellin, GA)是调控水稻种子休眠和萌发最重要的两种激素。ABA诱导和维持种子休眠,抑制种子萌发及幼苗生长,而GA作用相反。种子休眠和萌发受这两种内源激素含量和种胚对它们的敏感性共同调控。此外,茉莉酸(jasmonic acid, JA)和褪黑素(melatonin, MT)也参与调控。
ABA信号通路促进种子休眠的分子机制已研究得相对清晰。当ABA缺失时,蛋白磷酸酶OsPP2C30或OsPP2C51与激酶SAPK2在核中互作形成复合体;ABA存在时,ABA受体OsPYL/RCAR5与OsPP2C30、OsPP2C51结合,从而释放出SAPK2,磷酸化的SAPK2与bZIP蛋白如OREB1(即OsbZIP10)互作并将其激活,bZIP蛋白则进一步结合ABA应答元件(ABA-responsive elements, ABREs),诱导ABA应答基因如、和等表达,抑制种子萌发[15-16]。OsPP2C51在蛋白水平上能对OsbZIP10去磷酸化使其失活[15],AP2转录因子OsSAE1在转录水平上直接抑制的表达[17],因此,OsPP2C51和OsSAE1都能促进水稻种子萌发。此外,GF14h能与OsbZIP10互作降低其转录活性,而OsMFT2又能与OREB1-GF14h形成三元复合物,并破除GF14h对OsbZIP10的抑制作用[18]。类似地,TRAB1(OsbZIP66)与ABREs特异结合,并依赖于OsVP1调节ABA诱导的转录,控制水稻胚的成熟与休眠[77]。SAPK10能磷酸化OsbZIP66并将其激活[78]。研究还发现,OsbZIP23、OsbZIP66和OsbZIP72这三者在促进中花11种子休眠中的作用冗余,它们均正调控ABA应答基因表达,抑制水稻种子萌发;OsMFT2能分别与这3个bZIP转录因子互作,增强它们与启动子的结合[37]。过表达能清除敲除株系穗发芽的表型[37];双突材料的种子萌发提早,对ABA敏感性下降[79]。OsbZIP75和OsbZIP78能直接与启动子结合诱导其表达,OsDOG1L-3进一步上调ABA合成和信号传导相关基因表达,通过增强ABA信号通路促进水稻种子休眠[92]。
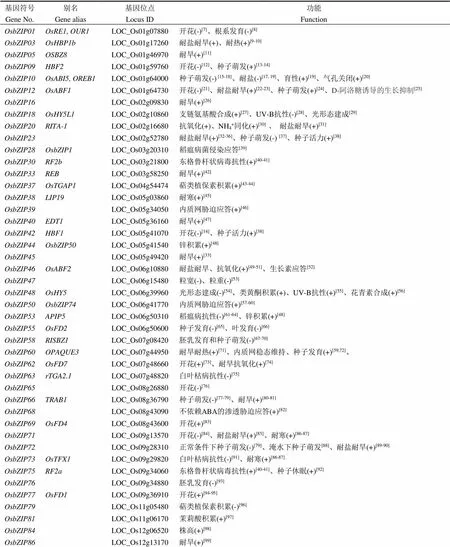
表1 已功能鉴定的水稻bZIP转录因子
(+)和(-)分别代表正调控和负调控。
有研究发现,ABA对种子萌发的抑制效应部分依赖于JA生物合成。SAPK10能磷酸化修饰OsbZIP72并增强其稳定性,磷酸化修饰增强了bZIP72结合JA合成途径关键基因启动子的能力,促进后者转录,从而提高内源JA水平并抑制种子萌发。外施JA合成抑制剂可缓解ABA对种子萌发的抑制也证实了这一点[79]。在过表达的转基因水稻中,茉莉酸水平也上调,这可能是因为OsbZIP81.1直接激活了基因表达所致[97]。
也有bZIP蛋白能抑制ABA积累和信号传导,正调控种子萌发。如OsbZIP09能直接与ABA分解基因启动子结合并增强其表达,降低ABA积累,也能直接与启动子结合并抑制其表达,减弱ABA信号通路,最终促进水稻种子发芽。突变体穗发芽现象较少[13-14]。
与ABA作用相反,GA能破除种子休眠,促进萌发。OsABF1(即OsbZIP12)通过抑制GA合成,负调控水稻种子萌发和节间伸长。OsABF1与多梳家族成员OsEMF2b互作,招募多梳蛋白抑制复合体PRC2到GA20氧化酶基因的启动子区域,实施H3K27me3甲基化,经表观修饰抑制的转录,进而维持水稻生长和种子萌发所需的GA平衡。过表达产生典型的GA缺失表型,半矮化和种子萌发迟缓,外施GA3可以恢复表型[25]。此外,C2C2型锌指蛋白OsLOL1与OsbZIP58互作,并经OsbZIP58激活贝壳杉烯氧化酶基因表达,促进GA生物合成,GA水平的上调促进了胚乳糊粉层细胞程序化死亡(programmed cell death, PCD)和种子萌发[70]。
有意思的是,如上文所述,OsbZIP10、OsbZIP23和OsbZIP72正调控ABA信号抑制种子萌发[15, 37],但在褪黑素、加速老化或淹水等非常规处理过程中,又能提高种子活力,表现出与常规条件下相反的作用。如低温和铬离子毒害会抑制种子萌发,而褪黑素处理能恢复种子活力。研究发现,OsbZIP10能增强褪黑素改善低温和铬离子胁迫下种子萌发的作用,褪黑素处理下,OsbZIP10通过直接上调过氧化氢酶基因和抗坏血酸过氧化物酶基因的表达,增加赤霉素合成,促进H2O2清除,从而提高低温和络离子毒害下的发芽率[105-106]。相对湿度80%、42℃的加速老化处理也能降低种子活力,但在Kasalath种子中,内源ABA含量增加促进表达,OsbZIP23进而激活下游靶标过氧化物还原酶基因表达,PER1A通过清除种子内活性氧正调控种子活力。突变体和种子中的H2O2含量均显著高于野生型,而过表达的Kasalath种子在加速老化处理15或18 d后发芽率显著高于对照[38]。该研究还发现OsbZIP42正调控水稻种子活力,但作用机制有待进一步研究[38]。在淹水条件下,OsbZIP72通过激活乙醇脱氢酶ADH1,随后产生更多参与酒精发酵和糖酵解途径的NAD+、NADH和ATP,提供必要的能量储备,促进水稻种子萌发和胚芽鞘伸长[88]。
2.3 bZIP蛋白调控水稻胚乳发育
粒重是决定水稻产量的重要性状,转录因子OsbZIP47参与调控水稻粒形和粒重。过表达的转基因植株籽粒细长,而突变体籽粒变宽。研究发现CC类谷氧还蛋白WG1能与OsbZIP47直接互作,并招募转录共抑制子ASP1来抑制OsbZIP47的转录活性;而E3泛素连接酶GW2可以泛素化WG1,并调控WG1的蛋白稳定性。因此,GW2-WG1-OsbZIP47构成了一个调控水稻种子发育的通路[53]。
OsCEN2-GF14f-OsFD2(即OsbZIP55)是另一个调控水稻籽粒发育的模块。实验表明OsCEN2能与GF14f发生互作,而GF14f能直接与OsFD2互作,OsFD2则通过控制颖壳中的细胞生长和分裂来影响水稻籽粒大小[65]。OsFD2还调控水稻叶片的发育,但需要形成FAC,Hd3a与14-3-3的互作会促进含有OsFD2的FAC的核运输。过表达会使水稻叶片变小,且先后两个叶原基形成的间隔期缩短[66]。
RISBZ1(即OsbZIP58)在水稻灌浆过程中起重要作用,影响种子中游离赖氨酸含量和储藏蛋白积累[67-68]。研究还发现,OsbZIP58能直接与6个淀粉合成基因、、、、和的启动子结合,调节其表达,从而调节胚乳中淀粉的生物合成。突变体种子形态异常,总淀粉和直链淀粉含量降低,且支链淀粉的短链比例较高、中链比例较低[69]。
OsbZIP76自身无转录激活活性,但它能分别与核因子Y家族转录因子OsNF-YB1和OsNF-YB9互作,调控水稻胚乳发育,共同参与储藏物质积累。突变体的胚乳细胞化进程提前,表现出和突变体类似的籽粒变小和直链淀粉含量降低等表型[93]。
2.4 bZIP蛋白调控水稻其他组织发育
发育良好的根系对于水稻有效吸水至关重要,特别是在干旱环境中。编码OsbZIP01,其突变通过抑制生长素信号促进水稻根系发育。突变体的种子根伸长加快,此外,与野生型相比,短而细的侧根数减少,而长而粗的侧根数增加[8]。此外,ABL1(即OsbZIP46)通过影响含ABRE元件基因实现对ABA和生长素应答的双重调控,突变体对外源吲哚乙酸超敏感,一些与生长素代谢或信号传导相关基因的表达量发生改变,且的表达能恢复拟南芥突变体的ABA 敏感性[52]。
支链氨基酸和5-羟色胺在植物生长发育方面发挥着重要作用。OsbZIP18通过直接与支链氨基酸生物合成基因和的启动子结合,正调控支链氨基酸的合成[27]。OsbZIP18还能直接与、和的启动子结合并激活表达,促进5-羟色胺的生物合成,5-羟色胺积累导致植株矮小、少分蘖、深棕色的表型,对中波紫外线(UV-B)抗性也下降[28]。此外,OsbZIP18(OsHY5L1)还能促进水稻在蓝光下的光形态建成[29]。
OsbZIP48在光调节的发育中发挥多种作用,过表达全长的转基因水稻,细胞分裂素水平升高,绿色期更长,半矮秆,节间长度和穗长缩短,茎秆薄;RNAi株系和T-DNA插入突变体表现为幼苗致死[54]。OsHY5(即OsbZIP48)还能与OsBBX14协同作用,直接激活或表达,精细调控花青素生物合成[56]。OsbZIP48被OsRLCK160磷酸化后,还能促进水稻中类黄酮的积累,参与UV-B抗性[55]。
OsbZIP84参与调控细胞伸长。上调表达导致植株变高,而表达量下调导致植株变矮,且这种矮化是由细胞变短造成。OsbZIP84可能通过调控赤霉素代谢路径中的合成调控胞内赤霉素的含量,并进一步调控水稻的生长发育[98]。
3 bZIP蛋白调控水稻非生物胁迫应答
非生物胁迫是一种广泛存在的环境胁迫,包括干旱、水涝、高温、低温、高盐、重金属和辐射等,严重威胁农作物生产。ABA是植物应答非生物胁迫的主要激素。环境胁迫能促进植物体内快速合成脱落酸,并激活ABA信号通路应答胁迫,从而增强植物的抗性[30]。bZIP转录因子被磷酸化激活后,通过与基因启动子中ABREs结合,参与ABA介导的转录调控。
3.1 bZIP蛋白调控水稻高低温胁迫应答
脱落酸和低温能诱导OsbZIP73表达,说明OsbZIP73 参与依赖ABA的低温信号通路。OsbZIP73Jap与OsbZIP71互作形成异源二聚体,抑制脱落酸生物合成,降低活性氧水平,从而提高水稻苗期对低温的耐受性[86]。生殖生长期,bZIP73Jap:bZIP71不仅能抑制花药中ABA水平,而且可促进可溶性糖从花药转运到花粉,提高了水稻结实率和产量;此外,bZIP73Jap:bZIP71还正调控qLTG3-1的表达,而qLTG3-1过表达株系的生殖期耐寒性大大提高[87]。(即)和(即)的表达分别受低温胁迫诱导和抑制。LIP19 没有bZIP蛋白常见的同源二聚化和结合DNA的能力,但LIP19能与OsOBF1互作形成异源二聚体,且这种结合比OsOBF1自身的互作要强。推测水稻中存在这样的一个感温模型:正常温度如25℃时,OsOBF1大量表达并形成同源二聚体结合到六聚体基序ACGTCA上从而诱导目标基因的表达;随着温度降低,OsOBF1 表达量下降而LIP19 表达量上升,至5℃时,LIP19大量表达而OsOBF1表达很低,LIP19和OsOBF1互作形成异源二聚体并结合到G/C序列上从而诱导耐冷性基因表达[45]。
内质网(endoplasmic reticulum, ER)内腔中未折叠或错折叠蛋白的积累会导致ER胁迫,并引发未折叠蛋白反应(unfolded protein response, UPR),包括减少翻译以缓解新生蛋白质折叠的需求,降解未折叠蛋白以减轻损伤,增加细胞伴侣蛋白表达以协助蛋白质折叠。ER胁迫传感因子IRE1通过介导一些特异响应ER胁迫的关键转录因子mRNA的非典型剪接,诱导它们的激活[57-58]。OsbZIP50(也称)在水稻胚乳发育过程中通过激活分子伴侣基因表达,影响种子贮藏蛋白和淀粉的积累[59]。研究发现,常规剪接下,蛋白不能转入细胞核,但ER胁迫或高温胁迫下,mRNA保守的双茎环结构被剪接,新产生的蛋白丢失了跨膜区,变成具有转录活性的核定位形式,从而将胁迫信号从内质网传递到细胞核,并调控胁迫应答基因表达。OsbZIP39功能和作用方式与OsbZIP50类似[46]。NAC转录因子OsNTL3能从质膜迁移到细胞核,直接与启动子结合并调控其表达[60]。过表达UPR响应基因、、和导致不同程度垩白,说明UPR反应会促进垩白形成。进一步研究发现,OsbZIP60 (OPAQUE3)能通过维持胚乳发育过程中内质网稳态来抑制过度激活,进而保证胚乳正常发育[59]。OsbZIP60在热胁迫和干旱胁迫应答中也具有重要作用,过量表达能增强水稻的抗热和抗旱能力[71]。OPAQUE3在水稻胚乳发育,特别是高温胁迫下胚乳的正常发育中起着核心的调控作用。自然高温环境下,突变体籽粒灌浆速率降低,成熟籽粒中总淀粉、直链淀粉和总蛋白质、谷蛋白含量显著降低,千粒重和单株产量均显著降低[72]。
3.2 bZIP蛋白调控水稻干旱和盐胁迫应答
干旱、盐和脱落酸处理能改变(、、和的表达水平,表明参与依赖ABA的干旱和高盐信号通路。过表达、或的转基因水稻,显著提高了对干旱和盐胁迫的耐受性;而突变体、表现相反[22-23, 31-33, 85]。研究发现OsSAPK10能与OsbZIP20互作并将其磷酸化激活,OsbZIP20进而与启动子的ABRE元件结合并诱导其转录,从而增强水稻的干旱和盐胁迫抗性[31]。OsbZIP23与组蛋白修饰协同调控水稻脱水蛋白基因表达,干旱胁迫下,组蛋白H3上第4位的赖氨酸三甲基化(H3K4me3)修饰水平提高,脱水蛋白基因表达水平增加,而OsbZIP23与脱水蛋白基因启动子的结合能力也提高;相反,突变体中的H3K4me3修饰和脱水蛋白基因表达水平均下调[34]。而SUMO蛋白酶OsOTS1能对OsbZIP23直接去SUMO化,影响其稳定性,从而负调控ABA依赖的干旱应答基因表达[35]。此外,SAPK2能磷酸化OsbZIP23从而将其激活,而OsPP2C49能与SAPK2互作使其失活从而抑制了OsbZIP23的激活,OsbZIP23又通过正调控和的表达,对ABA的信号转导和生物合成进行反馈调节(图2)[36]。过表达导致水稻对ABA敏感性降低,转基因幼苗置于空气中快速脱水,对干旱超敏感;而编码的9-顺式-环氧类胡萝卜素双加氧酶是控制ABA合成的关键酶。与OsbZIP23类似,OsbZIP86能与启动子结合,在干旱条件下激活表达,通过上调ABA合成来提高水稻耐旱性[99]。通过维持叶绿素含量和提高抗氧化能力,正调控水稻对盐和干旱胁迫的耐受性。与野生型相比,过表达的转基因水稻限制了活性氧的积累,表现出更高的存活率和更有利的渗透参数[9-10]。

图2 OsbZIP23参与调控ABA信号通路.
OsABF2(OsbZIP46)也是ABA 信号和非生物胁迫的正调控因子,纯合T-DNA插入突变体与野生型相比对干旱、高盐和氧化胁迫更敏感[49]。但OsbZIP46 含有的D 结构域对激活活性有负效应。因此,过量表达对耐旱性没有正效应,但过表达(去除D域的OsbZIP46) 显著提高了水稻对干旱和渗透的抗性[50]。有意思的是,MODD通过与OsTPR3-HDA702共抑制复合物互作来抑制OsbZIP46活性,下调OsbZIP46靶基因的组蛋白乙酰化水平;并通过与U-box型E3泛素连接酶OsPUB70互作,促进OsbZIP46降解。这一过程中,OsbZIP46的D结构域通过与MODD的互作,参与了OsbZIP46的去活化和降解[51]。与OsbZIP46类似,其他多个bZIP蛋白,包括OsBZIP10[20]、OsbZIP16[26]、OsbZIP33[42]、EDT1OsbZIP40[47]、OsbZIP45[33]、OsbZIP62[74]、OsbZIP66[80-81]和OsbZIP72[89]等,也都是ABREs结合因子,在干旱胁迫应答中作为转录激活因子发挥正调控作用。如晚期胚胎富集蛋白基因正调控水稻的耐旱性,且不影响产量[104];OsbZIP66和OsbZIP72通过直接与的启动子结合,上调表达水平,从而正调控水稻耐旱性[81, 89]。OsMFT1作为辅助激活因子能协调并增强OsbZIP66活性[81]。与野生型相比,干旱处理数天再复水后,过表达、、或的转基因水稻幼苗对ABA超敏感、存活率显著提高,抗旱性明显增强[46-47],而敲除株系的存活率显著降低,突变体的蒸腾速率和气孔导度显著高于野生型中花11[20]。OsbZIP72还能直接激活高亲和力钾转运蛋白基因的表达,参与ABA信号通路介导的耐盐途径[90]。
也有bZIP蛋白负调控水稻的干旱和盐胁迫应答。干旱胁迫下,RNAi株系的气孔开度和失水率相比野生型降低,生理指标如脯氨酸、叶绿素和丙二醛含量显著改善,存活率得到大幅提高,表明负调控水稻的耐旱性[13]。水稻中过量表达后对盐胁迫高度敏感,而抑制表达则提高了耐盐性,但同时导致育性降低[17, 19]。
3.3 bZIP蛋白调控其他非生物胁迫应答
OsbZIP20参与铵解毒和抗氧化。OsSAPK9能与OsbZIP20互作并将其磷酸化,从而激活OsbZIP20功能。高NH4+胁迫下,ABA能上调表达,OsSAPK9–OsbZIP20模块通过增强NH4+同化和抗氧化活性,降低活性氧和游离铵水平,减轻水稻铵中毒。突变体和对高NH4+高度敏感,伴随自由NH4+和H2O2在组织中积累[30]。
OsbZIP68参与调控渗透胁迫,且不依赖于ABA。GPX1作为氧化还原的传感元件,在暴露于渗透胁迫后很快被氧化,形成分子内二硫键,进而激活OsbZIP68。GPX1和OsbZIP68之间的二硫键交换诱导OsbZIP68形成同源四聚体,从而通过调节渗透应答基因的表达调节渗透胁迫反应[82]。
OsbZIP88参与调控水稻对3种不同类型除草剂的抗性,过表达的转基因水稻对除草剂敏感性下降,而敲除株系的敏感性增加[100]。
4 bZIP蛋白调控水稻生物胁迫应答
APIP5(即OsbZIP53)形成同源二聚体,与真菌效应子AvrPiz-t在细胞质中互作,AvrPiz-t能特异性抑制APIP5的转录活性。当宿主没有Piz-t时,AvrPiz-t通过作用于APIP5,增强效应子触发的坏死,帮助稻瘟菌进入死体营养阶段。当宿主包含Piz-t时,Piz-t通过与APIP5互作,稳定APIP5蛋白积累以阻止细胞坏死的发生,从而抑制稻瘟菌从活体营养阶段过渡到死体营养阶段[61]。除此,APIP5还有多种调控防卫反应的机制。APIP5能直接与羟基肉桂酰基转移酶基因/、、和的启动子结合,抑制它们的转录,负调控酚胺代谢物和木质素积累[62-63]。APIP5也能结合到细胞壁相关激酶基因和细胞色素P450基因的启动子上,抑制二者的表达;OsWAK5调节木质素积累,CYP72A1调控植保素合成和活性氧迸发,两者均正调控水稻基础免疫反应。有趣的是,APIP5能发生质核穿梭,并有RNA结合活性,稻瘟病菌侵染促使其在胞质RNA加工小体中富集,与细胞死亡和防卫相关基因和的3' UTR的poly(U)序列特异结合,促进和的mRNA降解,从而在转录后水平调控水稻基础免疫反应[64]。
白叶枯病抗性。研究发现白叶枯病菌Ⅲ型效应子基因诱导表达,而的异位表达增加了水稻对白叶枯病的易感性[90]。rTGA2.1OsbZIP63[74]和OsbZIP1OsbZIP28[39],也通过水杨酸途径分别调控水稻的白叶枯病抗性和稻瘟病抗性。RF2aOsbZIP75和RF2bOsbZIP30通过抑制东格鲁杆状病毒的复制,正调控水稻对该病毒的抗性[40-41]。
萜类植保素具有广谱抑菌活性,参与植物的生物防御响应。OsTGAP1OsbZIP37能上调效应子诱发的萜类植保素的积累[43-44],而OsbZIP79能与OsbZIP37互作,抑制萜类植保素的积累[96]。
5 问题与展望
bZIP蛋白是重要的转录因子,通过突变体表型分析、基因敲除(低)或过表达等技术,已从水稻中鉴定了45个bZIP 转录因子(表1)。它们广泛参与调控水稻各器官组织的发生发育、病虫害抗性和非生物胁迫应答等生物学过程。但是,这些进程的调节不是孤立的,需要上下游多个调控因子的参与。如我们发现OsbZIP76自身无转录激活活性,但它能分别与核因子Y家族转录因子OsNF-YB1和OsNF-YB9互作,调控水稻胚乳发育,共同参与储藏物质积累[93]。另外,多个信号间往往形成交叉,相互影响,而部分bZIP蛋白扮演中心节点的作用,如 OsbZIP12正调整水稻的耐盐耐旱性,也调节种子萌发、抽穗期以及D-阿洛糖诱导的生长抑制[21-25],OsbZIP23/66/72既促进种子休眠,也调控干旱胁迫应答,以及多种胁迫下种子萌发(图2)[37, 107]。因此,bZIP蛋白家族对水稻的生长发育和胁迫应答调控是复杂的,每个bZIP转录因子都扮演了调控网络中的一个或多个节点,我们仍需要通过鉴定更多的bZIP蛋白及其互作因子,为最终绘制出整个遗传网络填上一块块拼图。
[1] 王金英, 丁峰, 潘介春, 张树伟, 杨亚涵, 黄幸, 范志毅, 李琳, 王颖. 植物bZIP转录因子家族的研究进展[J]. 热带农业科学, 2019, 39(6): 39-45.
Wang J Y, Ding F, Pan J C, Zhang S W, Yang Y H, Huang X, Fan Z Y, Li L, Wang Y. Research progress of bZIP lineage transcription factors in plant[J]., 2019, 39(6): 39-45.
[2] 国家统计局. 中国统计年鉴[G]. 北京: 中国统计出版社, 2022: 385-387, 428-430.
National Bureau of Statistics of the People's Republic of China. Chinese Statistical Yearbook[G]. Beijing: China Statistics Press, 2022: 385-387,428-430. (in Chinese)
[3] Dröge-Laser W, Snoek B L, Snel B, Weiste C. ThebZIP transcription factor family: An update[J]., 2018, 45: 36-49.
[4] Nijhawan A, Jain M, Tyagi A K, Khurana J P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice[J]., 2008, 146(2): 333-350.
[5] Ji Q, Zhang L S, Wang Y F, Wang J. Genome-wide analysis of basic leucine zipper transcription factor families in,and[J]., 2009, 13(2): 174-182.
[6] Corrêa L G G, Riaño-Pachón D M, Schrago C G, dos Santos R V, Mueller-Roeber B, Vincentz M. The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes[J]., 2008, 3: e2944.
[7] Chai J T, Zhu S S, Li C N, Wang C M, Cai M H, Zheng X M, Zhou L, Zhang H, Sheng P K, Wu M M, Jin X, Cheng Z J, Zhang X, Lei C L, Ren Y L, Lin Q B, Zhou S R, Guo X P, Wang J, Zhao Z C, Wan J M. OsRE1 interacts with OsRIP1 to regulate rice heading date by finely modulatingexpression[J]., 2021, 19(2): 300-310.
[8] Hasegawa T, Lucob-Agustin N, Yasufuku K, Kojima T, Nishiuchi S, Ogawa A, Takahashi-Nosaka M, Kano-Nakata M, Inari-Ikeda M, Sato M, Tsuji H, Wainaina C M, Yamauchi A, Inukai Y. Mutation of/, which encodes a member of the basic leucine zipper transcription factor family, promotes root development in rice through repressing auxin signaling[J]., 2021, 306: 110861.
[9] Lakra N, Nutan K K, Das P, Anwar K, Singla-Pareek S L, Pareek A. A nuclear-localized histone-gene binding protein from rice (OsHBP1b) functions in salinity and drought stress tolerance by maintaining chlorophyll content and improving the antioxidant machinery[J]., 2015, 176: 36-46.
[10] Das P, Lakra N, Nutan K K, Singla-Pareek S L, Pareek A. A unique bZIP transcription factor imparting multiple stress tolerance in rice[J]., 2019, 12: 58.
[11] 仝宇, 王聪, 赵利利, 连娟, 刘晓梅, 赵宝存. 转录因子OsbZIP5负调控水稻的耐旱性[J]. 中国生物化学与分子生物学报, 2021, 37(6): 798-810.
Tong Y, Wang C, Zhao L L, Lian J, Liu X M, Zhao B C. Transcription factor OsbZIP5 negatively regulates drought-tolerance in rice[J]., 2021, 37(6): 798-810. (in Chinese with English abstract)
[12] Brambilla V, Martignago D, Goretti D, Cerise M, Somssich M, de Rosa M, Galbiati F, Shrestha R, Lazzaro F, Simon R, Fornara F. Antagonistic transcription factor complexes modulate the floral transition in rice[J]., 2017, 29(11): 2801-2816.
[13] Zhu C C, Wang C X, Lu C Y, Wang J D, Zhou Y, Xiong M, Zhang C Q, Liu Q Q, Li Q F. Genome-wide identification and expression analysis of OsbZIP09 target genes in rice reveal its mechanism of controlling seed germination[J]., 2021, 22(4): 1661.
[14] Wang C X, Zhu C C, Zhou Y, Xiong M, Wang J D, Bai H, Lu C Y, Zhang C Q, Liu Q Q, Li Q F. OsbZIP09, a unique OsbZIP transcription factor of rice, promotes rather than suppresses seed germination by attenuating abscisic acid pathway[J]., 2021, 28(4): 358-367.
[15] Bhatnagar N, Min M K, Choi E H, Kim N, Moon S J, Yoon I, Kwon T, Jung K H, Kim B G. The protein phosphatase 2C clade A proteinpositively regulates seed germination by directly inactivating[J]., 2017, 93(4): 389-401.
[16] Kim H, Hwang H, Hong J W, Lee Y N, Ahn I P, Yoon I S, Yoo S D, Lee S, Lee S C, Kim B G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth[J]., 2012, 63(2): 1013-1024.
[17] Li Y X, Zhou J H, Li Z, Qiao J Z, Quan R D, Wang J, Huang R F, Qin H. SALT AND ABA RESPONSE ERF1 improves seed germination and salt tolerance by repressing ABA signaling in rice [J]., 2022, 189(2): 1110-1127.
[18] Yoshida H, Hirano K, Yano K, Wang F, Mori M, Kawamura M, Koketsu E, Hattori M, Ordonio R L, Huang P, Yamamoto E, Matsuoka M. Genome-wide association study identifies a gene responsible for temperature-dependent rice germination[J]., 2022, 13: 5665.
[19] Zou M J, Guan Y C, Ren H B, Zhang F, Chen F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance[J]., 2008, 66(6): 675-683.
[20] Li Q, Zhou L Y, Chen Y N, Xiao N, Zhang D P, Zhang M J, Wang W G, Zhang C Q, Zhang A N, Li H, Chen J M, Gao Y. Phytochrome interacting factor regulates stomatal aperture by coordinating red light and abscisic acid[J]., 2022, 34(11): 4293-4312.
[21] Zhang C Y, Liu J, Zhao T, Gomez A, Li C, Yu C S, Li H Y, Lin J Z, Yang Y Z, Liu B, Lin C T. A drought-inducible transcription factor delays reproductive timing in rice[J]., 2016, 171(1): 334-343.
[22] Joo J, Lee Y H, Song S I. Overexpression of the rice basic leucine zipper transcription factor OsbZIP12 confers drought tolerance to rice and makes seedlings hypersensitive to ABA[J]., 2014, 8(6): 431-441.
[23] Hossain M A, Lee Y, Cho J I, Ahn C H, S K, Jeon J S, Kang H, Lee C H, An G, Park P B. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice[J]., 2010, 72(4-5): 557-566.
[24] Tang L Q, Xu H Y, Wang Y F, Wang H M, Li Z Y, Liu X X, Shu Y Z, Li G, Liu W N, Ying J Z, Tong X H, Yao J L, Xiao W F, Tang S Q, Ni S, Zhang J. OsABF1 represses gibberellin biosynthesis to regulate plant height and seed germination in rice (L.)[J]., 2021, 22(22): 12220.
[25] Fukumoto T, Kano A, Ohtani K, Inoue M, Yoshihara A, Izumori K, Tajima S, Shigematsu Y, Tanaka K, Ohkouchi T, Ishida Y, Nishizawa Y, Tada Y, Ichimura K, Gomi K, Yoo S D, Sheen J, Akimitsu K. Phosphorylation of d-allose by hexokinase involved in regulation ofexpression for growth inhibition inL.[J]., 2013, 237(5): 1379-1391.
[26] Chen H, Chen W, Zhou J L, He H, Chen L B, Chen H D, Deng X W. Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice[J]., 2012, 193-194: 8-17.
[27] Sun Y Y, Shi Y H, Liu G G, Yao F, Zhang Y Y, Yang C K, Guo H, Liu X Q, Jin C, Luo J. Natural variation in thepromoter contributes to branched-chain amino acid levels in rice[J]., 2020, 228(5): 1548-1558.
[28] Sun Y Y, Wang B, Ren J X, Zhou Y T, Han Y, Niu S Y, Zhang Y Y, Shi Y H, Zhou J J, Yang C K, Ma X M, Liu X Q, Luo Y H, Jin C, Luo J. OsbZIP18, a positive regulator of serotonin biosynthesis, negatively controls the UV-B tolerance in rice[J]., 2022, 23(6): 3215.
[29] Bai B, Lu N N, Li Y P, Guo S L, Yin H B, He Y N, Sun W, Li W, Xie X Z. OsBBX14 promotes photomorphogenesis in rice by activatingexpression under blue light conditions[J]., 2019, 284: 192-202.
[30] Sun L, Di D W, Li G J, Kronzucker H J, Wu X Y, Shi W M. Endogenous ABA alleviates rice ammonium toxicity by reducing ROS and free ammonium via regulation of the SAPK9-bZIP20 pathway[J]., 2020, 71(15): 4562-4577.
[31] Wang B X, Xu B, Liu Y, Li J F, Sun Z G, Chi M, Xing Y G, Yang B, Li J, Liu J B, Chen T M, Fang Z W, Lu B G, Xu D Y, Babatunde K B. A novel mechanisms of the signaling cascade associated with thesynergistic interaction to enhance tolerance of plant to abiotic stress in rice (L.)[J]., 2022, 323: 111393.
[32] Xiang Y, Tang N, Du H, Ye H, Xiong L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice[J]., 2008, 148(4): 1938-1952.
[33] Park Su H, Jeong J S, Lee K H, Kim Y S, Choi Y D, Kim J K. OsbZIP23 and OsbZIP45, members of the rice basic leucine zipper transcription factor family, are involved in drought tolerance[J]., 2015, 9(2): 89-96.
[34] Zong W, Yang J, Fu J, Xiong L. Synergistic regulation of drought-responsive genes by transcription factor OsbZIP23 and histone modification in rice[J]., 2020, 62(6): 723-729.
[35] Srivastava A K, Zhang C J, Caine R S, Gray J, Sadanandom A. Rice SUMO proteasetargets the transcription factor, OsbZIP23 to promote drought tolerance in rice[J]., 2017, 92(6): 1031-1043.
[36] Zong W, Tang N, Yang J, Peng L, Ma S, Xu Y, Li G, Xiong L. Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes[J]., 2016, 171(4): 2810-2825.
[37] Song S, Wang G, Wu H, Fan X, Liang L, Zhao H, Li S, Hu Y, Liu H, Ayaad M, Xing Y. OsMFT2 is involved in the regulation of ABA signaling mediated seed germination through interacting with OsbZIP23/66/72 in rice[J]., 2020, 103(2): 532-546.
[38] Wang WQ, Xu DY, Sui YP, Ding XH, Song XJ. A multiomic study uncovers a bZIP23-PER1A–mediated detoxification pathway to enhance seed vigor in rice[J]., 2022, 119(9): e2026355119.
[39] Meng X B, Zhao W S, Lin R M, Wang M, Peng Y L. Identification of a novel rice bZIP-type transcription factor gene,, involved in response to infection of[J]., 2005, 23(3): 301-302.
[40] Dai S H, Zhang Z H, Chen S Y, Beachy R N. RF2b, a rice bZIP transcription activator, interacts with RF2a and is involved in symptom development of rice tungro disease[J]., 2004, 101(2): 687-692.
[41] Dai S H, Wei X P, Alfonso A A, Pei L P, Duque U G, Zhang Z H, Babb G M, Beachy R N. Transgenic rice plants that overexpress transcription factors RF2a and RF2b are tolerant to rice tungro virus replication and disease[J]., 2008, 105(52): 21012-21016.
[42] Chen H, Dai X J, Gu Z Y. OsbZIP33 is an ABA-dependent enhancer of drought tolerance in rice[J]., 2015, 55(4): 1673-1685.
[43] Okada A, Okada K, Miyamoto K, Koga J, Shibuya N, Nojiri H, Yamane H. OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice[J]., 2009, 284(39): 26510-26518.
[44] Miyamoto K, Matsumoto T, Okada A, Komiyama K, Chujo T, Yoshikawa H, Nojiri H, Yamane H, Okada K. Identification of target genes of the bZIP transcription factor OsTGAP1, whose overexpression causes elicitor-induced hyperaccumulation of diterpenoid phytoalexins in rice cells[J]., 2014, 9(8): e105823.
[45] Shimizu H, Sato K, Berberich T, Miyazaki A, Ozaki R, Imai R, Kusano T. LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants., 2005, 46(10): 1623-1634.
[46] Takahashi H, Kawakatsu T, Wakasa Y, Hayashi S, Takaiwa F. A rice transmembrane bZIP transcription factor, OsbZIP39, regulates the endoplasmic reticulum stress response[J]., 2012, 53(1): 144-153.
[47] Wu T, Zhang M X, Zhang H J, Huang K, Chen M J, Chen C, Yang X, Li Z, Chen H Y, Ma Z M, Zhang X M, Jiang W Z, Du X L. Identification and characterization ofconferring drought tolerance in rice[J]., 2019, 62: 39-47.
[48] Lilay G H, Castro P H, Guedes J G, Almeida D M, Campilho A, Azevedo H, Aarts M G M, Saibo N J M, Assunção A G L. Rice F-bZIP transcription factors regulate the zinc deficiency response[J]., 2020, 71(12): 3664-3677.
[49] Hossain M A, Cho J I, Han M, Ahn C H, Jeon J S, An G, Park P B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice[J]., 2010, 167(17): 1512-1520.
[50] Tang N, Zhang H, Li X H, Xiao J H, Xiong L Z. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice[J]., 2012, 158(4): 1755-1768.
[51] Tang N, Ma S, Zong W, Yang N, Lv Y, Yan C, Guo Z, Li J, Li X, Xiang Y, Song H, Xiao J, Li X, Xiong L. MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice[J]., 2016, 28(9): 2161-2177.
[52] Yang X, Yang Y N, Xue L J, Zou M J, Liu J Y, Chen F, Xue H W. Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes[J]., 2011, 156(3): 1397-1409.
[53] Hao J Q, Wang D K, Wu Y B, Huang K, Duan P G, Li N, Xu R, Zeng D L, Dong G J, Zhang B L, Zhang L M, Inzé D, Qian Q, Li Y H. The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice[J]., 2021, 14(8): 1266-1280.
[54] Burman N, Bhatnagar A, Khurana J P. OsbZIP48, a HY5 transcription factor ortholog, exerts pleiotropic effects in light-regulated development[J]., 2018, 176(2): 1262-1285.
[55] Zhang F, Huang J C, Guo H, Yang C K, Li Y F, Shen S Q, Zhan C S, Qu L H, Liu X Q, Wang S C, Chen W, Luo J.contributes to flavonoid accumulation and UV-B tolerance by regulatingin rice[J]., 2022, 65(7): 1380-1394.
[56] Kim D H, Park S, Lee J Y, Ha S H, Lee J G, Lim S H. A rice B-box protein, OsBBX14, finely regulates anthocyanin biosynthesis in rice[J]., 2018, 19(8): 2190.
[57] Hayashi S, Wakasa Y, Takahashi H, Kawakatsu T, Takaiwa F. Signal transduction by IRE1-mediated splicing ofand other stress sensors in the endoplasmic reticulum stress response of rice[J]., 2012, 69(6): 946-956.
[58] Lu S J, Yang Z T, Sun L, Sun L, Song Z T, Liu J X. Conservation of IRE1-regulatedmRNA unconventional splicing in rice (L.) involved in ER stress responses[J]., 2012, 5(2): 504-514.
[59] Yang W, Xu P, Zhang J, Zhang S, Li Z, Yang K, Chang X, Li Y. OsbZIP60-mediated unfolded protein response regulates grain chalkiness in rice[J]., 2022, 49(5): 414-426.
[60] Liu X H, Lü Y S, Yang W, Yang Z T, Lu S J, Liu J X. A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice[J]., 2020, 18(5): 1317-1329.
[61] Wang R T, Ning Y S, Shi X T, He F, Zhang C Y, Fan J B, Jiang N, Zhang Y, Zhang T, Hu Y J, Bellizzi M, Wang G L. Immunity to rice blast disease by suppression of effector-triggered necrosis[J]., 2016, 26(18): 2399-2411.
[62] Fang H, Shen S Q, Wang D, Zhang F, Zhang C Y, Wang Z X, Zhou Q Q, Wang R Y, Tao H, He F, Yang C K, Peng M, Jing X Y, Hao Z Y, Liu X L, Luo J, Wang G L, Ning Y S. A monocot-specific hydroxycinnamoyl- putrescine gene cluster contributes to immunity and cell death in rice[J]., 2021, 66(23): 2381-2393.
[63] Fang H, Zhang F, Zhang C Y, Wang D, Shen S Q, He F, Tao H, Wang R Y, Wang M, Wang D B, Liu X L, Luo J, Wang G L, Ning Y S. Function of hydroxycinnamoyl transferases for the biosynthesis of phenolamides in rice resistance to[J]., 2022, 49(8): 776-786.
[64] Zhang F, Fang H, Wang M, He F, Tao H, Wang R Y, Long J W, Wang J Y, Wang G L, Ning Y S. APIP5 functions as a transcription factor and an RNA-binding protein to modulate cell death and immunity in rice[J]., 2022, 50(9): 5064-5079.
[65] He Y, Li L Y, Shi W B, Tan J H, Luo X X, Zheng S Y, Chen W T, Li J, Zhuang C X, Jiang D G. Florigen repression complexes involving rice CENTRORADIALIS2 regulate grain size[J]., 2022, 190(2): 1260-1274.
[66] Tsuji H, Nakamura H, Taoka K I, Shimamoto K. Functional diversification of FD transcription factors in rice, components of florigen activation complexes[J]., 2013, 54(3): 385-397.
[67] Kawakatsu T, Yamamoto M P, Touno S M, Yasuda H, Takaiwa F. Compensation and interaction between RISBZ1 and RPBF during grain filling in rice[J]., 2009, 59(6): 908-920
[68] Kawakatsu T and Takaiwa F. Differences in transcriptional regulatory mechanisms functioning for free lysine content and seed storage protein accumulation in rice grain[J]., 2010, 51(12): 1964-1974
[69] Wang J C, Xu H, Zhu Y, Liu Q Q, Cai X L. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm[J]., 2013, 64(11): 3453-3466.
[70] Wu J H, Zhu C F, Pang J H, Zhang X R, Yang C L, Xia G X, Tian Y C, He C Z. OsLOL1, a C2C2-type zinc finger protein, interacts with OsbZIP58 to promote seed germination through the modulation of gibberellin biosynthesis in[J]., 2014, 80(6): 1118-1130.
[71] 喻旭, 牛向丽, 杨盛慧, 李欲翔, 刘亮亮, 唐维, 刘永胜. 过量表达转录因子OsbZIP60对水稻抗热和抗旱能力的研究[J]. 中国农业科学, 2011, 44(20): 4142-4149.
Yu X, Niu X L, Yang S Hu, Li Y X, Liu L L, Tang W, Liu Y S. Research on heat and drought tolerance in rice (L.) by overexpressing transcription factor OsbZIP60 [J]., 2011, 44(20): 4142-4149. (in Chinese with English abstract)
[72] Cao R J, Zhao S L, Jiao G A, Duan Y Q, Ma L Y, Dong N N, Lu F F, Zhu M D, Shao G N, Hu S K, Sheng Z H, Zhang J, Tang S Q, Wei X J, Hu P S., encoding a transmembrane bZIP transcription factor, regulates endosperm storage protein and starch biosynthesis in rice [J]., 2022, 3(6): 100463.
[73] Kaur A, Nijhawan A, Yadav M, Khurana J P. OsbZIP62/OsFD7, a functional ortholog of FLOWERING LOCUS D, regulates floral transition and panicle development in rice[J]., 2021, 72(22): 7826-7845.
[74] Yang S Q, Xu K, Chen S J, Li T F, Xia H, Chen L, Liu H Y, Luo L J. A stress-responsive bZIP transcription factorimproves drought and oxidative tolerance in rice[J]., 2019, 19: 260.
[75] Fitzgerald H A, Canlas P E, Chern M S, Ronald P C. Alteration of TGA factor activity in rice results in enhanced tolerance topv.[J]., 2005, 43(3): 335-347.
[76] Pan T T, He M L, Liu H L, Tian X J, Wang Z Y, Yu X L, Miao X F, Li X F. Transcription factor bZIP65 delays flowering via suppressingexpression in rice[J]., 2022, 42(10): 63.
[77] Hobo T, Kowyama Y, Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription[J]., 1999, 96(26): 15348-15353.
[78] Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors[J]., 2005, 44(6): 939-949.
[79] Wang Y, Hou Y, Qiu J, Wang H, Wang S, Tang L, Tong X, Zhang J. Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’ pathway to synergistically inhibit seed germination in rice ()[J]., 2020, 228(4): 1336-1353.
[80] Yoon S, Lee D K, Yu I J, Kim Y S, Choi Y D, Kim J K. Overexpression of thetranscription factor enhances drought tolerance of rice plants[J]., 2017, 11(1): 53-62.
[81] Chen Y, Shen J, Zhang L, Qi H, Yang L, Wang H, Wang J, Wang Y, Du H, Tao Z, Zhao T, Deng P, Shu Q, Qian Q, Yu H, Song S. Nuclear translocation of OsMFT1 that is impeded by OsFTIP1 promotes drought tolerance in rice[J]., 2021, 14(8): 1297-1311.
[82] Zhou H, Zhang F, Zhai F C, Su Y, Zhou Y, Ge Z L, Tilak P, Eirich J, Finkemeier I, Fu L, Li Z M, Yang J, Shen W B, Yuan X X, Xie Y J. Rice GLUTATHIONE PEROXIDASE1-mediated oxidation of bZIP68 positively regulates ABA-independent osmotic stress signaling[J]., 2022, 15(4): 651-670.
[83] Cerise M, Giaume F, Galli M, Khahani B, Lucas J, Podico F, Tavakol E, Parcy F, Gallavotti A, Brambilla V, Fornara F. OsFD4 promotes the rice floral transition via florigen activation complex formation in the shoot apical meristem[J]., 2021, 229(1): 429-443.
[84] Li X, Tian X, He M, Liu X, Li Z, Tang J, Mei E, Xu M, Liu Y, Wang Z, Guan Q, Meng W, Fang J, Zhang J, Bu Q. bZIP71 delays flowering by suppressingexpression in rice[J]., 2022, 64(7): 1352-1363.
[85] Liu C T, Mao B G, Ou S J, Wang W, Liu L C, Wu Y B, Chu C C, Wang X P. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice[J]., 2014, 84(1-2): 19-36.
[86] Liu C, Ou S, Mao B, Tang J, Wang W, Wang H, Cao S, Schläppi M R, Zhao B, Xiao G, Wang X, Chu C. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates[J]., 2018, 9: 3302.
[87] Liu C, Schläppi M R, Mao B, Wang W, Wang A, Chu C. The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage[J]., 2019, 17(9): 1834-1849.
[88] Wang S, Liu W, He Y, Adegoke T V, Ying J, Tong X, Li Z, Tang L, Wang H, Zhang J, Tian Z, Wang Y. bZIP72 promotes submerged rice seed germination and coleoptile elongation by activating ADH1[J]., 2021, 169: 112-118.
[89] Lu G, Gao C, Zheng X, Han B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice[J]., 2009, 229(3): 605-615.
[90] Wang B, Liu Y, Wang Y, Li J, Sun Z, Chi M, Xing Y, Xu B, Yang B, Li J, Liu J, Chen T, Fang Z, Lu B, Xu D, Babatunde K B.is involved in transcriptional gene-regulation pathway of abscisic acid signal transduction by activating rice high-affinity potassium transporter[J]., 2021, 28(3): 257-267.
[91] Sugio A, Yang B, Zhu T, White F F. Two type III effector genes ofpv.control the induction of the host genesandduring bacterial blight of rice[J]., 2007, 104(25): 10720-10725.
[92] Wang Q, Lin Q B, Wu T, Duan E C, Huang Y S, Yang C Y, Mou C L, Lan J, Zhou C L, Xie K, Liu X, Zhang X, Guo X P, Wang J, Jiang L, Wan J M.regulates seed dormancy through the abscisic acid pathway in rice[J]., 2020, 298: 110570.
[93] Niu B, Deng H, Li T, Sharma S, Yun Q, Li Q, E Z, Chen C. OsbZIP76 interacts with OsNF-YBs and regulates endosperm cellularization in rice ()[J]., 2020, 62(12): 1983-1996.
[94] Taoka K I, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri Y A, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen[J]., 2011, 476(7360): 332-335.
[95] Peng Q, Zhu C, Liu T, Zhang S, Feng S, Wu C. Phosphorylation of OsFD1 by OsCIPK3 promotes the formation of RFT1-containing florigen activation complex for long-day flowering in rice[J]., 2021, 14(7): 1135-1148.
[96] Miyamoto K, Nishizawa Y, Minami E, Nojiri H, Yamane H, Okada K. Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells[J]., 2015, 173: 19-27.
[97] Liu D, Shi S, Hao Z, Xiong W, Luo M. OsbZIP81, a homologue ofVIP1, may positively regulate JA levels by directly targetting the genes in JA signaling and metabolism pathway in rice[J]., 2019, 20(9): 2360.
[98] 刘德芳. 水稻B-bZIP转录因子亚家族成员OsbZIP81和OsbZIP84的功能分析[D]. 武汉: 华中农业大学, 2019.
Liu D F. Functional analysis of rice B-bZIP subfamily members OsbZIP81 and OsbZIP84[D]. Wuhan: Huazhong Agricultural University, 2019.
[99] Gao W W, Li M K, Yang S G, Gao C Z, Su Y, Zeng X, Jiao Z L, Xu W J, Zhang M Y, Xia K F. miR2105 and the kinase OsSAPK10 co-regulate OsbZIP86 to mediate drought-induced ABA biosynthesis in rice[J]., 2022, 189(2): 889-905.
[100]Zhang Y H, Gao H T, Fang J P, Wang H, Chen J Y, Li J, Dong L Y. Up-regulation oftranscription factor is involved in resistance to three different herbicides in bothand[J]., 2022, 73(19): 6916-6930.
[101]Kaneko-Suzuki M, Kurihara-Ishikawa R, Okushita-Terakawa C, Kojima C, Nagano-Fujiwara M, Ohki I, Tsuji H, Shimamoto K, Taoka K I. TFL1-like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD[J]., 2018, 59(3): 458-468.
[102]Wang Y, Lu Y, Guo Z, Ding Y, Ding C., a-like gene, responses to drought stress and regulates rice flowering transition[J]., 2020, 13: 70.
[103]Cai M H, Zhu S S, Wu M M, Zheng X M, Wang J C, Zhou L, Zheng T H, Cui S, Zhou S R, Li C N, Zhang H, Chai J T, Zhang X Y, Jin X, Cheng Z J, Zhang X, Lei C L, Ren Y L, Wan J M. DHD4, a CONSTANS-like family transcription factor, delays heading date by affecting the formation of the FAC complex in rice[J]., 2021, 14(2): 330-343.
[104]Xiao B, Huang Y, Tang N, Xiong L. Over-expression of agene in rice improves drought resistance under the field conditions[J]., 2007, 115(1): 35-46.
[105]Li R Q, Zheng W Y, Yang R F, Hu Q W, Ma L Y, Zhang H L. OsSGT1 promotes melatonin-ameliorated seed tolerance to chromium stress by affecting the OsABI5–OsAPX1 transcriptional module in rice[J]., 2022, 112(1): 151-171.
[106]Li R Q, Jiang M, Song Y, Zhang H L. Melatonin alleviates low-temperature stress via ABI5-mediated signals during seed germination in rice (L.)[J]., 2021, 12: 727596.
[107]Yang L J, Chen Y, Xu L, Wang J X, Qi H Y, Guo J Z, Zhang L, Shen J, Wang H Y, Zhang F, Xie L J, Zhu W J, Lü P T, Qian Q, Yu H, Song S Y. The OsFTIP6-OsHB22-OsMYBR57 module regulates drought response in rice[J]., 2022, 15(7): 1227-1242.
Research Progress in the Function of Basic Leucine Zipper (bZIP) Protein Family in Rice
HAN Cong, HE Yuchang, WU Lijuan, JIA Lili, WANG Lei, E Zhiguo*
(China National Rice Research Institute, Hangzhou 310006, China;*Corresponding author, email: ezhiguo@caas.cn)
As a largefamily of transcriptional regulators, basic leucine zipper (bZIP) proteins are widespread in eukaryotes. The bZIP proteins characteristically harbor a bZIP domain composed of two closely adjacent structural features: a DNA-binding basic region and the leucine zipper region. Annotations to eighty-nine bZIP transcription factor-encoding genes are available in therice genome, 45 of which are identified. They are involved in regulating rice growth and development and responses to biotic and abiotic stress, including seed dormancy and germination, floral transition, and photomorphogenesis, andstress and hormone signaling pathway, etc.
rice; basic leucine zipper protein; bZIP transcription factor; gene function
10.16819/j.1001-7216.2023.221018
2022-11-17;
2023-02-02。
浙江省自然科学基金探索项目(LY21C130004);中央级公益性科研院所基本科研业务费专项(CPSIBRF-CNRRI-202202);中国农业科学院科技创新工程资助项目(CAAS-ASTIP-2021-CNRRI)。
