Characterization of the core microf lora and nutrient composition in packaged pasteurized milk products during storage
2023-01-03RuixueDingShnshnYngLijunGengYumengLiuBopingHeLiyunLiuXiqingYueRinWuJunruiWu
Ruixue Ding, Shnshn Yng, Lijun Geng, Yumeng Liu, Boping He,Liyun Liu, Xiqing Yue, Rin Wu,*, Junrui Wu,*
a Liaoning Engineering Research Center of Food Fermentation Technology, Shenyang Key Laboratory of Microbial Fermentation Technology Innovation,College of Food Science, Shenyang Agricultural University, Shenyang 110866, China
b Shenyang Institute for Food and Drug Control, Shenyang 110122, China
c Inner Mongolia Yili Industrial Group Co., Ltd., Hohhot 010080, China
d China Mengniu Dairy Co., Ltd., Hohhot 010000, China
Keywords:Pasteurized milk Storage conditions Microbiota High-throughput sequencing (HTS)
A B S T R A C T Pasteurized milk contains complex microbial communities affected by sterilization and storage conditions.This complex microflora may be the possible reason that pasteurized dairy products are highly prone to spoilage. In this study, packaged pasteurized milk products collected from dairy processing factories in China were stored at 0, 4, 10, 15, and 25 °C for 0-15 days and subjected to microbial identif ication using high-throughput sequencing. Accordingly, 6 phyla and 44 genera were identif ied as the dominant microbiota.Moreover, the changes in nutritional composition of the pasteurized milk, including in 16 free amino acids,7 taste values, and 8 chemical constituents, were analyzed using principal component and multi-factor analyses.The Pearson correlation analysis identif ied Pseudomonas, Aeromonas, Paenibacillus, and Serratia genera as the core functional microbiota that signif icantly affects the nutritional composition of pasteurized milk. Hence, the results provide a comprehensive understanding of the safety and shelf-life of stored pasteurized milk.© 2023 Beijing Academy of Food Sciences. Publishing services by Elsevier B.V. on behalf of KeAi Communications Co., Ltd. This is an open access article under the CC BY-NC-ND license(http://creativecommons.org/licenses/by-nc-nd/4.0/).* Corresponding authors at: College of Food Science, Shenyang Agricultural University,Shenyang 110866, China.
1. Introduction
Pasteurized milk is not only one of the most nutritious sources of dairy products, but also a complex biological f luid that serves as an excellent growth medium for many microorganisms [1]. Owing to the characteristics of pasteurized milk processing technology, some pathogenic microorganisms are retained, which then results in the growth of many different microorganisms under appropriate growth conditions [2]. Compared with the long shelf-life of thoroughly sterilized milk products [3], pasteurized milk is prone to deterioration if not stored properly during long-term or long-distance storage and transportation, respectively, compromising its quality as well as safety. Hence, the dilemma around selecting between nutrition and food safety has turned out to be a pressing technical problem that restricts the production and consumption of pasteurized dairy products in China.
In fact, recent studies have increasingly begun to focus on microorganisms present in the raw milk and their correlation with the quality of dairy products [4,5]. BothPaenibacillusandBacillushave been reported to be present in raw milk arriving at dairy processing plants [6]. Some of these bacteria comprise the dominant gram-positive spore-forming bacteria isolated from pasteurized cow milk [7]. MostPaenibacillusisolates from pasteurized milk have the ability to grow at low temperatures as compared to only a fewBacillusspecies that typically grow under similar conditions [8].Other studies have shown thatBacillusis the predominant population in the early shelf-life of pasteurized milk (7 days), withPaenibacilluscomprising the predominant bacteria and representing over 95%of the population during long storage intervals (10 days) [9,10].Additionally, the microbiota of dairy products is also influenced by product packaging parameters. During the storage of milk in cartons at 4 °C, the bacterial composition remains stable throughout the product shelf-life. However, storage at 8 °C significantly increases the abundance of operational taxonomic units (OTUs) belonging to the genusBacillusand the plate count levels of presumptiveBacillus cereus[11]. Additionally, Doyle et al. have examined the influence of refrigeration temperature (2, 4 and 6 °C) and storage duration (96 h)on the microbiota composition of raw bulk tank milk (BTM) [12].Although this capability is only observed in certain genera/species,includingPaenibacillusspp. andPseudomonasspp. [13-15], these strains seem to be the major cause of milk spoilage under refrigerated storage conditions. As these strains are not completely consistent among different studies, the microorganisms that remain unidentified in packaged pasteurized dairy products under different storage conditions might impact product quality and safety differently.Accordingly, a detailed investigation of the core strain population and their dynamic changes during storage might serve beneficial while addressing the issues related to food quality and safety in the dairy industry.
Recently, with the rapid development of high-throughput sequencing technology, the research field of microbial ecology has undergone revolutionary changes [16,17]. A large number of previously unknown non-cultivable microorganisms and their functions have been discovered. However, in the conventionally resultant data sets, culture-dependent techniques have always been employed to assess the milk microbiota, both in research and industry [18,19]. The results indicate that a vast majority of microorganisms cannot be isolated using existing culture techniques,which greatly limits the research on these microorganisms. Thus,it is only logical to assume that in the past, knowledge of the microorganisms in pasteurized dairy products and the dynamic changes in their population during storage may not be comprehensive and accurate enough. Hence, it is important to re-examine and re-evaluate the microbial dynamics of pasteurized dairy products during storage. High throughput-sequencing strategies have enabled the detection of the genera of bacteria previously unrelated to milk, such asPseudomonasandAeromonas, in milk [20]. Therefore, this research provides a more comprehensive analysis of microbial diversity in dairy products [21], making a remarkable leap in the research of microorganisms in dairy products. As very few studies have addressed this research area, investigating the correlation between the nutritional properties of pasteurized milk and the core microorganisms during storage will also provide a reference for preserving the nutritional quality of pasteurized milk.
Therefore, the aim of this study was to determine the dynamic changes in nutritional compounds and microbiota profiles in packaged pasteurized milk products, provide new insights into exploring the variances and similarities of the safety of storage conditions,and analyze the core functional microbiota related to the nutrient composition and safety of pasteurized milk.
2. Materials and methods
2.1 Milk sampling
Samples of packaged pasteurized milk (n= 78) produced on the same day were collected from Chinese dairy processing factories(Shenyang, Liaoning Province, China). Samples (78 × 500 mL, each)were immediately placed in liquid nitrogen tanks and sent back to the laboratory and stored in incubators at 0, 4, 10, 15 and 25 °C. The samples were collected from the dairy storage tank every 3 days until the end of the 15thday to prepare for the follow-up test. Three replicates of each sample from each pasteurized milk product were analyzed. For all the samples analyzed, the milk samples were also stored at -80 °C as backups.
2.2 Quantitative analysis of main components of pasteurized milk
The physicochemical and nutritional properties of the milk samples were analyzed using conventional laboratory methods with respect to the pH, acidity and chromatic aberrations such as brightness (L*), redness (a*), and yellowness (b*) values during the same measurement session. The fat, protein and solids-not-fat (SNF)contents of milk were determined using a 120 dairy comprehensive component index analyzer (FLOWSERVE milk analyzer, China) to verify the formula and process control.
The contents of free amino acids (FAAs) in pasteurized milk at different storage temperatures were analyzed according to von Neubeck et al. [22] method. Electronic tongue technology (L-8900,Japan) combined with Toko’s analysis method was applied to different storage times and temperatures of dairy products [23]. Each sample was measured 5 times and the average value was obtained.Quantitative analysis of the tested sample was carried out through the comprehensive taste information of the liquid sample.
2.3 Microbiological testing of milk samples
2.3.1 DNA extraction and pyrosequencing for 16S rRNA
The DNA extraction in pasteurized milk at different storage temperatures was performed as described by Ding et al. [3] method.In brief, the variable region V3 and V4 of the bacterial 16S rRNA gene was amplified, sequencing of the library was done using the Illumina Miseq platform (Shanghai Majorbio Bio-pharm Technology Co., Ltd., China) with a 300 bp paired-end sequencing kit. Three replicate samples were combined into one sample, and thus 26 such samples were subjected to 16S rRNA sequencing.
2.3.2 Sequencing data analysis
Denoising, chimera detection and clustering into OTUs (97%identity) were performed using the USEARCH software.αandβdiversities were generated in QIIME (v 1.9.0) and distinguished at the OTU level.
2.4 Statistical analysis
One-way ANOVA of the physical and chemical properties of pasteurized milk was performed using the SPSS software (version 22.0; SPSS, Armonk, NY, USA). Significance was calculated using Duncan’s multiple range test (P< 0.05).
Principal coordinate analysis (PCoA) plots were visualized in pasteurized milk at different storage temperatures. Redundancy analysis (RDA) analysis of physicochemical and nutritional properties was used to characterize the changes in bacterial communities upon storage of pasteurized milk samples. Correlation heatmap between core bacterial communities and the physicochemical parameters for cluster of orthologous groups (COG) functional classification analyses were generated using Majorbio Cloud Platform (www.majorbio.com).
3. Results
3.1 Analysis of physicochemical and nutritional properties of pasteurized milk samples during storage
The chemical composition of pasteurized milk samples was determined under varying storage conditions. The results of protein,fat, SNF, pH, acidity,L*,a*, andb* values in the milk samples are shown in Table 1. According to the routine standards (GB 19645-2010),the contents of protein, fat, and SNF in pasteurized milk were found to be 2.9, 3.1, and 8.1 g/100 g, respectively. Furthermore, the milk fat composition did not differ between the treatments at each storage condition (P> 0.05). No differences (P> 0.05) were observed in milk chromatic characteristics, while theL* values of milk samples decreased slowly at 0, 4, and 10 °C, which may be due to the influence of storage temperature, resulting in changes in color and refractive index of products. Moreover, acidity and pH were also affected, and the acidity of pasteurized milk during 0-3 days of storage at 15 and 25 °C exceeded the prescribed national standard (GB 5009.239-2016).This effect may be attributed to the fact that the acidity of pasteurized milk tends to decrease because of the influence of spoilage caused by microorganisms.

Table 1Chemical constituents of packaged pasteurized milk under different storage conditions.
As shown in Fig. 1, the content of essential amino acid (EAA)did not change significantly at 0, 4, 10, and 15 °C but increased significantly up to 268.25 mg/100 mL when the pasteurized milk samples were stored at 25 °C for 12-15 days. Noteworthily, glutamic acid (Glu) and aspartic acid (Asp) showed evident changes as the main EAA (data not shown). This phenomenon may be due to digestion of proteins during the growth of microorganisms, resulting in a rapid increase in EAA content. This speculation is also consistent with the rapid decline in protein content as compared to that of the previous time point. Fig. 2 shows that the taste value of pasteurized milk varied most significantly at different storage times at 0 °C. Our results showed that with the increase in storage temperature and time,the taste and bitterness of pasteurized milk decreased slightly, while the astringency was found to be augmented. The special sweetness and fragrance of pasteurized milk are the main change indices during storage. Therefore, according to the radar images, the sweetness of pasteurized milk decreased significantly after storage at 15 and 25 °C,indicating that microbial growth promotes fermentation.
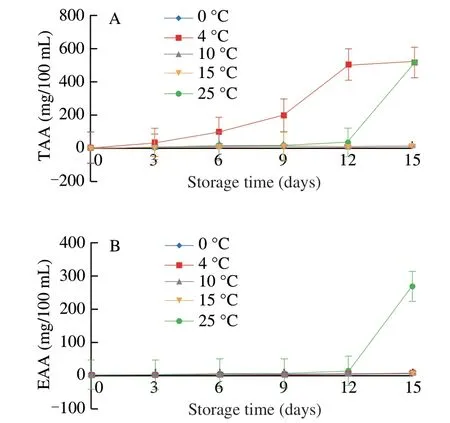
Fig. 1 Analysis of (a) total amino acids (TAAs), (b) EAAs in storage packaged pasteurized milk samples
3.2 Dynamics of the microbial population in pasteurized milk samples during storage
In order to investigate the microbial population dynamics during varying conditions of storage, all milk samples were subjected to 16S rRNA gene sequencing. OTU sequences were classified at a 97% similarity level, and 1 897 OTUs were found to be accumulated in pasteurized milk. A biplot of the PCA of stored pasteurized milk samples is shown in Fig. 3. The two-dimensional model (PC1 and PC2) of PCA explains 77.57% of the total variance, with the first and second principal components explaining the variances of 61.97%and 15.60%, respectively. According to the PCA cluster analysis of OTU levels, the samples were divided into 5 groups at 0, 4, 10,15, and 25 °C, all of which were clustered together within a group.However, the samples stored at 4 °C for 3 and 6 days and at 10 °C for 3 days were separated into one group and did not cluster into groups.Combined with the sample clustering analysis, the results showed that the storage temperatures had a significant effect on the abundance of microorganisms in pasteurized milk, which warrants further deliberation and analysis.
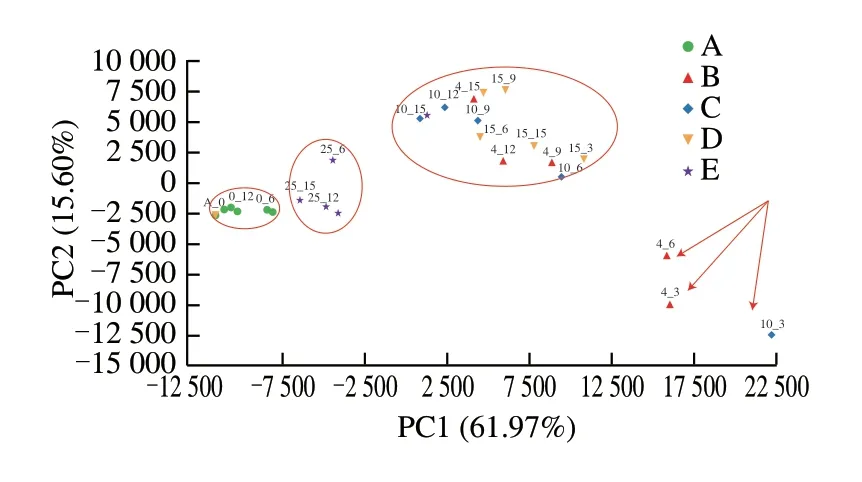
Fig. 3 Principal component analysis of different storage conditions and OTU level microbial count of pasteurized milk. Packaged pasteurized milk samples were storage condition at (A) 0, (B) 4, (C) 10, (D) 15, and (E) 25 °C for 0, 3, 6,9, 12 and 15 days as described. Same with Fig. 4.
Grouping samples at different storage temperatures and times revealed that they shared the same top abundant phyla (that is, phyla with over 1% of total sequences across all samples) but differed in their relative abundances (Fig. 4a). The results revealed that 6 dominant phyla were identified in the bacterial communities,including Proteobacteria, Firmicutes, Tenericutes, Bacteroidetes,Actinobacteria, and Saccharibacteria. Proteobacteria were the major

Fig. 2 Radar analysis of electronic tongue taste in pasteurized milk under different pasteurized storage conditions. Packaged pasteurized milk samples were storage condition at (A) 0, (B) 4, (C) 10, (D) 15, and (E) 25 °C for 0, 3, 6, 9, 12 and 15 days as described.
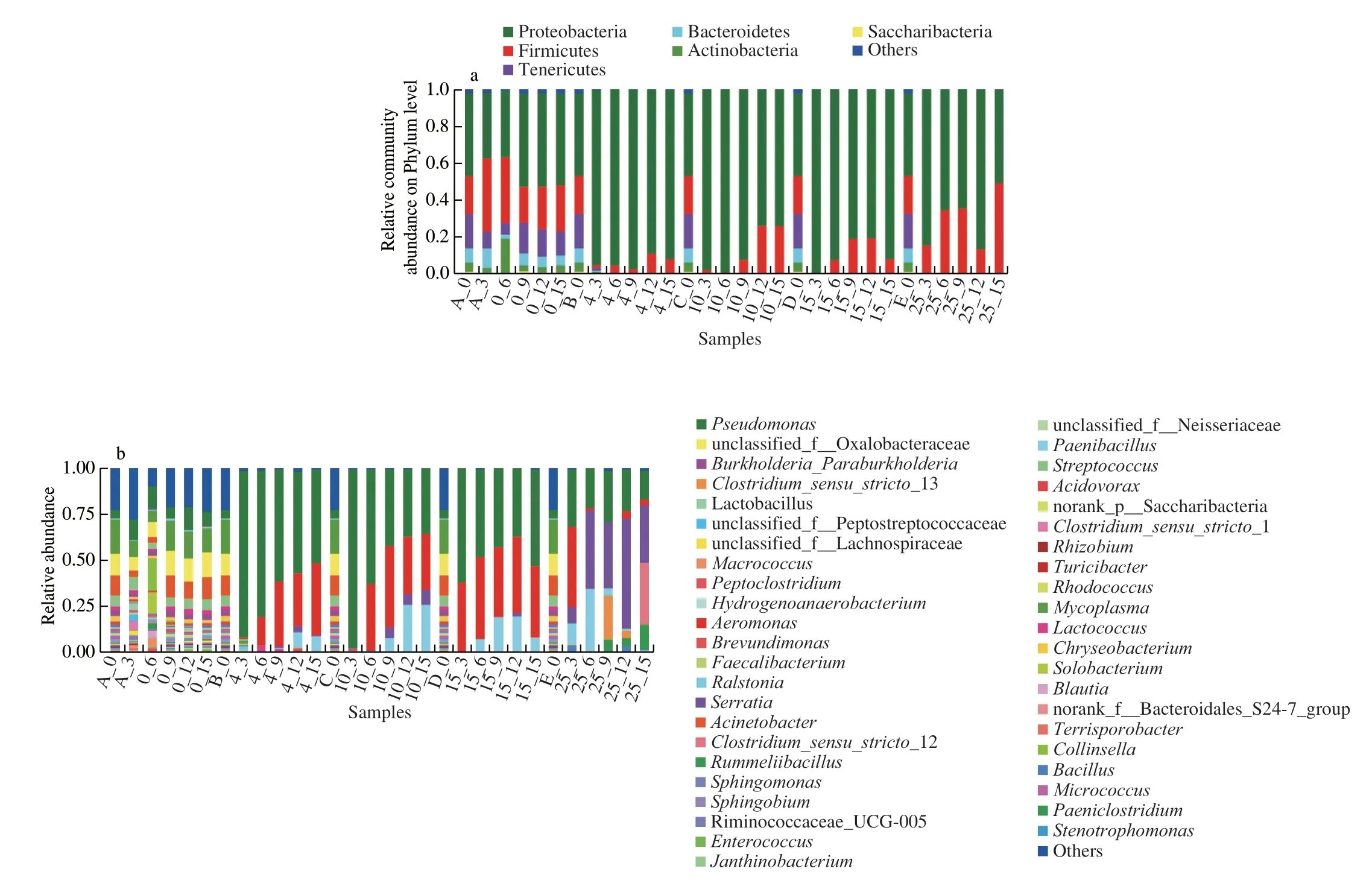
Fig. 4 Residue of microbial community in storage pasteurized milk samples. The relative abundance species of bacteria at the (a) phylum and (b) genus level.Taxonomic profiles of sample groups on the phylum level. Top abundant phyla were defined as those that were > 1% of total sequences across all samples combined; the remainder were combined and lumped in a category designated as ‘Others’.
bacteria among the identified phyla (P< 0.01), accounting for more than 50% of the bacterial populations. Similar studies have shown that raw milk is dominated by bacteria within the Firmicutes and Proteobacteria phyla, with each comprising more than 40% of the total population [24]. At 4, 10, and 15 °C, the bacterial populations became significantly less diverse over time, with the Proteobacteria population increasing as a unique phylum, whereas the populations of Tenericutes and Firmicutes declined gradually. Furthermore,the abundance of Proteobacteria was almost the same as that of Firmicutes at 25 °C for 15 days, presumably because of the growth of psychrotrophic bacteria, with the population differences observed among the samples being statistically significant (Fig. 4a).
3.3 Diversity of bacterial community in pasteurized milk
At the genus level, the high-throughput sequencing results showed the presence of 44 diverse genera in the pasteurized milk samples. Interestingly, microorganisms in the samples stored at 0 °C showed rich diversity and remained generally consistent for 15 days.At 0 °C, the four dominant populations wereMycoplasma(12.97%),unclassified-f-Oxalobacteraceae (10.97%),Pseudomonas(9.08%),andAcinetobacter(8.71%). Furthermore, the microorganisms in pasteurized milk stored at 4, 10, and 15 °C varied greatly with storage temperatures and times. At 4 °C,PseudomonasandAeromonaspopulations increased significantly (P< 0.05), with their presence comprising 57.15% and 19.95% of the total microflora, respectively(Fig. 4b).Pseudomonasis reported to be one of the most harmful psychrotrophic bacteria in raw milk. It can easily become a dominant bacterial group under cold storage conditions [25,26]. Relevant studies have shown that long-term storage can straightforwardly lead to population transformation, in which psychrophilic microorganisms,especiallyPseudomonasandAcinetobacter, become the predominant microorganisms [27]. The presence of a large number of psychrophilic bacteria is considered to be the most common cause of pasteurized milk deterioration [28-30]. At the same time, this study had found that the abundance ofAeromonas increased significantly after 6 days of storage at 4 °C, 9 days storage at 10 °C, and 6 days storage at 15 °C.Previous studies have shown thatAeromonasis widespread in the environment and has been isolated from a variety of foods,including raw milk [31]. However, previous data have shown that it has been associated with spoilage of refrigerated foods of animal origin other than raw milk [32], which coincides with the results of our storage experiment. Additionally, recent studies have shown that the genusSerratiahas also been detected in dairy processing factories [33], raw milk samples stored at 4 °C [34-36],and in bulk milk storage tanks [37]. In our study,Serratiawas detected in the pasteurized milk samples stored at 4 °C. Furthermore,the diversity of pasteurized milk bacterial flora changed significantly when the storage temperature was raised at 25 °C. After 6 days of storage, the population of the predominantAeromonasdecreased significantly and that ofSerratiabacteria increased sharply to become the dominant bacteria which lasted until the 15thday. Additionally,two species of anaerobicBacillus(Clostridium sensu stricto-12 andC. sensu stricto-13) andRummeliibacilluswere found during storage of the pasteurized milk samples for 9 days. These two bacteria have been known to cause inflammation, septicemia, and other diseases in humans and animals [31].
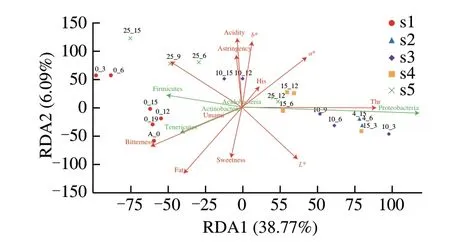
Fig. 5 RDA analysis of physicochemical and nutritional properties characteristics to changes in bacterial communities of storage pasteurized milk samples. Packaged pasteurized milk samples were storage condition at 0, 4,10, 15, and 25 °C (S1, S2, S3, S4 and S5, respectively) for 0, 3, 6, 9, 12 and 15 days as described.
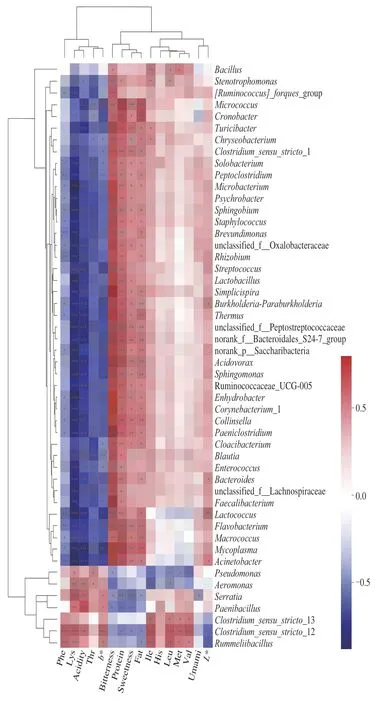
Fig. 6 Heatmap analysis of the core bacterial communities related to the nutrient composition. Presents clustering trees of species and environmental factors (such as left and upper). *P < 0.05, ** P < 0.01, *** P < 0.001.
3.4 Core functional microbiota associated with nutritional composition of pasteurized milk samples in storage
The effects of the core nutritional compounds in milk on the microbial community composition under different storage conditions were analyzed using the RDA and correlation thermography(Figs. 5 and 6). The results showed that the bacterial community in the samples were markedly affected by nutrients, amino acids and taste values. Some bacteria showed a positive relationship with the abundance of Phe, Lys, and Thr, acidity andb* values, such asPseudomonas, Aeromonas, Serratia,andPaenibacillus. According to the previous basic indices for milk and physical and chemical indices under different storage conditions (Figs. 1 and 2), the influence of milk on related microorganisms under different storage conditions was determined. A previous study has reported similar results. The dominant bacteria detected in milk werePseudomonasandDelftiaat 7 days [38]. This may be due to the prolonged storage of dairy products resulting in fat rancidity, gelation and bitterness [39-41]. The lipolytic ability ofPseudomonasdepends on lipases secreted by these microorganisms that act mainly on α portions of triglycerides [42,43],conferring a bitter taste and an unpleasant aroma. We also analyzed the correlation between taste and colony structure, and identified 4 bacteria as core microorganisms that negatively correlated with levels of protein and fat, bitterness, and sweetness and participated in the microbial functional features of our research, promoting sugar metabolism, proteolysis, and the production of important metabolites. Furthermore, the abundance ofC. sensu-stricto-12,C. sensu-stricto-13, andRummeliibacillubacteria increased significantly at 25 °C, which then became the new dominant bacteria and positively correlated with the contents of 5 amino acids, Met,Leu, Ile, Val, and His. Therefore, analysis of microbial growth dynamics and physicochemical nutrient composition of pasteurized milk has shown that dairy products undergo reasonable deterioration during long-term storage.
4. Conclusions
Physicochemical parameters, nutritional components (16 FAAs, 7 taste values, and 8 chemical constituents), and microbial profiles were investigated, and their correlations were explored in pasteurized milk under storage in our study. The results of this study revealed that the storage temperature and duration of pasteurized milk had a significant effect on the physicochemical parameters, nutritional components,and microbiota profiles of the milk samples. Additionally, we also found that microorganisms includingPseudomonas,Aeromonas,Paenibacillus,andSerratiademonstrated the most significant changes in the abundance of the bacterial genera. Moreover, they negatively correlated with the protein content and taste value and participated in the microbial functional features of our research, which promoted sugar metabolism, proteolysis, and the production of important metabolites. Lastly, our results revealed that pasteurized milk stored at 4 °C for 0-9 days was indeed of the best quality with low microbial abundances. However, microbial abundance, after storage at 10 °C for 3 and 6 days, similar to that at 4 °C maintained good quality of the pasteurized milk. These findings, thus, provide new insights into the safety of packaged pasteurized milk under storage in addition to enhancing the knowledge about the quality control parameters of milk products in dairy industry.
Declaration of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31871831, 32172279), Liaoning Provincial Natural Science Foundation regional Joint Fund project (2020-MZLH-34),Shenyang City Youth Science and Technology Innovation Leading Talent Project (RC200495); Shenyang Science and technology innovation platform project (21-103-0-14, 21-104-0-28).
杂志排行
食品科学与人类健康(英文)的其它文章
- Emerging natural hemp seed proteins and their functions for nutraceutical applications
- A narrative review on inhibitory effects of edible mushrooms against malaria and tuberculosis-the world’s deadliest diseases
- Modulatory effects of Lactiplantibacillus plantarum on chronic metabolic diseases
- The role of f lavonoids in mitigating food originated heterocyclic aromatic amines that concerns human wellness
- The hypoglycemic potential of phenolics from functional foods and their mechanisms
- Insights on the molecular mechanism of neuroprotection exerted by edible bird’s nest and its bioactive constituents
