Complete Mitochondrial Genome Analysis of Daphniopsis tibetana (Branchiopoda: Diplostraca): New Insights into the Taxonomy of the Genus and Its Phylogenetic Implications for Branchiopoda
2022-12-27LIUBingjianPENGYingLIUYifanLIJiashengZHANGKunCHENJianGONGLiLIULiqinZhenmingandZHANGChi
LIU Bingjian , PENG Ying , LIU Yifan , LI Jiasheng , ZHANG Kun ,CHEN Jian GONG Li , LIU Liqin , LÜ Zhenming , and ZHANG Chi
1)National Engineering Laboratory of Marine Germplasm Resources Exploration and Utilization, Zhejiang Ocean University,Zhoushan 316022, China
2) Institute of Fisheries Science, Tibet Academy of Agricultural and Animal Husbandry Sciences, Lhasa 850002, China
3) Key Laboratory of Tropical Marine Bio-Resources and Ecology, Chinese Academy of Sciences, Guangzhou 510301, China
Abstract Daphniopsis tibetana is widely distributed in Xinjiang, Qinghai, Tibet of China, as well as in Russia and India, which is the dominant zooplankton in many high-altitude (4000 m)salt lakes. D. tibetana can adapt to saline waters, whereas the other species of the superorder Cladocera can only inhabit in freshwater. However, the phylogenetic status of D. tibetana in Branchiopoda remains unclear primarily because of limited mitogenome. In this study, complete mitochondrial genome sequences of D. tibetana were sequenced and annotated for the first time to obtain a comprehensive understanding of its phylogenetic status. The complete mitogenome of D. tibetana is 16196 bp in length. It contains 37 genes, including two ribosomal RNAs (12S and 16S rRNAs)genes, 22 transfer RNA (tRNA)genes, 13 protein-coding genes, and one non-coding region. The overall base composition is 29.6% A, 33.2% T,19.0% G, and 18.2% C with a high AT bias (62.8%). Except for trnS1 (GCT), most tRNAs have a typical cloverleaf secondary structure.Phylogenetic analysis based on maximum likelihood and Bayesian inference generates identical phylogenetic topology and shows the phylogenetic status of D. tibetana, which reconfirm the distinction between the genera Daphniopsis and Daphnia. Meanwhile, the class Branchiopoda is clustered into three clades (Anostraca, Notostraca, and Diplostraca)with high support values. These results provide not only a comprehensive understanding of the characteristics of D. tibetana mitogenome and its phylogenetic position in Diplostraca, but also information for future research on the phylogeny of Branchiopoda.
Key words Daphniopsis tibetana; mitogenome; phylogenetic analysis; Branchiopoda; Diplostraca
1 Introduction
Mitochondria are a subcellular organelle with important biochemical functions, which is the powerhouse of the eukaryotic cell. The mitochondrial genome is located within the organelle, which is independent of the nuclear genome but with a close relationship with each other (Wolstenholme,1992). The typical animal mitogenome is a closed circular molecule of 14 – 20 kilobases (kb)containing 13 proteincoding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (12Sand16S), and an AT-rich region(also known as the control region, CR)(Boore, 1999). At present, mitochondrial DNA is considered an efficient and reliable molecular marker for phylogenetic studies (Wanget al., 2008). Given that amplification is non-recombining,fast evolving, and relatively easy, the circular mitochondrial genomes have been widely used in the studies of phylogenetics, population genetics, and evolutionary genomics(Neet al., 2011; Tanet al., 2015). In addition, the number of complete mitochondrial genomes increases rapidly on the basis of next-generation sequencing technology, which allows a comprehensive understanding of the complete mitogenome (Fendtet al., 2009; Wanget al., 2020).
Branchiopoda, derived from the Cambrian of the Paleozoic, comprises two subclasses and three orders: Sarsostraca, including Anostraca; Phyllopoda, including Notostraca and Diplostraca (Joel, 1995). In addition, Brendoncket al. (2008)found that several Branchiopod species were endangered because of the reduced quantity and quality of temporary wetlands. Meanwhile, the inter-ordinal and evolutionary relationships of many Branchiopods with lowlevel taxa remain unclear (Brendoncket al., 2008). Previously, considerable research preferred to construct taxonomy and phylogenetics of Branchiopods using the morphology of sex symbols rather than mitogenomes (Linder, 1941).These factors have posed a great challenge to researchers in the study of Branchiopod species. Recently, several researchers have attempted to establish phylogenetic relationships to solve particularly systematic problems of high-level taxa in Branchiopods using mitogenomes (Olesen, 1998;Liuet al., 2015; Tokishitaet al., 2017; Bellecet al., 2019;Tladiet al., 2020). However, these mitogenome phylogenetic studies have included only a few representatives of the diversity of Branchiopoda because of the meager mitogenome sequences for most order-level taxa in Branchiopoda. Therefore, re-establishing Branchiopod phylogenetic relationship using suitable species is necessary.
Daphniopsis tibetanabelongs to Crustacea, Branchiopoda, Diplostraca, Cladocera, and Daphniidae. It is the dominant zooplankton and apex species in many high-altitude(4000 m)salt lakes (Zhaoet al., 2016; Wanget al., 2019).In addition,D. tibetanacan adapt to saline water, whereas other species in Cladocera can only inhabit in freshwater(Zhaoet al., 2004). Therefore, it plays a role in the study of Cladocerans (Zhao and Li, 2012). The phylogenetic status ofD. tibetanawill provide a comprehensive understanding of the lake ecosystem. At present, the ecological distribution (Zhaoet al., 2002), morphological characterization (Zhao and Wang, 2005), nutritional analysis (Zhaoet al., 2006), oxygen consumption rate (Zhaoet al., 2007),and chromosome karyotype (Zhaoet al., 2004)ofD. tibetanawere investigated. Moreover, the molecular approach of investigation has focused onD. tibetana. Colbourneet al. (2006)examined the molecular genetic divergence and evolution ofDaphniaandDaphniopsison the basis ofCOI,16S rDNA, and12S rDNAsequences, indicating thatD. tibetanashould be classified in the genusDaphnia.However, Zhaoet al. (2011)constructed a phylogenetic tree based on18S rDNAand found that the taxonomic level ofD. tibetanawas closer toDaphniopsisthan toDaphnia. Since then, the systematic classification ofD. tibetanahas been controversial for a long time.
As the largest order of the Branchiopoda class, the relationship among branches in Diplostraca is unclear (Olesen, 1998; Brabandet al., 2002; Dewaardet al., 2006).Moreover, the phylogenetic significance based on the complete mitochondrial sequence ofD. tibetanahas not been investigated because of the lack of mitogenome sequences in Diplostraca. In this study, the first complete mitogenome sequences ofD. tibetanawere sequenced, annotated, and characterized. The phylogenetic tree (Bayesian inference(BI)and maximum likelihood (ML))was constructed on the basis of 13 PCGs to assess the systematic position ofD. tibetanain Diplostraca and provide new insights into the phylogenetic implications for Branchiopoda. Therefore,the complete mitogenome ofD. tibetanawill provide a comprehensive understanding of the phylogenetic relationships within Branchiopoda.
2 Materials and Methods
2.1 Sample Collection and DNA Extraction
An adult specimen ofD. tibetanawas collected from a saline lake in Tibet, China (30˚44′13.1′′N, 90˚33′33′′E).The specimen was then stored in 95% ethanol until DNA extraction. Whole genomic DNA was extracted from a single specimen using an improved method and multi-well plates according to the manufacturer’s instructions (Yue and Orban, 2010). Finally, the extracted DNA was stored at -20℃until used for PCR amplification.
2.2 Mitogenome Sequencing and Assembly
The complete mitogenome ofD. tibetanawas sequenced using next-generation sequencing. First, iTru adaptors and primers and the KAPA Hyper Plus kit were used to construct an Illumin library following the manufacturer’s protocol (Glennet al., 2019). Then, the library was sequenced on an Illumina HisSeq 4000 PE150. In addition, the raw reads were assessed and cleaned using FastQC and Trimmomatic, respectively (Bolgeret al., 2014; Brandine and Smith, 2019). Finally, NOVOPlasty (Dierckxsenset al.,2017)was used to assemble the paired reads. Moreover,De NovoAssemble, Map to Reference, and BLAST were used in accordance with Geneious 11.1.2 to verify the resulting mitogenome (Altschulet al., 1997; Ripmaet al.,2014).
2.3 Sequence Annotation and Analysis
For 37 mitochondrial genes, 13 PCGs were annotated using Sequin version 15.10 (http://www.ncbi.nlm.nih.gov/Sequin)and MITOS2 (Berntet al., 2013)and compared with other Diplostraca mitochondrial genomes from Genbank. NCBI-BLAST (http://blast.ncbi.nlm.nih.gov)and tRNAscan-SE 1.21 (Schattneret al., 2005)were used to identify the boundaries of rRNA and tRNA genes, respectively. The mitochondrial genome map was constructed using OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html)(Lohseet al., 2007). The secondary tRNA structure was predicted through the MITOS webserver(Berntet al., 2013). MEGA (version 10.1.8)(Sudhiret al.,2018)was used to analyze the nucleotide composition and relative synonymous codon usage (RSCU). The bias of nucleotide composition was calculated using the following formulas (Perna and Kocher, 1995):
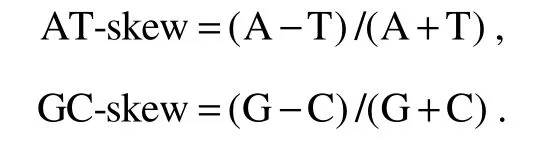
2.4 Phylogenetic Analysis
Phylogenetic analysis was conducted on the basis of the nucleotide sequences of 13 PCGs using BI and ML to assess the systematic position ofD. tibetana. 27 complete mitogenome sequences of Branchiopoda downloaded from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank)and one newly determined sequence (D. tibetana)were used to reconstruct the phylogenetic relationships among Branchiopoda (Table 1). In addition, two species from Archosauria were used as outgroups (Table 1). First, Phylo-Suite (Zhanget al., 2020)was used to extract nucleotide sequences of 13 PCGs for each species from GenBank files.Then, the MAFFT program (Toh, 2010)integrated into PhyloSuite was used to align the multiple sequences in normalalignment mode. Moreover, the alignment results were imported to Gblocks (Gerard and Jose, 2007)to identify and remove ambiguously aligned regions. Afterward, the sequences were concatenated and used to generate input files(phylip and nexus format)for phylogenetic analyses. ModelFinder (Kalyaanamoorthyet al., 2017)was used to select the best-fit model based on the Bayesian information criterion. The best-fit models, namely, TVM + F + R5 and GTR + F + I + G4, were selected to perform ML and BI analyses, respectively. Next, ML analysis was conducted in IQTREE (Lamet al., 2015)using an ML + rapid bootstrap algorithm with 1000 replicates (Guindonet al., 2010). Furthermore, BI analysis was performed in MrBayes 3.2.6 (Nylanderet al., 2004)using default parameters and 3 × 106Metropolis-coupled Markov Chain Monte Carlo generations and sampled every 100 generations with a burn-in of 25%.The average standard deviation of split frequencies below 0.01 was considered to reach convergence. Finally, the resulting phylogenetic trees were visualized through iTOL(https://itol.embl.de/)(Letunic and Bork, 1988).
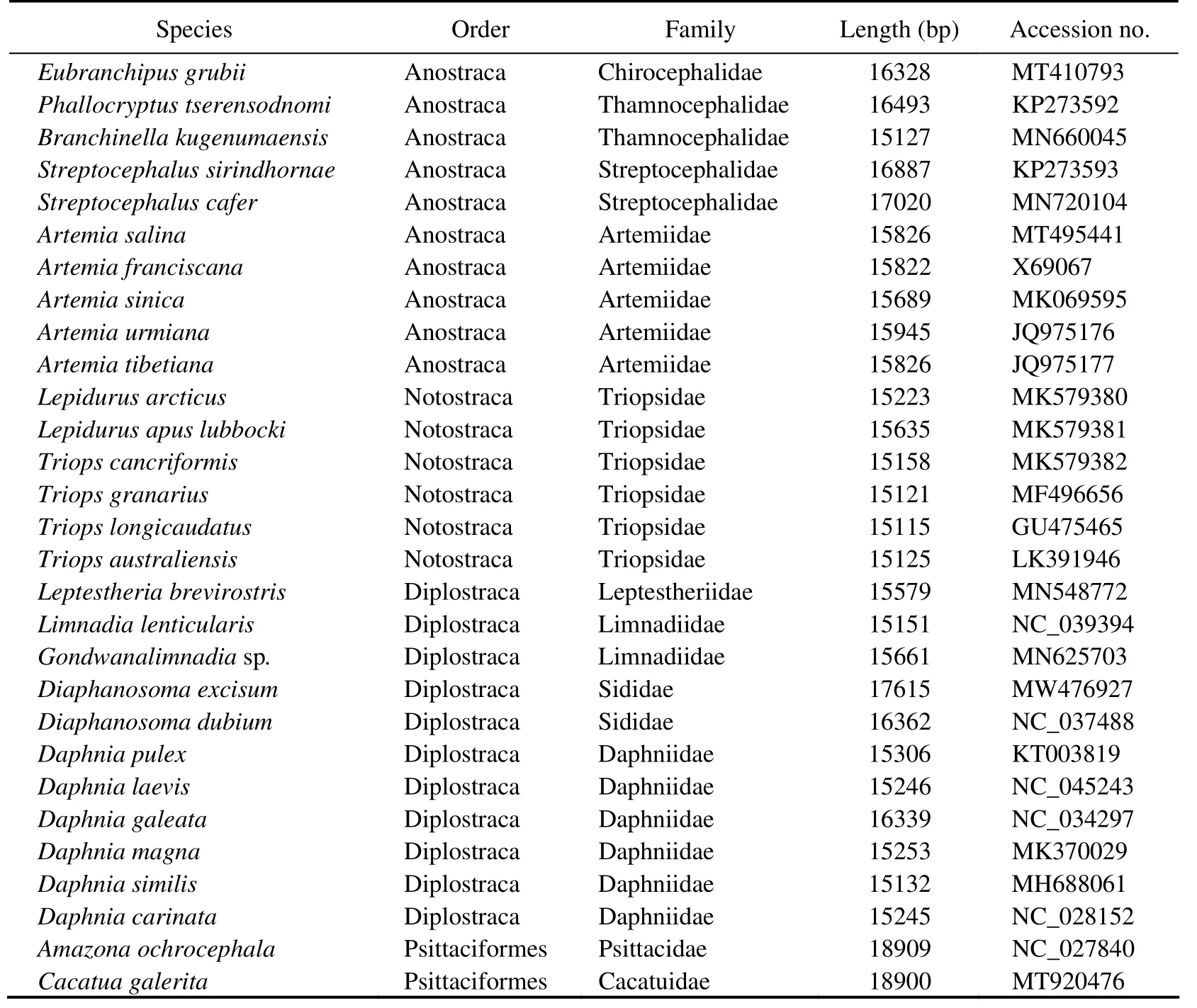
Table 1 List of 27 Branchiopoda species and two outgroups used in this paper
3 Results and Discussion
3.1 Genome Organization and Base Composition
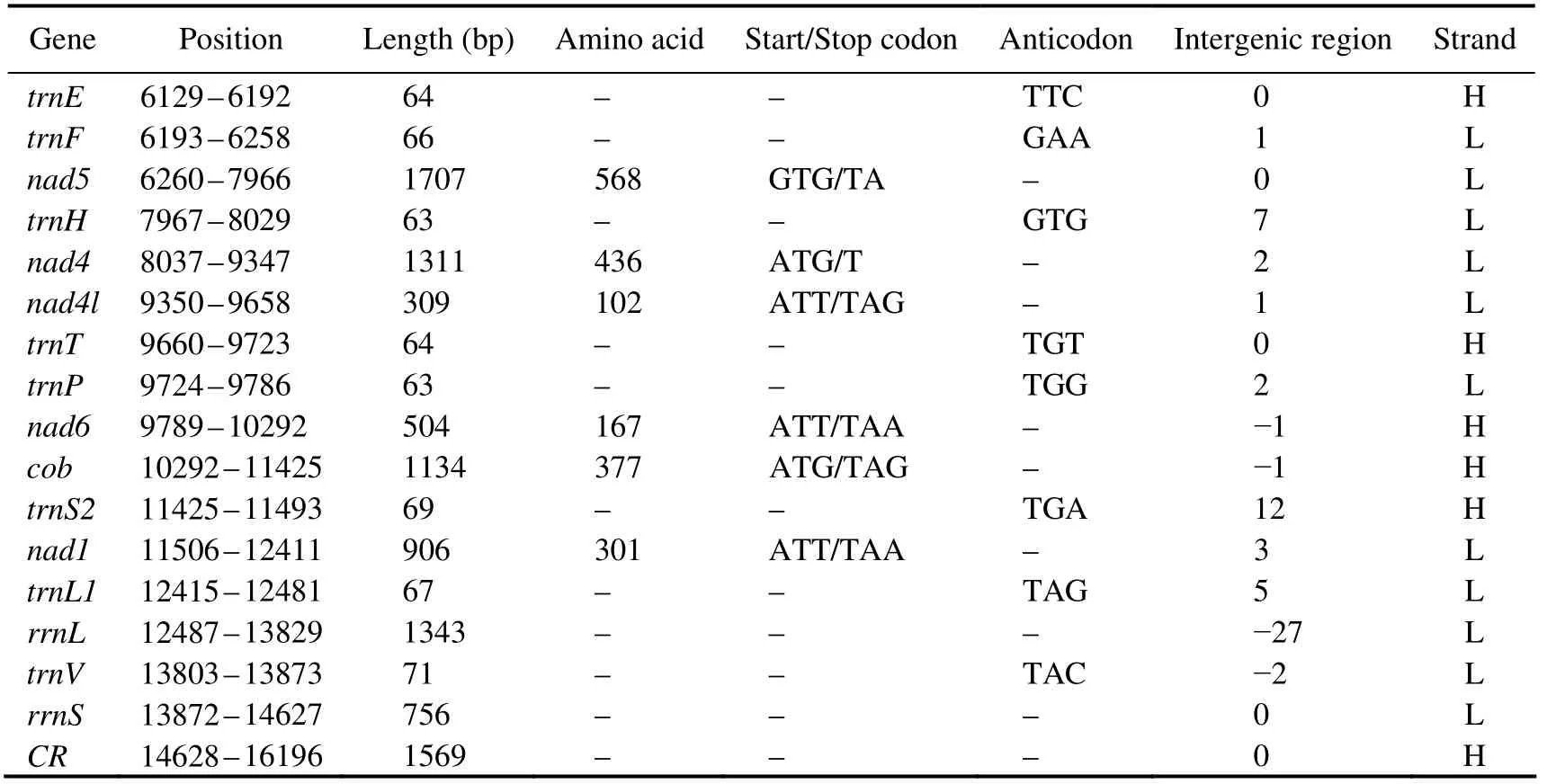
The complete mitogenome ofD. tibetanais a closed circular molecule, which is 16196 bp long (GenBank accession number MW981579). This mitogenome contains 13 PCGs, two rRNAs, 22 tRNAs, and a putative CR, which is consistent with most published studies on Diplostraca (Fig.1,Table 2)(Crease, 1999; Liuet al., 2017). In addition, most of the mitochondrial genes are distributed on the heavy(H-)strand except for four PCGs (nad5,nad4,nad4l, andnad1), eight tRNAs (trnQ,trnC,trnY,trnF,trnH,trnP,trnL1, andtrnV), and two rRNAs, which are encoded on the light (L-)strand (Table 2). Similar to other invertebrate mtDNAs, overlapping (nine overlaps totaling 48 bp)and non-coding bases among genes are found (Table 2)(Wanget al., 2020). The overall base composition ofD. tibetanais 29.6% A, 33.2% T, 18.2% C, and 19.0% G. Calculations show that the GC-skew value and AT-skew value are 0.021 and -0.059, respectively (Table 3). The results suggest that AT base migration primarily occurs in the hysteretic chain,whereas GC base migration primarily occurs in the leading chain, which is similar to other Diplostraca species (Bellecet al., 2019). Moreover, the composition of nucleotide is highly A + T biased (62.80%), as exhibited in other Diplostraca mitogenomes (Tokishitaet al., 2017). Furthermore,the A + T contents of PCGs, tRNAs, and rRNAs are 61.2%,65.6%, and 68.2%, respectively (Table 3).
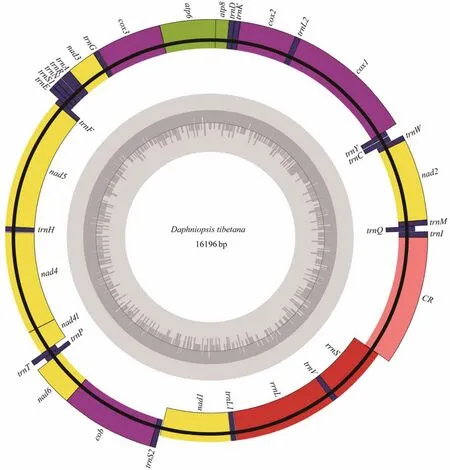
Fig.1 Gene map of the D. tibetana mitogenome. Genes encoded on the heavy or light strands are shown at the outside or inside of the circular gene map, respectively.
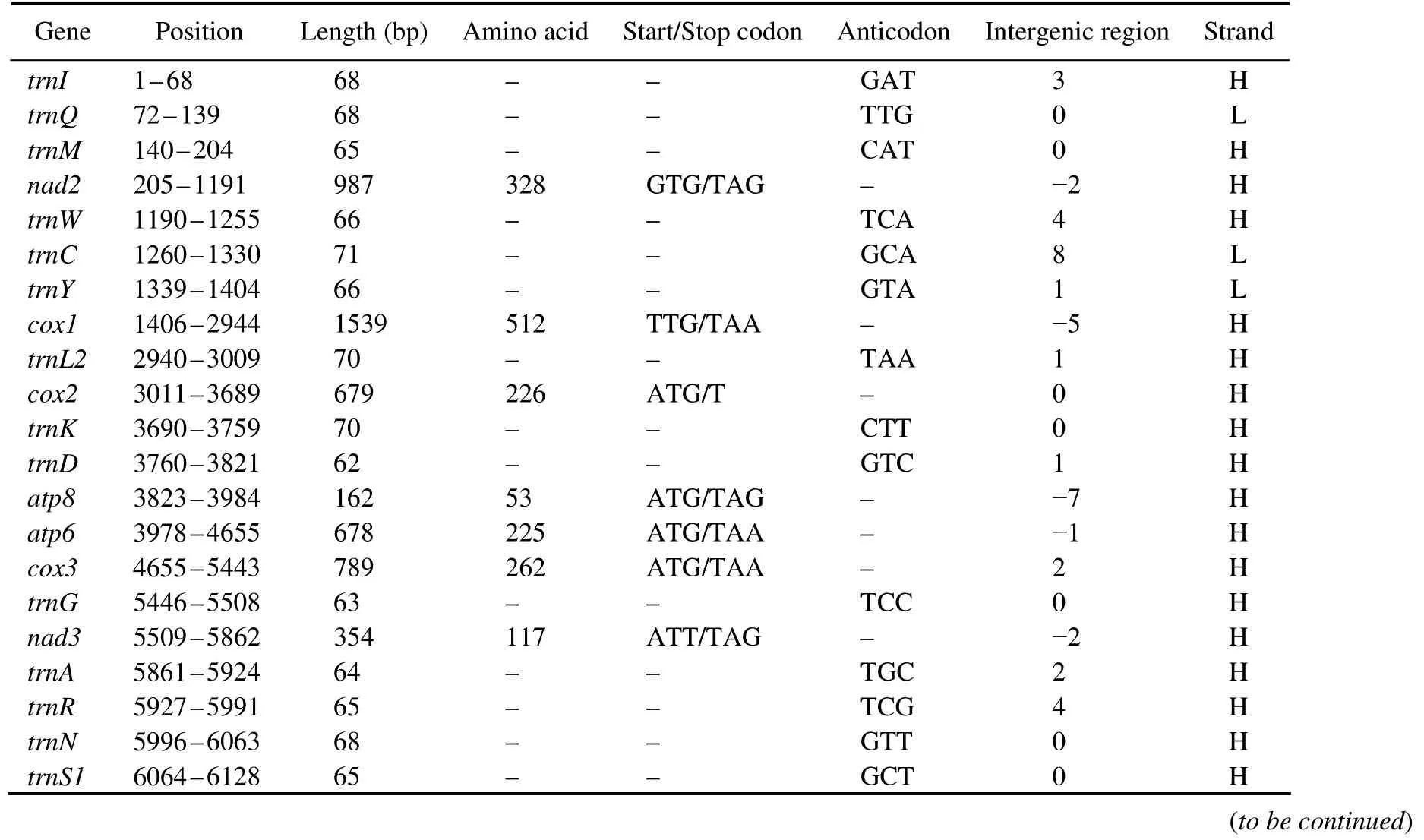
Table 2 Features of the mitochondrial genome of D. tibetana
3.2 Protein-Coding Genes and Codon Usage
(continued)
The 13 PCGs of this sequence is 11059 bp long, containing seven NADH dehydrogenases (nad1–nad6andnad4l),three cytochrome c oxidases (cox1–cox3), two ATPases(atp6andatp8), and one cytochrome b (cob), which were also found in previous studies on Diplostraca species (Table 2)(Crease, 1999; Liuet al., 2017). The length of the PCGs ranges from 162 bp (atp8)to 1707 bp (nad5)(Table 2), encoding a total of 3674 amino acids. The total AT bias of 13 PCGs inD. tibetanais 62.80%, ranging from 59.0%(cox1)to 71.0% (atp8)(Table 3). Furthermore, all AT-skew of 13 PCGs show notable negative values, and most GCskew values are positive, indicating that Ts and Gs are more than As and Cs (Table 3). Of the 13 PCGs, six are initiated by the canonical start codon ATG (cox2,atp8,atp6,cox3,nad4, andcob), while two use GTG (nad2andnad5), four use ATT (nad1,nad3,nad4l, andnad6), and one use TTG(cox1). These rare start codons have been found in other animals (Wolstenholme, 1992), and they are similar to some Daphnia species (Tokishitaet al., 2017). For the stop codon, 10 PCGs perform the routine termination codon (TAA or TAG), whereas three other PCGs (cox2,nad5, andnad4)are with an incomplete stop codon T or TA (Table 2). Stop codons are consistent with other Diplostraca species (Liuet al., 2017).
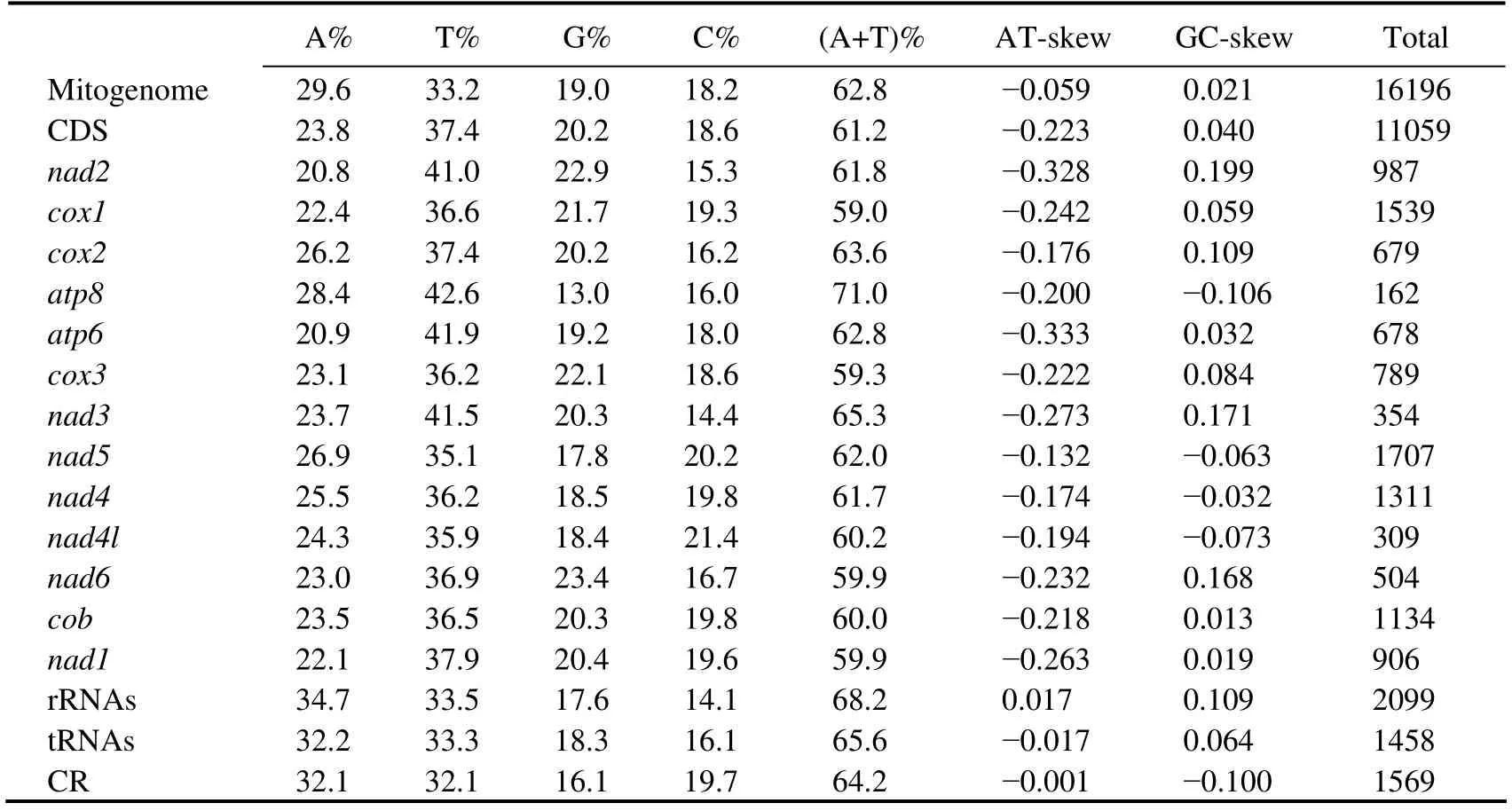
Table 3 Composition and skewness of D. tibetana mitogenome
Among the 3674 amino acids encoded by 13 PCGs, the frequently used amino acids include Leu(16.03%), Ser(9.03%), Phe(8.60%), and Val(8.21%). By contrast, the least common amino acids include Glu(2.06%), Arg(1.79%),Gln(1.79%), and Cys(0.10%)(Fig.2A, Table 4). In addition, RSCU analysis shows that the frequently used codons are UCU(S), UUA(L), and CCU(P), whereas CUC(L),ACG(T), CCG(P), and AGG(S)are the least often used codons (Fig.2B, Table 4). The preference for NNU codons can be found in the mitochondrial PCGs, which is similar to other invertebrate animals (Wanget al., 2015; Zhanget al.,2020).
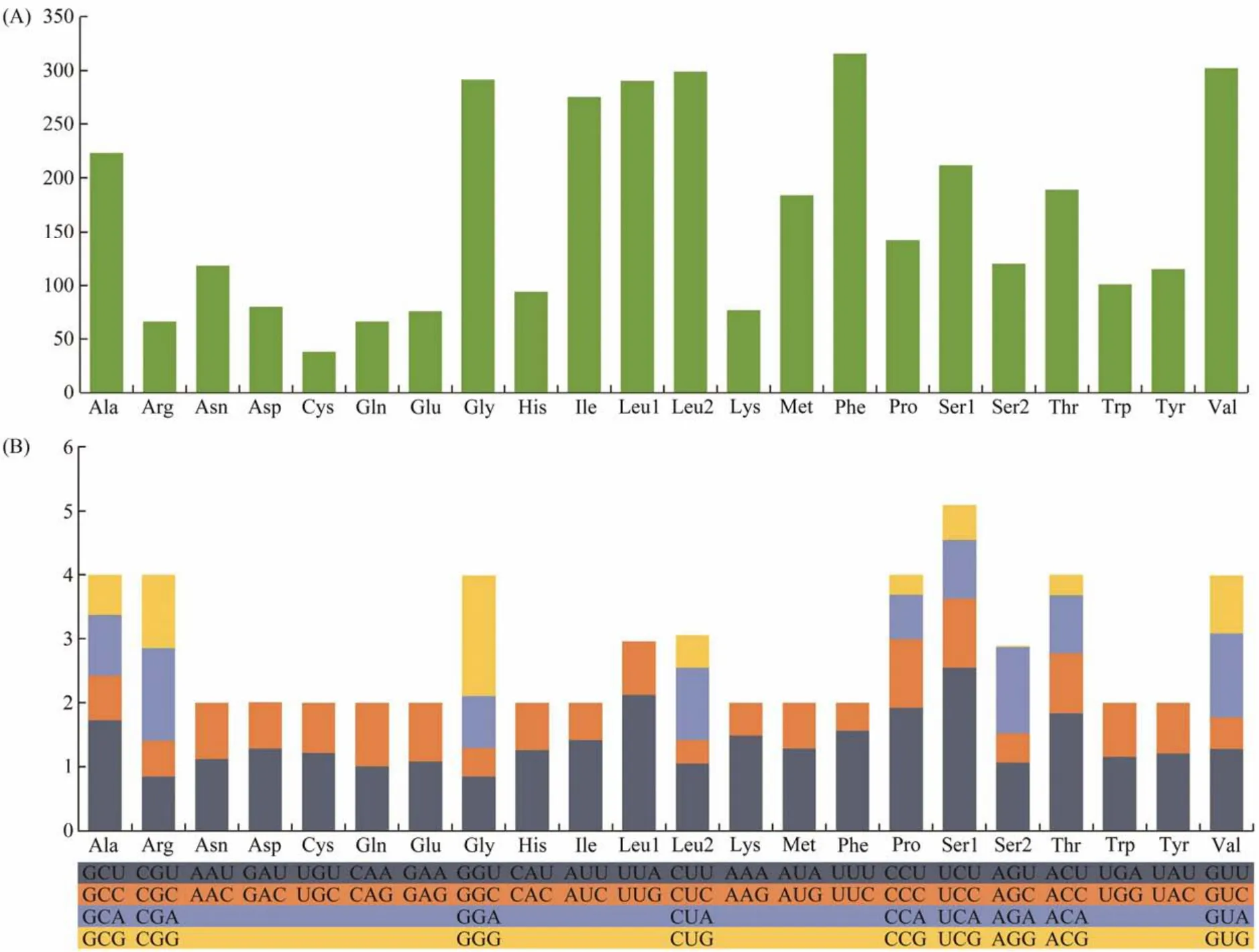
Fig.2 Amino acid composition (A)and relative synonymous codon usage (B)in the mitogenome of D. tibetana.
3.3 Transfer RNAs (tRNA), Ribosomal RNAs(rRNA), and Control Region
A total of 22 tRNAs (total length 1458 bp)are identified in the mitogenome ofD. tibetana, ranging from 63 bp (trnG)to 71 bp (trnCandtrnV)in length (Table 2). The AT bias of 22 tRNAs is high (65.6%). The AT-skew (-0.017)of tRNAs is lower than the GC-skew (0.064), indicating that Ts and Gs are more than As and Cs (Table 3). Eight tRNAs(trnQ,trnC,trnY,trnF,trnH,trnP,trnL1, andtrnV)are encoded by the L-strand, whereas the other tRNAs are encoded by the H-strand. Moreover, onlytrnS1(TCT)lacks a dihydrouridine arm when most tRNAs can fold into the typical cloverleaf structure (Fig.3), which is a common feature in vertebrate and invertebrate mitogenomes (Fig.3)(Zhanget al., 2019; Luet al., 2020). Based on the Watson-Crick base pair (A– T and G– C)matches (Holbrooket al.,1991), a total of 15 unmatched base pairs (G– U pairs)are found in theD. tibetanamitochondrial tRNA genes (trnA,trnQ,trnR,trnD,trnC,trnQ,trnG,trnH,trnI,trnL1,trnM,trnP,trnS1,trnW,trnY, andtrnV; Fig.3), forming a weak bond.
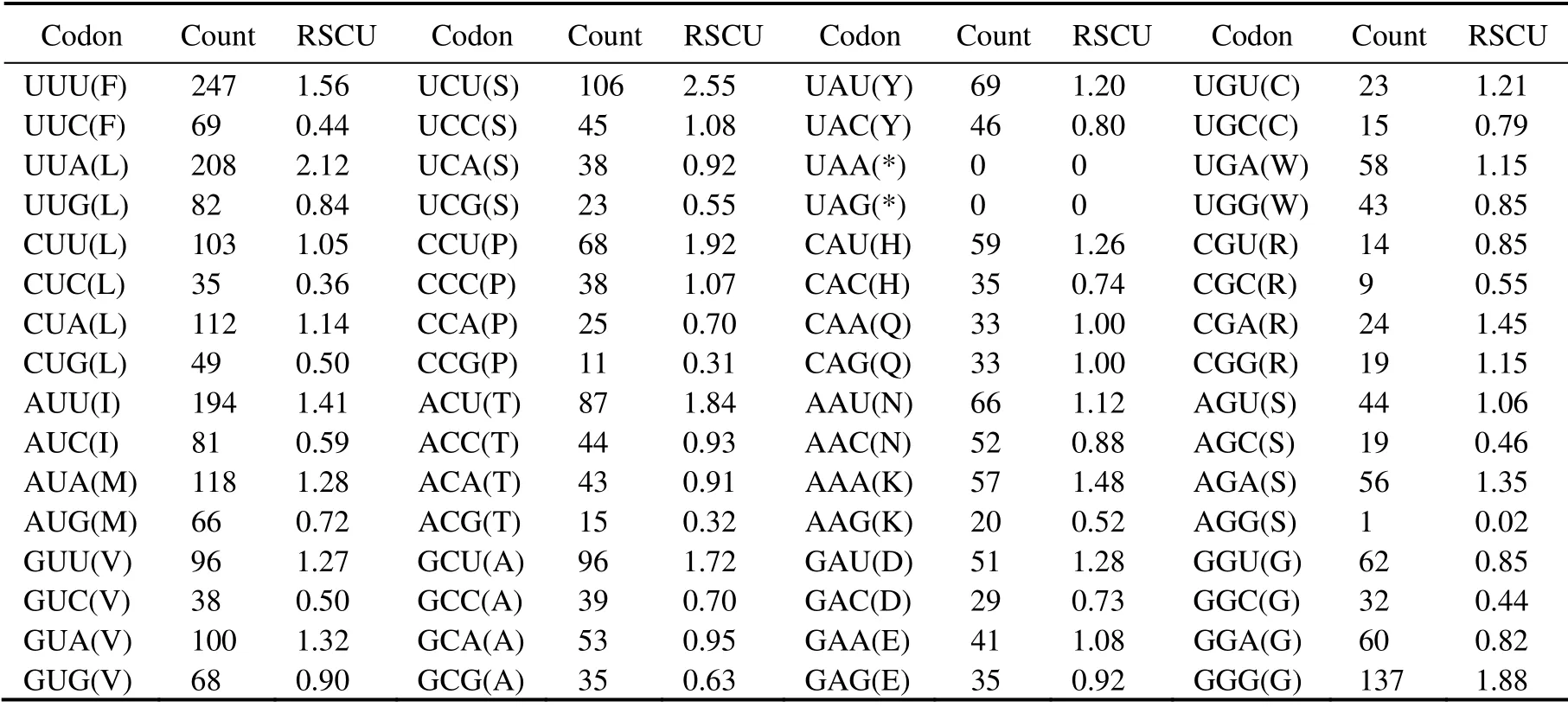
Table 4 Codon number and relative synonymous codon usage (RSCU)of 13 PCGs in D. tibetana mitogenome
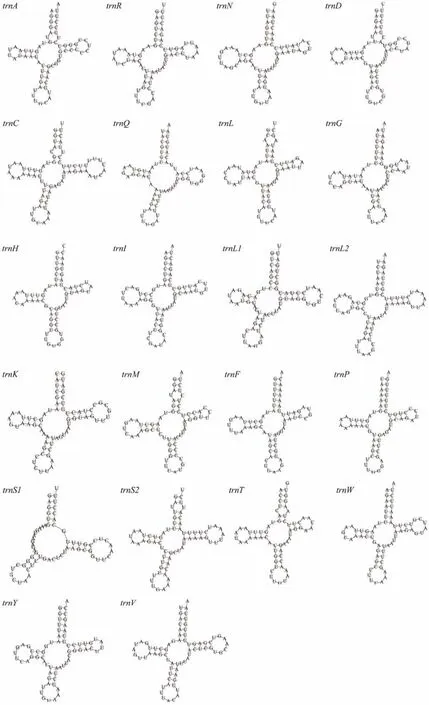
Fig.3 Potential secondary structures of 22 inferred tRNAs in D. tibetana mitogenome.
As shown in Table 2, the mitogenome ofD. tibetanacontains two rRNAs,rrnLandrrnS,which are 1343 and 756 bp long.Theyare typically separated bytrnV,and both rRNAs are located on the L-strand (Table 2), which are consistent with most invertebrate mitogenomes (Kimet al.,2016; Luet al., 2020). The total A + T content of two rRNAs is 68.2%, and the AT-skew and GC-skew exhibit a positive value (0.017 and 0.109, Table 3), indicating that As and Gs are more than Ts and Cs.
The CR of the mitogenome is located betweenrrnSandtrnI. In comparison with rRNA and PCGs genes, CR is the dominant region for evaluating intraspecies genetic variations caused by the high variation and mutation rates throughout the mitogenome (Parsonset al., 1997; Normanet al.,2010). The 1569 bp CR is evidently AT-biased (64.2%), and the AT-skew and GC-skew in CR are -0.001 and -0.100,respectively, indicating an evident bias toward the use of Ts and Cs.
3.4 Phylogenetic Analysis
Two phylogenetic trees (ML and BI)were constructed on the basis of the sequences of 13 PCGs, including 28 species of Branchiopoda and two outgroup species, to investigate the phylogenetic position ofD. tibetanawithin Branchiopoda. ML and BI trees show an identical topology; thus,only one topology (ML)with both support values is displayed. However, the BI tree has a higher support value than the ML tree (Fig.4). The phylogenetic trees show that the class Branchiopoda is divided into three major clades(Diplostraca, Anostraca, and Notostraca)and nine families.First, the order Anostraca, includingA. urmiana,A. tibetiana,A. salina,A. franciscana,A. sinica,B. kugenumaensis,S.cafer,S. sirindhornae,P. tserensodnomi, andE. grubii, belong to four families (Chirocephalidae, Thamnocephalidae,Streptocephalidae, and Artemiidae), which show a sister relationship with Notostraca and Diplostraca. Meanwhile, the relationship among families Chirocephalidae, Thamnocephalidae, and Streptocephalidae is closer than the relationship with Artemiidae. In this clade of Anostraca, most terminal relationships are well resolved, which is consistent with the previous studies (Danielset al., 2015; Bellecet al.,2019). Then, Notostraca with its two representative generaTriops(Schrank, 1803)andLepidurus(Leach, 1819), belonging to the family Triopsidae, is considered as a monophyletic group in the present mitogenomic phylogeny, which was also proven by Longhurst (1955)and Brendoncket al.(2008).
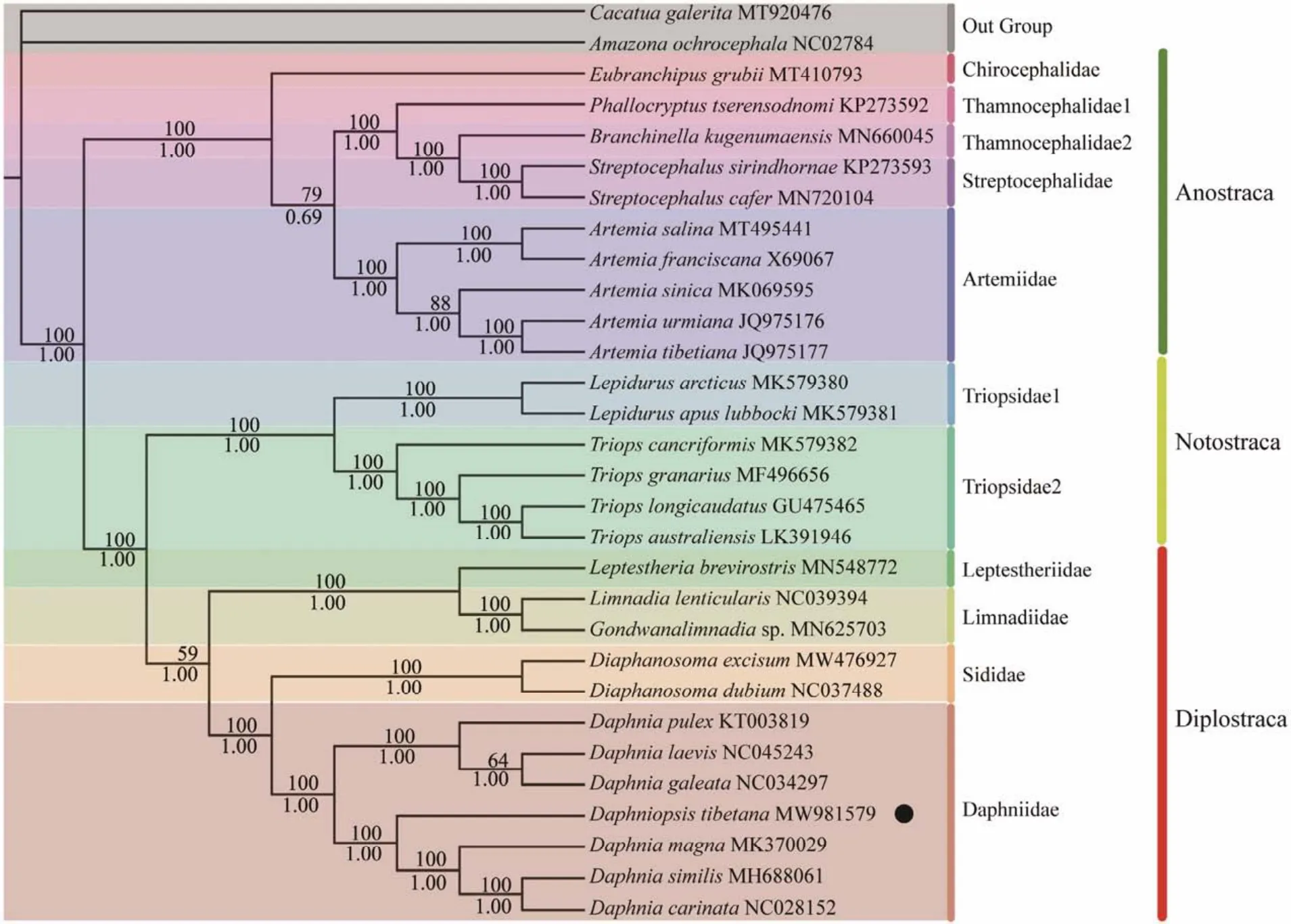
Fig.4 Phylogenetic tree of Branchiopoda species inferred from the nucleotide sequences of 13 PCGs based on maximum likelihood (ML)and Bayesian inference (BI)analyses. The dots indicate the species studied in this paper.
In addition, three clades (Anostraca, Notostraca, and Diplostraca)in this phylogenetic tree show high support values except for Diplostraca (posterior probabilities = 1; bootstrap = 59; Fig.4), indicating that the target species (D. tibetana)in Diplostraca is not highly differentiated. At the bottom of the tree, there are 12 Diplostraca species, includingL. brevirostris,L. lenticularis,Gondwanalimnadiasp.,D. excisum,D. dubium,D. pulex,D. laevis,D. galeata,D.tibetana,D. magna,D. similis, andD. carinata. They are separated into four clades, Daphniidae, Sididae, Limnadiidae, and Leptestheriidae. Seven species (D. pulex,D. laevis,D. galeata,D. tibetana,D. magna,D. similis, andD. carinata)cluster together as the main clade Daphniidae species. The results are similar to the previous phylogenetic tree of Bellecet al. (2019). BI and ML trees show thatD.tibetanais close toD. magna,D. similis,andD. carinata,forming a sister group at the end of the trees, which belongs to Diplostraca and Daphniidae. On the contrary, Sars(1903)erected the genusDaphniopsisthrough his description ofD. tibetanaand found thatDaphniopsiswas intermediate toSimocephalusandDaphnia. In 1936, Wagler assigned the genus toDaphniaand not toDaphniopsis, and a taxonomic debate on the designation of this genus began (Zhao and Wang, 2005). Later, other researchers recognized that the validity of the genusDaphniopsiswas closer toDaphniathan toSimocephalus(Hann, 1986).Moreover, Zhao and Li (2012)found that the differences in mitochondrial12S rRNAindicated thatDaphniopsistibetanaandDaphniatibetanawere different species and that the two genera were highly similar, which is consistent with the results of this study using whole mitogenomes ofD. tibetana. However, considering only 12 mitogenomes of this order, more sequences of Branchiopoda are needed to confirm the phylogenetic position of Diplostraca. In future studies, more taxa samplings are required to conclusively resolve the origin and evolutionary relationships among Branchiopoda.
4 Conclusions
In this study, the complete mitogenome ofD. tibetanawas sequenced and characterized, which is the first complete mitogenome of the genusDaphniopsiswithin Branchiopoda. The mitogenome ofD. tibetanais 16196 bp long,which is a typical mitogenome of Branchiopoda with 37 genes and an AT-rich region. The overall base composition is 29.6% A, 33.2% T, 19.0% G, and 18.2% C, which contains a high AT bias (62.8%)and exhibits a negative ATskew (-0.059)and a positive GC-skew (0.021). ML and BI phylogenetic trees show that Branchiopoda species are classified into three major clades (Anostraca, Notostraca, and Diplostraca)with high support values. Meanwhile, the tree shows thatD. tibetanahas a close relationship withD. magna,D. similis,andD. carinata,but it belongs to the genusDaphniopsis, which is consistent with previous studies based on morphology. These results not only reconfirm the phylogenetic positionofD. tibetanain Diplostraca, but also reconstruct the phylogenetic relationship between Diplostraca and other orders in Branchiopoda.
Acknowledgements
This study was supported by the Zhejiang Provincial Natural Science Foundation of China (Nos. LY22D060001,LY20C190008), the National Natural Science Foundation of China (Nos. 41806156, 31702321), the Fund of Guangdong Provincial Key Laboratory of Fishery Ecology and Environment (FEEL-2021-8), the Open Foundation from Key Laboratory of Tropical Marine Bio-resources and Ecology, Chinese Academy of Sciences (No. LMB20201005),the Science and Technology Project of Zhoushan (No. 2020 C21016), the Open Foundation from Marine Sciences in the First-Class Subjects of Zhejiang (Nos. 20200201, 2020 0202), and the Starting Research Fund from the Zhejiang Ocean University.
杂志排行
Journal of Ocean University of China的其它文章
- Comparison of Flavor Substances in Dried Shrimp Products Processed by Litopenaeus Vannamei from Two Aquaculture Patterns
- Identifying Summer/Autumn Habitat Hotspots of Jumbo Flying Squid (Dosidicus gigas)off Chile
- Allelopathic Interactions Between the Tropical Macrophyte Enhalus acoroides and Epibenthic HAB Dinoflagellate Prorocentrum concavum
- Removal of Arsenic from Chlamys farreri with Different Methods
- Optimal Culture Capacity of White Shrimp (Litopenaeus vannamei)and Razor Clam (Sinonovacula constricta)in a New Series-Connection Culture Model
- Identification of Non-Coding RNAs Based on Alignment-Free Features in Crassostrea gigas (Pacific Oyster)Transcriptome
