A highly effective polycyclic aromatic hydrocarbon-degrading bacterium,Paracoccus sp.HPD-2,shows opposite remediation potential in two soil types
2022-11-01WeiCHENYingTENGWenjieRENYongmingLUOandYaoYU
Wei CHENYing TENGWenjie RENYongming LUO and Yao YU
1KeyLaboratoryof Soil Environment and Pollution Remediation,Institute of Soil Science,Chinese Academyof Sciences,Nanjing 210018(China)
2Institute of Agricultural Resources and Environment,Jiangsu Academyof Agricultural Sciences,Nanjing 210014(China)
3School of Resources and Environmental Engineering,Hefei Universityof Technology,Hefei 230009(China)
ABSTRACT Bioaugmentation is an efficient and eco-friendly strategy for the bioremediation of polycyclic aromatic hydrocarbons(PAHs).Since the degrading abilities of soils can greatly alter the abilities of PAH-degrading bacteria,illustrating the potential and mechanism of highly efficient degrading bacteria in different soil environments is of great importance for bioremediation.A PAH-degrading bacterium,Paracoccus aminovorans HPD-2,and two soil types,red and paddy soils,with distinct PAH-degrading abilities,were selected for this study.A soil microcosm experiment was performed by adding pyrene(PYR)and benzo[a]pyrene(B[a]P).Illumina sequencing was used to examine bacterial community structure.The results showed that inoculation with HPD-2 significantly elevated PYR and B[a]P degradation rates by 44.7%and 30.7%,respectively,in the red soil,while it only improved the degradation rates by 1.9%and 11%,respectively,in the paddy soil.To investigate the underlying mechanism,the fate of strain HPD-2 and the response of the indigenous bacterial communities were determined.Strain HPD-2 occupied certain niches in both soils,and the addition of the bacterium changed the native community structure more noticeably in the red soil than in the paddy soil.The addition of PAHs and strain HPD-2 significantly changed the abundances of 7 phyla among the 15 detected phyla in the red soil.In the paddy soil,5 of the 12 dominant phyla were significantly affected by PAHs and the inoculation of HPD-2,while 6 new phyla were detected in the low-abundance phyla(<0.1%).The abundances of Massilia,Burkholderia,and Rhodococcus genera with PAH degradation efficiency were significantly increased by the inoculation of HPD-2 in the red soil during 42 d of incubation.Meanwhile,in the paddy soil,the most dominant effective genus,Massilia,was reduced by HPD-2 inoculation.This research revealed the remediation ability and inherent mechanism of the highly effective PAH-degrading strain HPD-2 in two different soil types,which would provide a theoretical basis for the application of degrading bacteria in different soils.KeyWords: bacterial inoculation,benzo[a]pyrene,bioremediation,organic contaminant,paddy soil,Paracoccus aminovorans,pyrene,red soil
INTRODUCTION
Polycyclic aromatic hydrocarbons(PAHs)are a group of persistent,semi-volatile,organic contaminants(Chauhanet al.,2008;Alegbeleyeet al.,2017).Polycyclic aromatic hydrocarbons are ubiquitous in the environment,and they enter environmental matricesviaboth natural and anthropogenic processes,including certain industrial and agricultural activities.Since PAHs are lipophilic and barely soluble in water,they are easily dissolved and transported by cell membrane lipoproteins and therefore accumulate in the fatty tissues of living organisms. The accumulation of PAHs can lead to carcinogenic effects,malformation,and gene mutations in humans(Choiet al.,2006).
The stability and hydrophobicity of PAH molecules are two primary factors that define the persistence of PAHs in the environment(Kanaly and Harayama,2000;El Amraniet al.,2015). Polycyclic aromatic hydrocarbons are classified as low-molecular-weight(LMW)-and high-molecular-weight(HMW)-PAHs(Kanaly and Harayama,2000; Sohnet al.,2004;Mallicket al.,2011).The LMW-PAHs refer to PAHs with 2–3 rings,such as naphthalene,fluorene,phenanthrene,and anthracene.As the molecular weight of PAHs increases,the solubility decreases,and the toxicity increases(Haritash and Kaushik,2009).Therefore,HMW-PAHs,which contain 4–7 rings,are not only less degradable in natural matrices because of their enhanced persistence(Haritash and Kaushik,2009;Kuppusamyet al.,2017),they also have significantly higher toxicity,recalcitrance,and carcinogenicity in humans.
Studies have shown that 90%of the total global environmental PAH load is present in the soil(Kuppusamyet al.,2017).With the growing concern about the potential adverse effects of PAHs on public health and the environment,remediation technologies are receiving increasing attention,and some of the approaches(physical,chemical,or biological)have been successful(Rancet al., 2016; Kuppusamyet al., 2017; Alegbeleyeet al., 2017). Among these approaches,bioremediation is an appealing option because it is relatively cost-effective,environmentally sustainable,and publicly accepted(Mohanet al.,2006;Kadriet al.,2017).Bioaugmentation is one of the most efficient bioremediation strategies,and is performed by inoculating contaminated soil with degrading microbes(Tenget al.,2010;Luet al.,2011;Kuppusamyet al.,2017).This strategy is often used when the native soil microbiota does not have a strong ability to efficiently degrade HMW-PAHs.
An increasing number of soil microorganisms have been found to degrade HMW-PAHs and include the generaMycobacterium,Rhodococcus,Sphingomonas,Alcaligenes,Agrobacterium,Bacillus,Burkholderia,Flavobacterium,Phanerochaete,Pleurotus,Bjerkandera,etc.(Andreoniet al.,2004;Baboshinet al.,2008;Seoet al.,2009;Gallegoet al.,2014; Zhaoet al., 2017). These microbes were screened by enrichment cultivation and selected based on their ability to utilize PAHs as the sole carbon or energy source(Baboshinet al., 2008; Chenet al., 2016). The pathways for the biodegradation of HMW-PAHs have already been well explored(Kimet al.,2007;Fuchset al.,2011;Mallicket al.,2011;Gaoet al.,2013).The initial step in the aerobic catabolism of a PAH molecule by microbes is the oxidation of a PAH to a dihydrodiol by a multicomponent enzyme system (Kanaly and Harayama, 2000; Barry and Challis,2013).
In practical situations,bioremediation can be effective under appropriate environmental conditions,and microbes can grow,colonize and express activated enzymes for successful enzymatic attacks on PAHs(Barry and Challis,2013;Gallegoet al.,2014).Various abiotic and biotic factors can influence the process of bioremediation,such as pH,temperature,oxygen levels,nutrient availability,and bioavailability of pollutants (Haritash and Kaushik, 2009; Ghosalet al.,2016). Additionally, indigenous microorganisms play an important role in bioremediation by competing with exogenous degrading microbes (Kuppusamyet al., 2017). The density,biomass,composition,and structure of native microbial communities are associated with PAH degradation.Nevertheless, it has been reported that different soil type show distinct degradation capacities,as each soil type has a specific native microbial community(Renet al.,2016;Chen
et al.,2016).
We isolated a bacterial strain, HPD-2, from a heavily PAH-contaminated soil(Maoet al.,2008;Tenget al.,2010).It has been proven that this strain can utilize HMW-PAHs,including fluoranthene,pyrene(PYR),and benzo[a]pyrene(B[a]P),as its sole carbon and energy source(Tenget al.,2010; Ganet al.,2018).According to its 16S rRNA gene sequence, strain HPD-2 was identified asParacoccussp.After inoculation with the strainParacoccussp.HPD-2 to an aged PAH-contaminated soil collected from the Yangtze Delta of East China,35.1%,20.7%,and 24.3%of the total 3-,4-,and 5(+6)-ring PAHs were degraded in 28 d,respectively(Tenget al., 2010). However, the potential of this highly effective degrading bacterium for application in different regions and different soil types remains unclear.
Renet al.(2016)found that the dissipation rate of PYR varied greatly with soil type.Their results showed that among four test soils, the paddy soil (Anthrosol) had the highest potential for PYR degradation,whereas no reduction in PYR was observed in the red soil during 42 d of incubation.Therefore, it could be speculated that the highly efficient degrading bacterium might exhibit diverse abilities in different soils, as the exogenous bacteria functioning may be controlled by the soil properties and the characteristics of the indigenous microbial community.Paracoccussp.HPD-2 has previously been isolated from PAH-contaminated soil(Maoet al.,2008).The degradation rates of PYR in liquid culture at 5 and 50 mg L-1were 88.4%and 57.1%in 7 d,respectively. For B[a]P,the degradation rate of 3 mg L-1by the strain HPD-2 was 89.7%in the liquid culture after 5 d(Maoet al.,2008;Liuet al.,2010).Accordingly,paddy and red soils,which had the highest and lowest capacities for degrading HMW-PAH, respectively, were selected to evaluate the bioremediation potential and mechanisms of the degrading bacteriumParacoccussp.HPD-2 therein.This research provides a theoretical basis for the application of degrading bacteria in different soils.
MATERIALS AND METHODS
Bacterial strain
The test bacterium,Paracoccussp.HPD-2,was isolated from a historically PAH-contaminated soil collected from Wuxi,Jiangsu,East China(Maoet al.,2008).The strain was screened for its ability to degrade HMW-PAHs,including fluoranthene,PYR,and B[a]P.
Tested soils
The test soils were red soil and paddy soil, collected from the Red Earth Ecological Experimental Station of Chinese Academy Science,Yingtan,Jiangxi,China(28°15′N,116°55′E)and the National Field Observation and Research Station for Agricultural Ecological Systems, Changshu,Jiangsu,China(31°33′N,120°38′E),respectively.The soil was collected from the 0–10 cm layer,sieved(2 mm),and stored at 4°C.The chemical and physical properties of the test soils are listed in Table I.
According to the soil classification system of Food and Agricultural Organization of the United Nations (FAO),the red soil was an Ferric Acrisol,and the paddy soil was anthropogenic.The properties of the two soils were distinct:the red soil was acidic with relatively low levels of organic matter,N,P,and K,while the paddy soil was alkaline with no less than 7 times higher levels of organic matter, N,P,and K than the red soil.According to the PAH thresholds in soil issued by European countries,when sum of 16 kinds of PAHs(∑16PAHs)are below 200 μg kg-1dry weight soil(d.w.s.),the soil is considered not contaminated with PAHs(Maliszewska-Kordybach,1996),as were the two test soils.
Microcosm experiment
Before the experiment,sterile water was added to both the test soils and left for equilibrium for 7 d.The levels of PAHs were 30 mg kg-1d.w.s.The soils(30 g)were thoroughly mixed with 3 mL of either PYR or B[a]P dissolved in acetone(3 000 mg L-1)to prepare PYR and B[a]P at 300 mg kg-1d.w.s . In the control group, 3 mL of acetone was mixed with 30 g of soil before the three groups of soils(PYR or B[a]P contaminated group and the control group)were left overnight to volatilize acetone. The three soils were then mixed with 270 g of the test soil to obtain the soils directly used in the experiment,that is,the test soils with 30 mg kg-1d.w.s.of PYR or B[a]P and the control soils without PAH addition.The three groups of soils(each 30 g)were loaded in 120-mL serum bottles.
The HPD-2 strain was activated and transferred to 200 mL of Luria-Bertani(LB)medium at an inoculation amount of 2%before being shaken at 28°C overnight.The strain was incubated until the late period of logarithmic growth(17–18 h after inoculation).Subsequently,the bacterial fluid was centrifuged at 4 000 r min-1for 10 min, thoroughly washed with sterile water, and re-suspended twice before being used as the seed fluid.
The seed fluid of strain HPD-2 was added to the experimental soils at 5%of the inoculation amount,and deionized water was added to maintain the soil moisture at 60% of maximum water-holding capacity.Additionally,treatment with sterile water was also included.The serum bottles were sealed, and the soils were incubated at 28°C in the dark for 42 d,during which time sterile water was supplemented under sterile conditions every 7 d to maintain soil moisture.On days 0,7,14,21,28,35,and 42 of incubation,10 g of soil was collected from each bottle and freeze-dried for the determination of PYR.Additionally,5 g of soil was stored at-80°C for DNA extraction.
Quantitative analysis of HMW-PAHs
The HMW-PAHs in the soil were extracted and purified according to previously reported methods(Maoet al.,2012; Renet al.,2016)with slight modifications.Briefly,soils were freeze-dried and then passed through a 0.25-mm mesh.Pyrene-treated soil(2.00 g)was continuously Soxhletextracted with 70 mL of dichloromethane for 24 h at 53°C,and the extract was collected in a rotary evaporator to dryness at 38°C and 41.3 kPa.Then,2 mL of cyclohexane was added to dissolve the residue,and the solution(0.5 mL)was then transferred to a silica gel column(220 mm length×8 mm inner diameter)and eluted with a hexane-dichloromethane mixture(1:1,volume/volume)for purification.First,1 mL of eluate was discarded,and the following 2 mL of eluate was collected and dried under a stream of N2in a graduated test tube.Two mL of acetonitrile was added to the tube to re-dissolve the residue. The solution was filtered through a 0.45-μm syringe filter membrane. The PYR and B[a]P concentrations were then analyzed using a Shimadzu Class-VP HPLC system (Shimadzu, Japan) with a fluorescence detector(RF-10 AXL).The separation of PAHs was carried out on a C18reversed phase column (VP-ODS 150 mm length×4.6 mm inner diameter,particle size 5 μm).The mobile phase was acetonitrile-water(4:1,volume/volume)at a constant solvent flow rate of 2 mL min-1.The excitation and emission wavelengths were 296 and 404 nm, respectively.The remaining time of PYR and B[a]P was 6.12 and 14.4 min,respectively.
DNA extraction and PCR amplification
The incubated soil in the microcosm was used to measure bacterial structure.Total DNA was extracted from 500 mg of soil from each sample using the FastDNA kit(Qbiogene Inc.,USA).Briefly,vigorous shaking was performed using a FastPrep®instrument at a speed setting of 6.0,for 40 s for cell lysis with sodium phosphate buffer.Soil and cell debris were removed by centrifugation(14 000 r min-1,10 min,4°C).Soil DNA was purified by binding to the matrix and then washing with SEWS-M solution,before 100 μL of elution buffer was added to dissolve the purified DNA.Soil DNA was visualized on 1.0% agarose gels by electrophoresis,while the quantity and purity of DNA were determined using a Nanodrop® ND-1000 UV-vis spectrophotometer.The primers 515 F(5′-GTGCCAGCMGCCGCGG-3′)and 907 R(5′-CCGTCAATTCMTTTRAGTTT-3′)targeting the V4-V5 hypervariable regions of microbial 16S rRNA genes were selected.The forward and reverse primers were tagged with adapter,pad,and linker sequences(Renet al.,2016).Each barcode sequence(12 mer)was added to the reverse primer for pooling multiple samples in one run of MiSeq sequencing.The PCR reaction(20 μL)mixture contained 4 μL of 5×FastPfu buffer(TransGen Biotech,Beijing,China),2 μL of dNTPs(2.5 mmol L-1),0.8 μL of each primer(5 μmol L-1),0.4 μL of FastPfu polymerase,10 ng of template DNA,and ddH2O.The PCR reaction was amplified using ABI GeneAmp®9700,and the conditions were as follows:94°C for 5 min,followed by 32 cycles of 94°C for 30 s,55°C for 30 s,and 72°C for 45 s,with an extension at 72°C for 5 min.The PCR product(2 μL)was used for agarose gel(1.0%)detection after amplification.The triplicate PCR reactions for each sample preparation were combined and purified using the AxyPrep DNA Gel extraction kit(Axygen,USA)and then quantified using the QuantiFluor™-ST system(Promega,USA).
Illumina Miseq sequencing and data processing
The DNA samples were sequenced at the Shanghai Majorbio Bio-Pharm Technology Co.Ltd.(Shanghai,China)using paired-end sequencing on an Illumina MiSeq PE300 platform.
Raw fastq files were demultiplexed and quality-filtered using QIIME(version 1.17)with the following proposals:first,the 300 bp reads were truncated at any site receiving an average quality score<20 over a 50-bp sliding window,discarding the truncated reads that were shorter than 50 bp;second,exact barcode matching,2 nucleotide mismatch in primer matching,and reads were removed if they contained ambiguous characters;third,only sequences that overlapped by more than 10 bp were assembled according to their overlap sequence.Reads that could not be assembled were discarded.
Operational taxonomic units (OTUs) were clustered with a 97%similarity cutoffusing UPARSE(version 7.1,http://drive5.com/uparse/), and chimeric sequences were identified and removed using UCHIME(http://drive5.com/u chime/).The taxonomy of each 16S rRNA gene sequence was analyzed by the Ribosomal Database Project(RDP)classifier(http://rdp.cme.msu.edu/) against the silva (SSU115)16S rRNA database using a confidence threshold of 70%(Wanget al.,2007).
Statistical analysis
One-way analysis of variance and Fisher’s least significant difference test(P <0.05 andP <0.01)were used to compare the average physical and chemical properties of the soils and the abundance of bacteria at various levels.Statistical analyses were performed using SPSS(version 17.0,SPSS Inc.,Chicago,USA).Redundancy analysis(RDA)of bacterial abundance and soil properties was calculated and drawn using Canoco for Windows 4.5.
RESULTS
Dynamics of PAHs concentration in soil
Generally, the degradation of PAHs in the paddy soil was more profound than that in the red soil(Fig.1).Without the HPD-2 strain,PAHs were barely degraded in the red soil,which is consistent with the results of Renet al.(2016).On the other hand,greater degradation of PYR with four fused benzene rings was observed in the paddy soil; 28 d after the incubation,95%of PYR was removed from the paddy soil.At the same time,B[a]P with five benzene rings less degraded (53.5%), indicating that PAHs with fewer rings were more readily degraded in the paddy soil.
The inoculation of the soils with HPD-2 significantly enhanced the degradation of PAHs.When originally added at 30 mg kg-1d.w.s.,44.7%of PYR and 30.7%of B[a]P had degraded in the red soil 21 d after incubation(Fig.1a,c). However, in the paddy soil, HPD-2 did not obviously stimulate the degradation of either PAH,and it significantly decreased the degradation rate of PYR during 14–28 d.Nonetheless,at the end of the incubation,the degradation rate of PYR still reached 97.3% with HPD-2; only 1.9%less than with no inoculation(Fig.1b).Meanwhile,HPD-2 inhibited the degradation of B[a]P in the paddy soil after the fourth sampling on day 21(by 11%compared to that without HPD-2),even when it slightly elevated the degradation rate before 14 d(Fig.1d).
The inhibition of PAH degradation induced by HPD-2 could be attributed to the strong degradation ability of indigenous bacteria originally in the paddy soil where microbial populations had already established an ecological balance.Based on this,the inoculation of HPD-2 may interfere with the balance and alter the abundance of some existing microbial populations in the paddy soil,including the degradation-effective bacteria of PAHs.Moreover,the degrading function of HPD-2 could also be impaired by indigenous microbes; therefore, the degradation of PAHs was inhibited in the paddy soil with HPD-2 inoculation.
Bacterial communityvariation over incubation time
Bacterial community structure is an indicator of both the composition and abundance of bacterial populations.Therefore,the UPGMA method was applied in this study to classify the treatment groups(two soils with two PAHs and
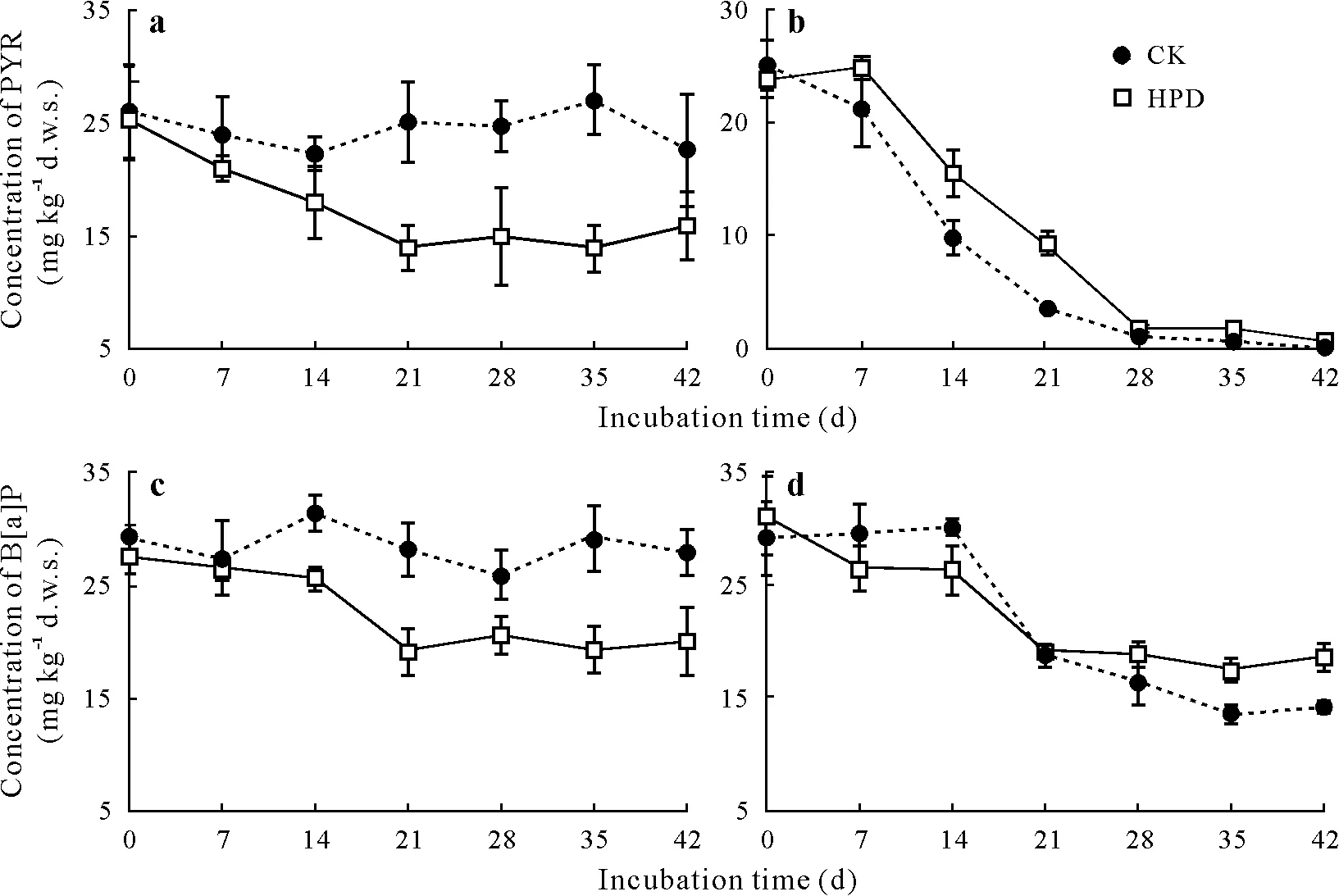
Fig.1 Pyrene(PYR)(a and b)and benzo[a]pyrene(B[a]P)(c and d)concentration dynamics in the red soil(a and c)and paddy soil(b and d)during the incubation period.Bars are standard deviation of means(n=3).CK=the control group without bacterial inoculation;HPD=the group inoculated with strain HPD-2;d.w.s.=dry weight soil.
two bacterial treatments on days 0,14,and 42 of incubation)into 4 different types. Types 1 and 2 contained red soil and paddy soil with HPD-2 at the first sampling (0 d),respectively.Type 3 included the rest of the red soil groups,and Type 4 the rest of the paddy soil groups (on days 14 and 42;Fig.S1,see Supplementary Material for Fig.S1).According to the cluster analysis,inoculation of HPD-2 led to significant alteration of the bacterial community structure in the red soil,since the variation was the highest between the red soil group with HPD-2 on Day 0 and the original red soil sample group(before incubation).As incubation progressed,the original bacterial community structures recovered to the original conditions in both the red soil and paddy soil(Fig.S1).The results showed a distinct difference in community structure of native bacteria between the paddy soil and red soil,as found by Renet al.(2016).Additionally,the addition of PAHs and highly effective degradation bacteria could change the community structure more remarkably in the red soil than in the paddy soil,indicating that red soil was more susceptible to variations in environmental conditions.
Differential analysis of bacterial groups at phylum/genus level
To further identify which bacterial populations responded to the HPD-2 inoculation and the relationship of these populations to PAH degradation, the variations in the abundances of the major microbial populations(15 most abundant populations)in the two soils were analyzed at the phylum level.In the red soil,bacteria from 19 phyla were observed, including relatively abundant Chloroflexi(20.0%–27.8%),Proteobacteria(21.3%–23.6%),Actinobacteria (12.8%–26.0%), Firmicutes (13.3%–25.0%),Acidobacteria(9.2%–12.2%),and Planctomycetes(2.4%–3.3%;Fig.2a).The addition of PAHs and the inoculation of HPD-2 significantly changed the abundance of 7 phyla of bacteria(P <0.05), 5 of which were highly significantly affected (P <0.01). Furthermore, PAHs and HPD-2 reduced the abundance of the native Deferribacteres in the red soil to below the detection limit,but new phyla(WS6 and Parcubacteria)were observed(Fig.2a).
Compared to that in the red soil, the microbial community in the paddy soil varied (Fig. 2b). The bacteria identified in the paddy soil were from 33 phyla: the major species (>0.1%) represented 17 phyla, with the less abundant species from the remaining 16 phyla (Fig. 2c).The major taxa included Proteobacteria (26.6%–41.0%),Firmicutes(27.9%–35.3%),Actinobacteria(8.9%–11.8%),Bacteroidetes(6.6%–10.4%),Acidobacteria(2.7%–7.9%),Gemmatimonadetes(4.5%–5.2%),and Chloroflexi(1.5%–4.4%),which were significantly different from those in the red soil.The PAH addition and HPD-2 inoculation significantly changed the abundances of 12 phyla(P <0.05),and highly significant differences were observed in 5 of these(P <0.01;Fig.2b).In addition,among the less abundant bacterial populations,PAHs and strain HPD-2 increased the abundances of most native populations in the paddy soil,and led to the identification of 6 new phyla with low abundances(<0.1%)(Berkelbacteria,Microgenomates,Omnitrophica,FBP,Ignavibacteriae,and Nitrospinae).
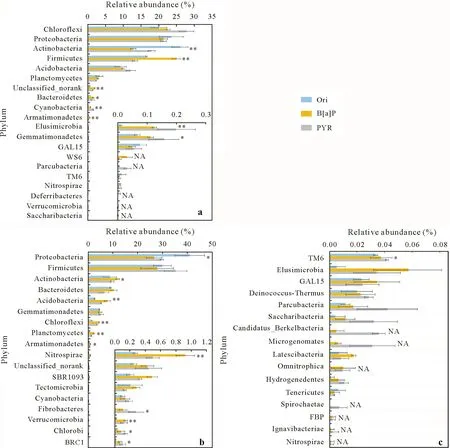
Fig. 2 Comparison of microbial groups at the phylum level in original soils (Ori, before incubation) and the soils treated with pyrene (PYR) and benzo[a]pyrene(B[a]P)and inoculated with Paracoccus sp.HPD-2 after 42-d incubation:microbial groups in the red soil(a),major microbial groups(relative abundance >0.1%)in the paddy soil(b),and the microbial groups with low abundance(relative abundance <0.1%)in the paddy soil(c).Asterisks*and**represent significant differences at P <0.05 and P <0.01,respectively.NA=not detected in one or more treatments.
The test strain,Paracoccussp. HPD-2, belonged to Proteobacteria,and this phylum was ecologically dominant in both test soils;however,it did not cause much difference in the abundances of other Proteobacteria in the red soil after 42 d of incubation(Fig.2a,b).More importantly,the abundances of Proteobacteria were significantly reduced in the paddy soil at the same time, probably because of the PAH-induced inhibition of such microorganisms.
The fate of strain HPD-2 in the soils and its effects on the dynamics of the native microbial community were investigated at the genus level(Fig.3).Strain HPD-2 was defined as theParacoccusgenus,which had relatively low abundances in the original soils(0.07%in the red soil and 0.01%in the paddy soil).
In the B[a]P addition group, the abundances ofParacoccuswere 76.8% and 49.1% in the red soil and paddy soil,respectively,on day 0 after HPD-2 inoculation(Fig.3).Because of the same inoculum amount of HPD-2(5%)before the incubation, the total abundance of bacteria in the red soil was significantly lower than that in the paddy soil,and therefore, the exogenous inoculation of HPD-2 led to greater effects on the microbial community in the red soil.On day 14,the abundances ofParacoccusbacteria significantly decreased to 1.4%and 3.1%in the red soil and paddy soil,respectively.When the incubation time was extended to 42 d,their abundances were further reduced to 0.37%and 0.41%in the two soils.In the group without HPD-2 inoculation,the abundance of theParacoccusgenus was not observed in either of the test soils on days 14 and 42.
When PYR was added,the alteration of the microbial community was generally consistent with that in the B[a]Ptreated soils(Fig.3).In the group with HPD-2 inoculation on day 0,the abundances ofParacoccusreached 84.0%and 48.7%in the red soil and paddy soil,respectively,but on day 14,the abundances were substantially reduced to 4.1%and 1.1%,respectively.Paracoccusstill occupied a certain niche in both soils after 42 d of incubation, the abundances of which were 0.34%and 0.43%,respectively.In contrast,after 42 d of incubation,Paracoccusabundances were extremely low in soils without HPD-2 inoculation(0.001%in paddy soil and below the detection limit in red soil).
The dynamics of bacterial structures showed that at the genus level,HPD-2 was not able to ecologically dominate within 42 d; nonetheless, it could still occupy a niche in the habitat and alter the microbial community in soil to some extent.On day 42,other bacterial species responded to HPD-2,as shown in Fig.4.Based on the common and specific populations in different communities at the level of OTUs on day 42,the inoculation of HPD-2 induced 62 and 61 new OTUs in the B[a]P- and PYR-contaminated red soil, respectively, and 210 and 201 new OTUs in the B[a]P-and PYR-contaminated paddy soil,respectively.At the same time,according to Venn diagrams(Fig.4),PAHs and exogenous HPD-2 significantly altered indigenous bacterial communities in the soils.The additions of B[a]P and PYR decreased 154 and 163 OTUs, respectively, to below the detection limits in the red soil,and for the paddy soil,the undetected OTUs were 147 and 156, respectively. Thus,the toxicity of PAHs to the microbial community was more profound in the red soil.
Responses of functional bacteria to HPD-2
Bacteria that are effective in degrading PAHs,such as B[a]P and PYR, are not widespread, including genera ofMassilia,Burkholderia,Sphingomonas,Mycobacterium,Pseudomonas,Rhodococcus,Stenotrophomonas,Sphingobium,Novosphingobium,andFlavobacterium,according to previous studies(Kanaly and Harayama,2000;NíChadhainet al., 2006; Maigari and Maigari, 2015). Therefore, the changes in the functional groups of such genera were investigated in this study,and because of their low abundances(mostly below 1%),we mainly focused on the trends(Figs.5 and 6).
The efficient generaMassilia,Burkholderia,Sphingomonas,Mycobacterium, Pseudomonas,Rhodococcus,Stenotrophomonas,andNovosphingobiumwere observed in the red soil(Fig.5). The most variable genera wereMassiliaandSphingomonas.In the B[a]P-treated red soil,the abundance ofMassiliaincreased by 37 times on day 42(0.17%)compared to that on day 0(0.002%)when HPD-2 was inoculated; no significant differences were found in the PYR-treated red soil.Meanwhile,both B[a]P and PYR significantly reduced the abundance of nativeSphingomonas,from 0.9%and 0.2%,respectively,on day 0 to 0.04%and 0.01%,respectively,on day 42.Inoculation with HPD-2 also had significant effects on theBurkholderiaabundance.On day 14,HPD-2 inoculation resulted in significantly higher abundances ofBurkholderia.In the B[a]P-or PYR-treated red soil,the abundance of this genus was 1.1%and 2.5%,respectively.Rhodococcusabundance significantly increased on day14 of inoculation,with no significant differences at any other sampling.The abundance ofStenotrophomonasalso significantly increased in the PYR-treated red soil;however,it declined below the detection limit in the B[a]P-treated group.Moreover,HPD-2 inoculation reduced the abundances ofNovosphingobiumandMycobacterium(the former with significant differences),whereasPseudomonasabundance was not significantly affected by any of the treatments.
In the paddy soil,the number of PAH-degrading bacteria was much larger than that in the red soil. Moreover,the abundances of all the genera were significantly different from those in the red soil, and 2 new genera,SphingobiumandFlavobacterium, were identified. Among the degrading-effective genera,Massiliawas the most abundant,but its abundance was significantly decreased by HPD-2 inoculation.At the same time,the abundances ofSphingomonas,Mycobacterium,Rhodococcus, andFlavobacteriumwere all elevated by the inoculation of HPD-2 at the end of the incubation compared to the groups with no inoculation,even when no significant differences were observed on day 14.
Correlation between PAH degradation and major microbial groups
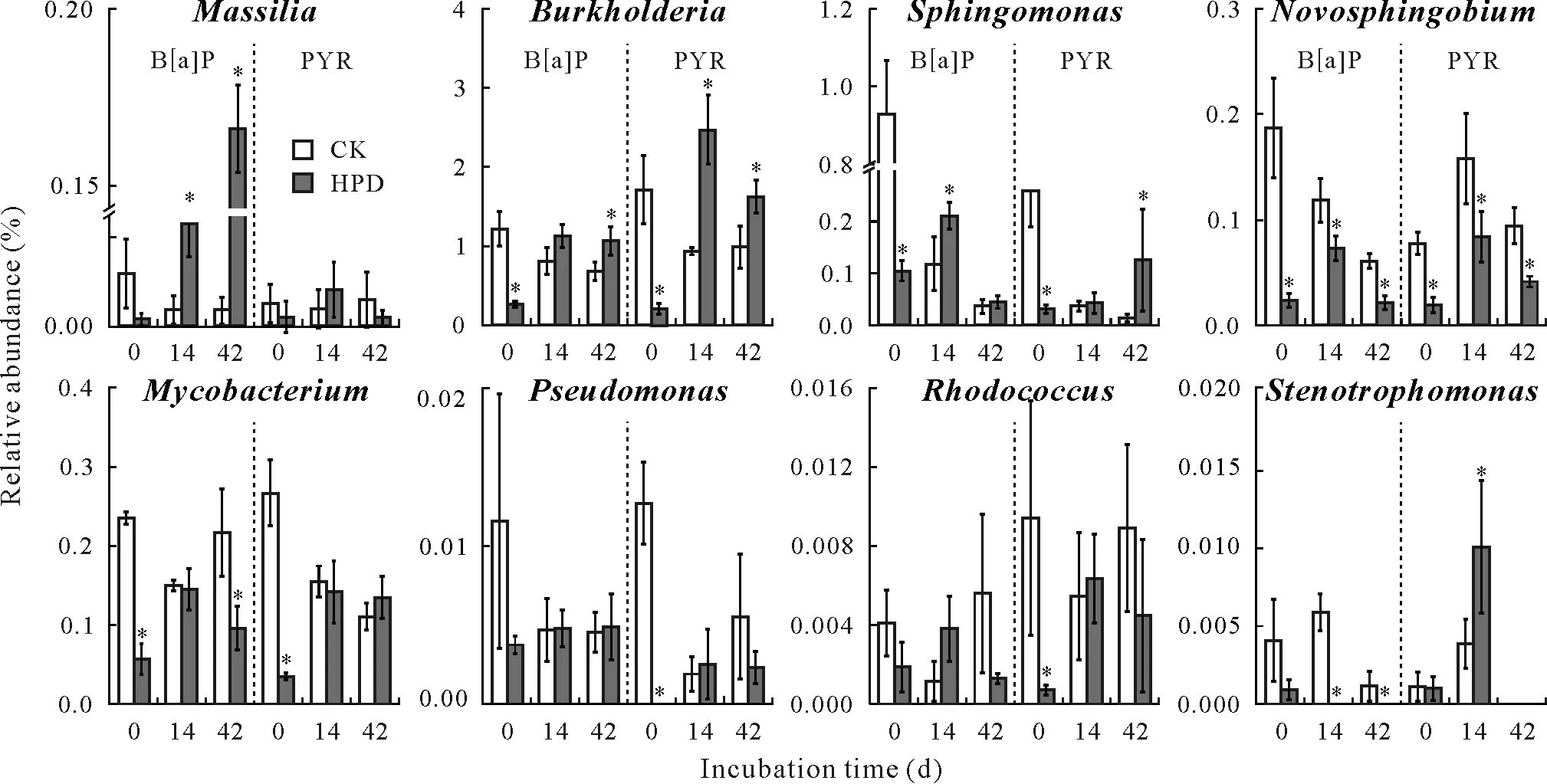
Fig.5 Relative abundances of the genera with high-molecular-weight polycyclic aromatic hydrocarbon(PAH)-degrading capacity in the red soil with pyrene(PYR)and benzo[a]pyrene(B[a]P)addition during the incubation period.Vertical bars are standard deviations of means(n=3).An asterisk*indicates significant differences at P <0.05.CK=the control group without bacterial inoculation;HPD=the group inoculated with strain HPD-2.
To determine which populations were related to (or responsible for)PAH degradation in soils,the correlations between PAH degradation efficiency and major microbial communities at the phylum level were analyzed based on the RDA analysis(Fig.7).The results showed that there were significant differences between PAH degradation-related microbial communities in the red soil and paddy soil.
In the red soil, the distributions of Proteobacteria,Cyanobacteria,Bacteroidetes,Actinobacteria,and Acidobacteria were significantly correlated with the PAH-degrading ability of soil;highly significant positive correlations were found between the community distributions of Cyanobacteria and Bacteroidetes and the degrading ability,whereas that of Actinobacteria was negatively correlated(P <0.01)(Table II).In contrast,the community distributions of Planctomycetes,Nitrospirae,Firmicutes,Chloroflexi,Bacteroidetes,Actinobacteria,and Acidobacteria were all significantly positively correlated with the degradation of PAHs in the paddy soil,except for Firmicutes,and highly significant correlations were found for Planctomycetes,Nitrospirae,and Acidobacteria(P <0.01).Furthermore,the community distribution of the phylum Proteobacteria,to which strain HPD-2 belonged,was significantly positively correlated with PAH degradation in the red soil.However,such effects were not observed in paddy soil.This result was consistent with the assumption that the inoculation of HPD-2 only significantly enhanced PAH degradation in the red soil,and did not reinforce the degradation efficiency in the paddy soil,possibly due to the significantly distinct native microbial communities in the two soils.
DISCUSSION
Nutrient levels directly regulate the existence,growth,flourishing, and, therefore, the functions of indigenous microorganisms in soil. Previous studies have shown that the native microbial community structures are distinct in the red soil and paddy soil, probably attributable to the different soil properties.Additionally,the supplementation of macronutrients in soil(e.g.,elevated levels of N and P)could increase the abundances of heterotrophic microorganism groups,thereby stimulating the biodegradation of PAHs(Ataganaet al., 2003; Konget al., 2018). Breedveld and Sparrevik(2000)reported that the addition of inorganic N and P to soil enhanced PAH degradation,and more importantly,only when N and P were supplied at sufficient levels could PAHs with four benzene rings be degraded in soil with relatively low levels of available N and P.In our study,the nutrient levels in the red soil were greatly lower than those in the paddy soil,mostly by orders of magnitude,and organic matter in the red soil was only 15.6%as much as that in the paddy soil(Table I).Presumably,the insufficient nutrients could limit the growth and reproduction of microorganisms in the red soil, and therefore, the microbial abundances were relatively low,making the microbial environment more susceptible to fluctuations of environmental conditions.
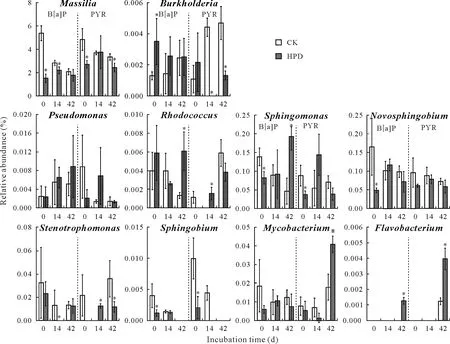
Fig.6 Relative abundances of the genera with high-molecular-weight polycyclic aromatic hydrocarbon(PAH)degradation capacity in the paddy soil with pyrene(PYR)and benzo[a]pyrene(B[a]P)addition during the incubation period.Vertical bars are standard deviations of means(n =3).An asterisk*indicates significant differences at P <0.05.CK=the control group without bacterial inoculation;HPD=the group inoculated with strain HPD-2.
Biological stimulation is an effective strategy for removing PAHs from soil,especially for HMW-PAHs.Daviset al. (1993) found that thePhanerochaete sordidastrain could enhance the degradation of four-ring PAHs,opposed to those with 3, 5, or 6 rings, when soil was under the combined contamination of PAHs and pentachlorophenol.Konget al.(2018)reported the degradation of total PAHs in soil,especially HWM-PAHs,induced byRhodococcus ruberEm1,a PAH-degrading bacterium.In our study,when strain HPD-2 was inoculated to different soils,it exhibited distinct degradation abilities.Inoculation significantly elevated the degradation rate in the red soil that could not degrade PAHs primitively.In contrast,for the paddy soil that was effective in degrading PAHs,the inoculation clearly inhibited PAH degradation.Based on the bacterial community structure analysis,it could be inferred that the exogenous HPD-2 resulted in significant alterations to the native microbial community in soil, impaired the existing balance of degrading-active bacteria in paddy soil,and reduced the degradation abilities.This speculation was confirmed by the abundance analysis of microorganisms with PAH-degradation abilities(Figs.5–7).In the red soil,most of the functional bacteria had elevated abundances because of HPD-2 inoculation, especially forMassiliaandBurkholderia,which were already relatively abundant, which could also mediate the HPD-2-induced enhancement of PAH degradation in the red soil.In contrast,exogenous HPD-2 reduced the most abundant genus,Massilia,in the paddy soil.Furthermore,significantly increased abundances(Sphingomonas,Mycobacterium,Rhodococcus,andFlavobacterium)were only observed on day 42,and no significant changes were found on day 14.Therefore,during most stages of the incubation, the amounts of functional microorganisms were probably declined in HPD-2 inoculated paddy soil, which in turn caused a decrease in the degradation ability therein.This was then restored at the end of the incubation period.
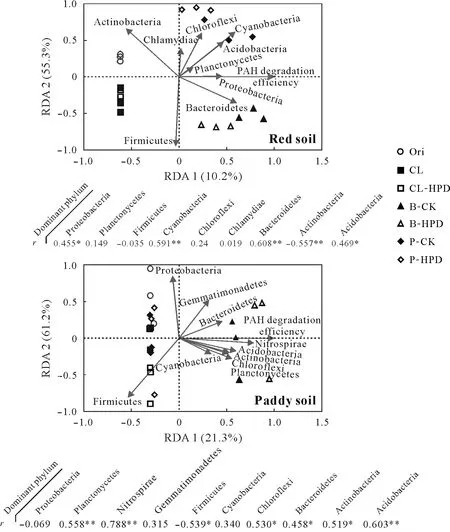
Fig.7 Redundancy analysis(RDA)of the major microbial groups and the polycyclic aromatic hydrocarbon(PAH)degradation efficiency of the red soil and paddy soil with pyrene(PYR)and benzo[a]pyrene(B[a]P)addition after 42-d incubation.The correlations between the major bacterial groups(at phylum level)and the efficiency of PAH degradation of different soils are illustrated below the figures.Asterisks*and**indicate significant differences at P <0.05 and P <0.01,respectively.Ori=original soils before incubation;CL=clean soils;CL-HPD=clean soils with strain HPD-2;B-CK=B[a]P-contaminated control soils without strain HPD-2;B-HPD=B[a]P-contaminated soils with strain HPD-2;P-CK=PYR-contaminated control soils without strain HPD-2;P-HPD=PYR-contaminated soils with strain HPD-2.
As shown in Fig. 8, we proposed a model for the alteration of the microbiomes during PAH degradation after HPD-2 inoculation in the red soil and paddy soil.As a source of environmental disturbance,exogenous HPD-2 could influence the community composition and even PAH degradation functioning in the soils.Strain HPD-2 and indigenous communities,which might already be degrading active,have to deal with their interactions once HPD-2 is inoculated.Bacterial communication includes heterogeneous patterns,such as competing,cooperating,cell-to-cell contact,exchange of dedicated signal molecules,and sharing of nutrients(Shonget al., 2012; Wanget al., 2018). In this study, one of the most important patterns of bacterial communication was competition for nich and nutrients.Exogenous HPD-2 must occupy a certain niche in the soil for colonization.The living spaces occupied by HPD-2 interfere with the local microbial community.In addition,the cooperation of HPD-2 and indigenous groups altered the steady state of the soil microbiomes.Competing for nutrients and co-metabolizing of the HMW-PAHs are probably the two main ways for bacterial mutual benefits.Furthermore,PAH-degrading bacteria are commonly considered to contain key genes for degradation,such as genes of PAH dioxygenase.The degrading genes of strain HPD-2 could be transferred to other bacterial groups as a result of horizontal gene transfer.This could stimulate the function of microorganisms using PAHs as a carbon source for energy metabolism and enhance the degradation of PAHs(DeBruynet al.,2012).
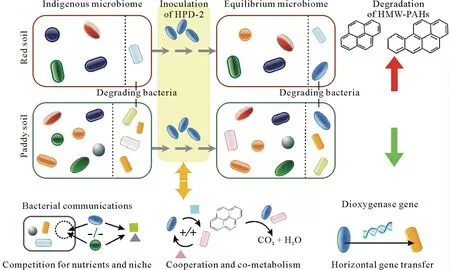
Fig.8 Proposed model for the alteration of microbiomes during polycyclic aromatic hydrocarbon(PAH)degradation after the strain HPD-2 inoculation in the red soil and paddy soil tested.HMW-PAHs=high-molecular-weight PAHs.
The abundance of the HPD-2 strain sharply declined after 14 d of incubation,and could not ecologically dominate in either test soil.This is attributable to the influence of the soil environment,soil components,and native microorganisms on the existence of HPD-2(Adamset al.,2015;Konget al.,2018).However,the bacterial microbiomes of different soils gradually reached an equilibrium state through the interactions between HPD-2 and the local microbes.When a new microbiome is formed, its function changes simultaneously. As a result, the degrading ability of the primitively effective paddy soil was inhibited,while the red soil with low degrading ability was significantly elevated by the inoculated HPD-2.This research provides a practical basis for the application of biostimulation and also helps elucidate the mechanisms of microbial function alterations by bacterial interactions in different soil types.
ACKNOWLEDGEMENTS
This study was funded by the National Key R&D Program of China (2019YFC1803705), the Projects of the National Natural Science Foundation of China(41991335 and 42130718),and the Key Research Program of Frontier Sciences, Chinese Academy of Sciences (QYZDJ-SSWDQC035).
SUPPLEMENTARY MATERIAL
Supplementary material for this article can be found in the online version.
杂志排行
Pedosphere的其它文章
- Effect of lignite amendment on carbon and nitrogen mineralization from raw and composted manure during incubation with soil
- Magnesium-fortified phosphate fertilizers improve nutrient uptake and plant growth without reducing phosphorus availability
- Recycling of isabgol(Plantago ovata Forsk.)straw biomass and mineral powder with bio-inoculants as an effective soil amendment for isabgol cultivation
- Enduring legacy of coal mining on the fungal community in a High Arctic soil after five decades
- Elevated atmospheric CO2 reduces CH4 and N2O emissions under two contrasting rice cultivars from a subtropical paddy field in China
- Improvement of phosphorus uptake,phosphorus use efficiency,and grain yield of upland rice(Oryza sativa L.)in response to phosphate-solubilizing bacteria blended with phosphorus fertilizer
