Regulation of LncRNA00067110 on Proliferation, Apoptosis and Melanins Production of B16-F10 Cells by Targeting Cabyr
2022-09-06YANGWanYunAJABKhanJIAQiongHUShiXiongJIAODingXingYULeiTaoFANRuiWen
YANG Wan-Yun, AJAB Khan, JIA Qiong, HU Shi-Xiong, JIAO Ding-Xing, YU Lei-Tao, FAN Rui-Wen*
(1)College of Veterinary Medicine, Shanxi Agricultural University, Taigu 030801, Shanxi, China;2)Faculty of Veterinary and Animal Sciences, The University of Agriculture, Dera Ismail Khan 29050, Khyber Pakhtunkhwa, Pakistan)
Abstract Long non-coding RNA (lncRNA) is a type of non-coding RNA with the more than 200 nucleotides. Several lncRNAs have been identified as the potential targets for cancer therapy. LncRNA00067110 is one of the differentially expressed genes in the transcriptome profiles of melanoma B16-F10 cells compared to normal mice melanocytes. To investigate whether lncRNA00067110 regulates the proliferation, apoptosis and melanogenesis of B16-F10 cells, the calcium-binding tyrosine phosphorylation regulated protein (Cabyr) target gene was predicted by LncTar and verified by dual luciferase activities. The regulating function of lncRNA00067110 was investigated by the analysis of transcriptome profiles and to detect the proliferation, apoptosis and melanin production of B16-F10 cells transfected by the overexpression plasmids of lncRNA00067110. The results showed that the relationship of lncRNA00067110 targeting Cabyr, the mRNA and protein levels of proliferation (MEK/ERK/ MNK/ CREB) and melanogenesis-related genes (TYR family and CREB) were significantly down-regulated, while the mRNA and protein levels of apoptosis-related genes (AKT and Bcl-2) were up-regulated in B16-F10 cells with lncRNA00067110 overexpression. The transcriptome profile of B16-F10 cells with lncRNA00067110 overexpression showed that 17 genes were differentially expressed, among which Cabyr was up-regulated. Furthermore, the effect of lncRNA00067110 on the phenotypes of cell proliferation and apoptosis were verified. The results suggested that lncRNA00067110 might be a novel target for the treatment of melanoma by targeting Cabyr, which regulate the expression of related genes to inhibit the proliferation and melanogenesis, as well as to induce the apoptosis of B16-F10 cells.
Key words lncRNA00067110; B16-F10; proliferation; apoptosis; melanin production; calcium-binding tyrosine phosphorylation regulated protein (Cabyr)
Melanoma is the most aggressive skin cancer, which is characterized by rapid progression[1]. The incidence of malignant melanoma is increasing worldwide[2]. Although, early primary melanomas can be cured by surgery, but the development of melanoma is usually accompanied by tumor metastasis to regional lymph nodes or even organs, which lacks radical treatment[3]. However, the molecular mechanisms underlying melanoma progression and tumor metastasis remained poorly understood. Therefore, the understanding of tumorigenesis and progression is essential for the development of diagnostic markers and novel effective therapies for melanoma patients.
Long non-coding RNAs (lncRNAs) are class of ncRNAs having more than 200 nucleotides lacking protein-coding ability[4]. LncRNAs have many specific functional features and are involved in many diverse biological processes in cells, such as RNA processing[5], modulation of apoptosis and invasion[6], marker of cell fate[7], chromatin modification[8]and so on. Several studies have showed that lncRNAs expression is tissues specific and are frequently dysregulated in various types of cancers and some lncRNAs are correlated with cancer recurrence and poor prognosis[9]. These lncRNAs are involved in tumorigenesis and cancer metastasis through silencing tumor suppressors or activation of oncogenes via different mechanisms including epigenetic modification, alternative splicing, RNA decay and regulation of post-translational modification[10]. Regulation of lncRNAs expression has been proven as potential therapeutic target for cancer treatment. However, function of most lncRNAs remained unclear.
LncRNA00067110 is one of the differentially expressed genes found in the transcriptome of melanoma cells (B16-F10) versus normal mice melanocytes. LncRNA00067110 contained hairpin loops and multiloops, with the size of 711 bp. It is predicted by RNAPLEX that calcium-binding tyrosine phosphorylation regulated protein (Cabyr) might be one of the target genes. According to the National Biotechnology Information Reference Sequence,Cabyris isolated from human sperm, which encodes 5 protein isoforms (Cabyr-a,Cabyr-b,Cabyr-c,Cabyr-d, andCabyr-e) by 6 transcript variants.Cabyrprotein with protein tyrosine kinases activity is high expressed in testicular and a variety of tumor tissues but shows no or low expression in normal tissues, inducing specific humoral immune response[11]. Thus,Cabyrprotein has been identified as a cancer/testis antigen in lungs cancer, playing an important role in the proliferation, metastasis and anti-tumor drug sensitivity[12]. Whether lncRNA00067110 had a role in melanoma by regulatingCabyris unknown. Here, the function and potential molecular mechanism will be investigated.
1 Materials and Methods
1.1 Cell culture
The melanoma cells (B16-F10 and HEK-293T) used in this study were purchased from Zhongqiao Xinzhou Biological Technology Co., Ltd. For the luciferase assay, the melanoma cells were cultured in Roswell Park Memorial Institute (RMPI; Gibco, Grand Island, NY, USA) and Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; ScienCell Research Laboratories).
1.2 Plasmid construction and cell transfection
The full sequence of lncRNA00067110 was amplified by PCR (primers listed in Table 1) and inserted into Pmscv-puro vectors. The empty Pmscv-puro plasmid was used as a negative control (NC). B16-F10 cells and HEK-293T cells were added in 6-well plates at a density of 5×106cells/well and cultured to reach 80%-90% confluence for transfection using Lipofectamine 2000 (Promega, USA) according to the manufacturer’s instructions. Then the cells were collected and lysed to obtain the total RNA, protein and melanins for subsequent analysis.
1.3 RNA-seq
Trizol reagent (Invitrogen, USA) was used for total RNA isolation and RNA-seq was conducted by The Beijing Genomics Institute (Beijing, China).
1.4 Luciferase assay
LncTar URL was used to predict LncRNA target genes by complementary base pairing and luciferase reporter gene assay was performed to verify the binding of lncRNA00067110 toCabyr. The dual fluorescent reporter vector ofCabyrwas constructed. HEK-293T cells were cultured in DMEM (Gibco, New York, USA) supplemented with 10% fetal bovine serum (FBS), which were co-transfected with pmirGLO-Cabyrand Pmscv-LncRNA or pmirGLO-Cabyrand Pmscv or pmirGLO and Pmscv-LncRNA expression plasmids. After 48 hours of co-transfection, cells were lysed using passive lysis buffer of the dual luciferase assay kit (Promega), and firefly luciferase activity was normalized to Renilla luciferase activity.
1.5 Quantitative real-time PCR (qPCR)
According to the manufacturer’s instructions, total RNA was extracted from melanoma cells using TRIzol reagent (Invitrogen, Shanghai, China). A universal primer and cDNA synthesis kit (TaKaRa, Dalian, China) were used for qPCR. The reaction was carried out using SYBR Green PCR Master Mix (TaKaRa, Dalian, China) with the analysis of the melting curve to verify the amplification specificity for each sample. All samples were repeated three times, and the relative mRNA expressions were separately detected by comparing the threshold cycle (CT) method to detect the comparative expression of mRNA.
1.6 Western blotting
The total protein of cells was extracted using protein extracts (Boster, Wuhan, China) and their concentration was determined by the BSA method. Proteins were separated by SDS-PAGE and analyzed by Western blotting. The following primary antibodies were used: MEK1, pMEK1, ERK, pERK, MNK1, CREB, AKT, pAKT (1∶500; Proteintech, Wuhan, China), TYR, TYRP1, TYRP2, MITF (1∶1 000; Abcam, Cambridge, MA, USA) and β-actin (1∶1 000; Boster). The protein bands intensity was measured by the optical density method using Image-Pro Plus software (Olympus, Tokyo, Japan). X-ray film (Kodak, Rochester, New York, USA) was exposed for less than 5 minutes. The quantitative protein expression was normalized to β-actin level.
1.7 Proliferation of melanoma cells
Cell proliferation was monitored using XCELLigence RTCA DP instrument and Cell Counting Kit-8 (CCK-8), and all operations were performed in accordance with instrument requirements. XCELLigence RTCA DP instrument measures electrical impedance across interdigitated micro-electrodes incorporated into the bottom of tissue culture E-Plates. Electrical impedance was indicated as a cell index (CI) value, and it provides quantitative information about the condition of the cells, including cell number, viability and morphology. The optical density (A) value of CCK-8 was measured using a microplate spectrophotometer (BioTek Instruments, USA) at a wavelength of 450 nm. Each experiment was performed in triplicate and were repeated three times independently.
1.8 Tyrosinase activity assays
Post 72 hours of transfection, melanocytes were transfected, tyrosinase activity was analyzed by the oxidation of L-dopa to dopachrome and measured by absorbance at 460 nm using a Multiscan Spectrum microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
1.9 Melanins measurements
Post 72 hours of transfection, melanocytes were collected and rinsed three times with PBS. Melanin production was analyzed using the total alkali soluble melanin (ASM) assay. Melanocytes were lysed in 0.2 mol/L NaOH at 85 ℃ for 5 min. The absorbance of solution was measured by using a Multiscan Spectrum microplate reader at 450 nm (Thermo Fisher Scientific, Waltham, MA, USA). Eumelanin production was analyzed using the eumelanin (EM) assay. Melanocytes were hydrolyzed in 30% hypophosphoric acid and hydriodic acid at 85 ℃. After cooling, 50% ethanol was added and the cell samples were centrifuged at 2 234 g for 10 min. Insoluble eumelanis were selectively solubilized in sodium hydroxide and hydrogen peroxide at 85 ℃. The absorbance of the supernatant was measured at 350 nm. Pheomelanin production was analyzed using the pheomelanin (PM) assay. Melanocytes were solubilized in phosphate buffer (pH 10.5) and chloroform. The solution was centrifuged at 10 700 g for 10 min. The absorbance of supernatant was measured at 400 nm. The melanin production was normalized to the total number of cells. All experiments were performed in triplicate.
1.10 Apoptosis assays
Apoptosis was determined by the terminal deoxyribonucleotidyl transferase-mediated terminal deoxyribonucleotidyl transferase mediated dUTP-digoxigenin nick end labeling (TUNEL) using a kit from Roche. The detection procedures were performed in accordance with the manufacturer’s instructions.
1.11 Statistical analysis
Data were analyzed using SPSS 11.5 software (SPSS, Chicago, IL, USA). All data were presented as mean ± standard error of at least three replicates. ANOVA and Student’st-test were used for multiple and two-group comparisons, respectively.P< 0.001 was considered as highly significant, andP< 0.05 as significant.
2 Results
2.1 LncRNA00067110 targeted Cabyr and potentially regulated the expression of related genes
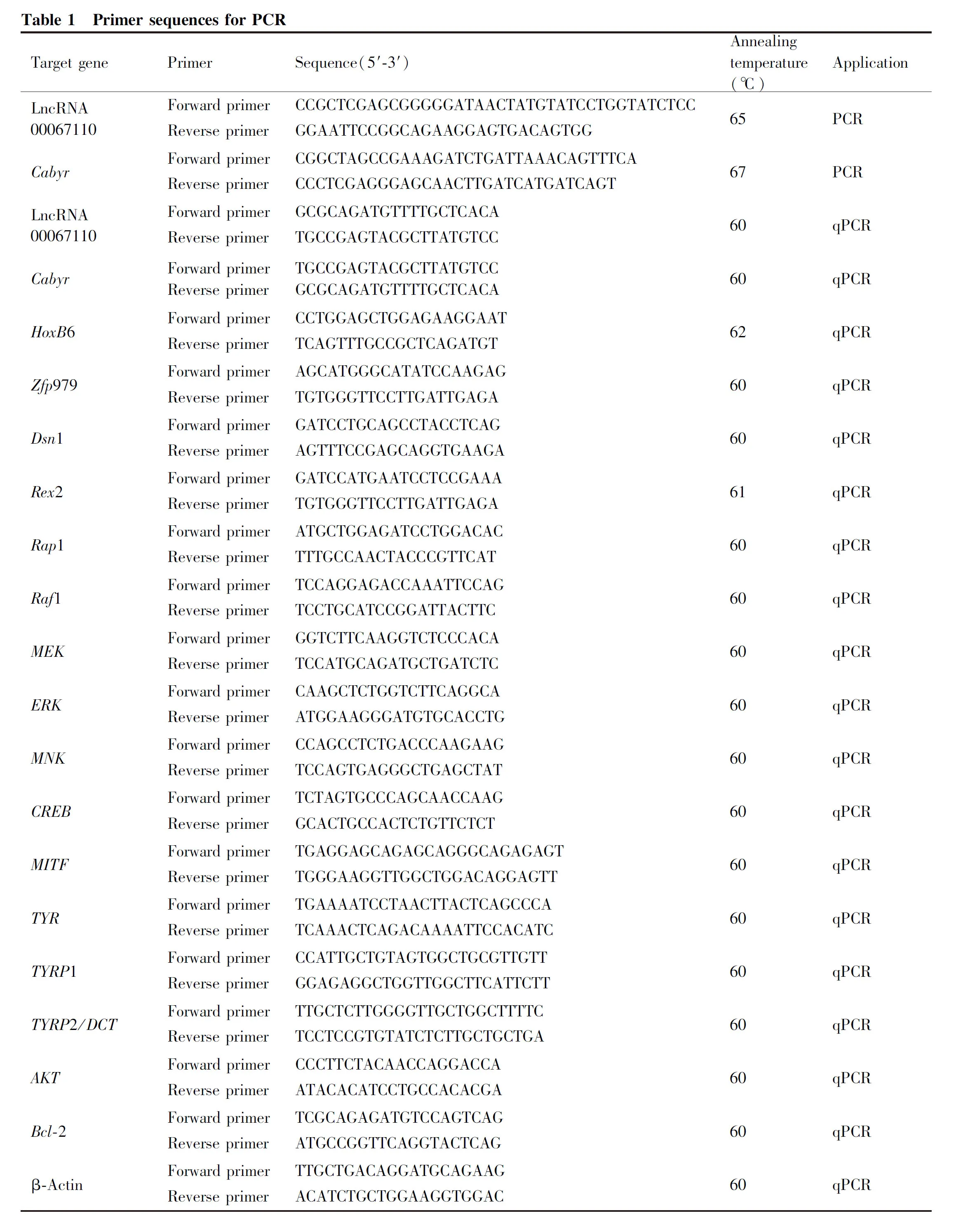
The qPCR results showed that overexpression of LncRNA00067110 in B16 cells increased the mRNA expression of LncRNA000671110 (Fig.1B). The target gene of lncRNA00067110 wasCabyrwhich was predicted by analyzing the location of the genes and lncRNAs using the LncTar software (Fig.1A). Complementary base pairing suggested thatCabyrwas a potential target for lncRNA00067110. The result of luciferase activity showed that it was significantly decreased in 293T cells co-transfected with lncRNA00067110 andCabyr(Fig.1D).
The high-throughput sequencing analysis of RNA from B16-F10 cells with overexpression of lnc-RNA00067110 was conducted to further analyze the potential genes regulated by lncRNA00067110, In total, 17 genes were significantly expressed, among which 10 genes were upregulated and 7 genes were downregulated. qPCR was performed to verify 5 differentially expressed genes that showed the similar expression trend as sequencing data and the results showed thatCabyr, homeobox B6 (Hoxb6), Zinc finger protein 979 (zfp979) andDsn1 were upregulated and Reduced expression 2 (Rex2) was downregulated (Fig.1C).

Table 2 Differential gene expression between the experiment group (LncRNA00067110) and the control group (NC)
2.2 LncRNA00067110 inhibited B16-F10 cell proliferation
The RTCA system and CCK-8 were used to analyze the effect of lncRNA00067110 on the cell proliferation of B16-F10 cells. The results showed that lnc-RNA00067110 could inhibit B16-F10 cells proliferation (Fig.2A, B, C). Based on the PKA domain ofCabyr(from NCBI), it was speculated that lncRNA00067110 might regulate PKA and participate in the MAPK pathway to regulate cell proliferation. The mRNA and protein expressions of Rap1, Raf1, MEK1, ERK1, MNK1 and CREB of the MAPK pathway, were down-regulated (Fig.2D, E, F). Since phosphorylation is key to control the cell cycle, the down-regulation of pMEK and pERK proteins were detected, while the expression of pCREB protein was unchanged (Fig.2G).
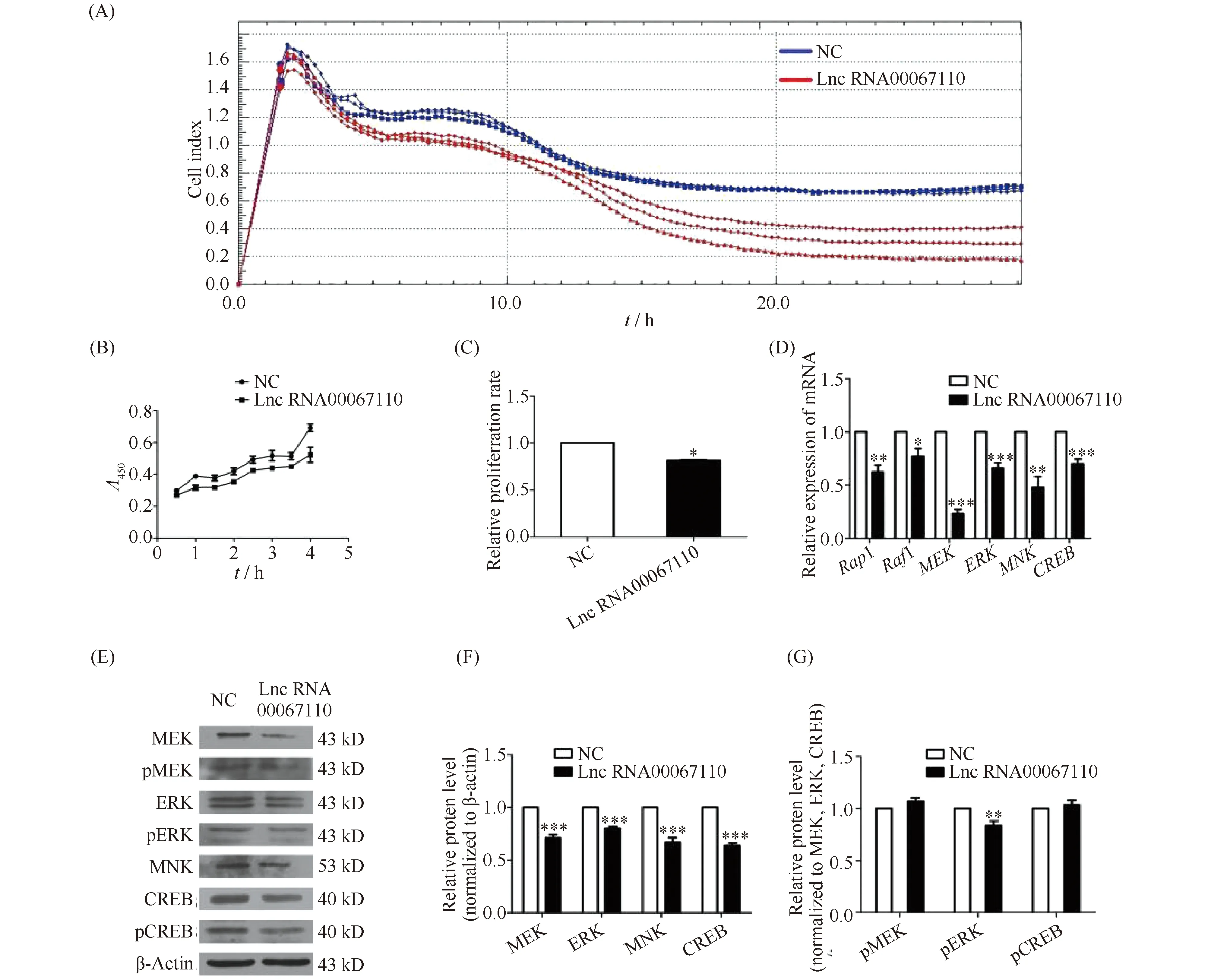
Fig.2 LncRNA00067110 regulated the proliferation of B16-F10 melanoma cells (A) The RTCA system was used to detect the proliferation of B16-F10 cells with lncRNA00067110 overexpression. (B, C) CCK-8 detected the proliferation of B16-F10 cells with lncRNA00067110 overexpression (n = 8);*P <0.05. (D) The relative expression levels of Rap1, Raf1, MEK1, ERK1, MNK1 and CREB in B16-F10 melanoma cells were significantly reduced by qPCR analysis. The data were represented as the mean ± standard error (n = 4).*P <0.05,**P <0.01,***P <0.001. (E, F, G) Western blotting was used to detect the relative expression of MEK, ERK, MNK, CREB and pMEK, pERK, pCREB in B16-F10 cells with lncRNA00067110 overexpression. The data were represented as the mean ± standard error (n = 4).*P <0.05,**P <0.01,***P <0.001
2.3 LncRNA00067110 inhibited melanins production in B16-F10 cells
The tyrosinase activity and the production of total melanins (ASM), eumelanin (EM) and pheomelanin (PM) were measured at the absorbance of 460 nm, 450 nm, 350 nm and 400 nm, respectively. Compared with the NC group, lncRNA00067110 overexpression in B16-F10 cells significantly reduced the tyrosinase activity and the production of total melanin, eumelanin and pheomelanin content (Fig.3A). Also, the mRNA and protein expression of melanogenic genes TYR, MITF, TYRP1 and TYRP2 in B16-F10 cells transfected with lncRNA00067110 were down-regulated (Fig.3B, C, D).
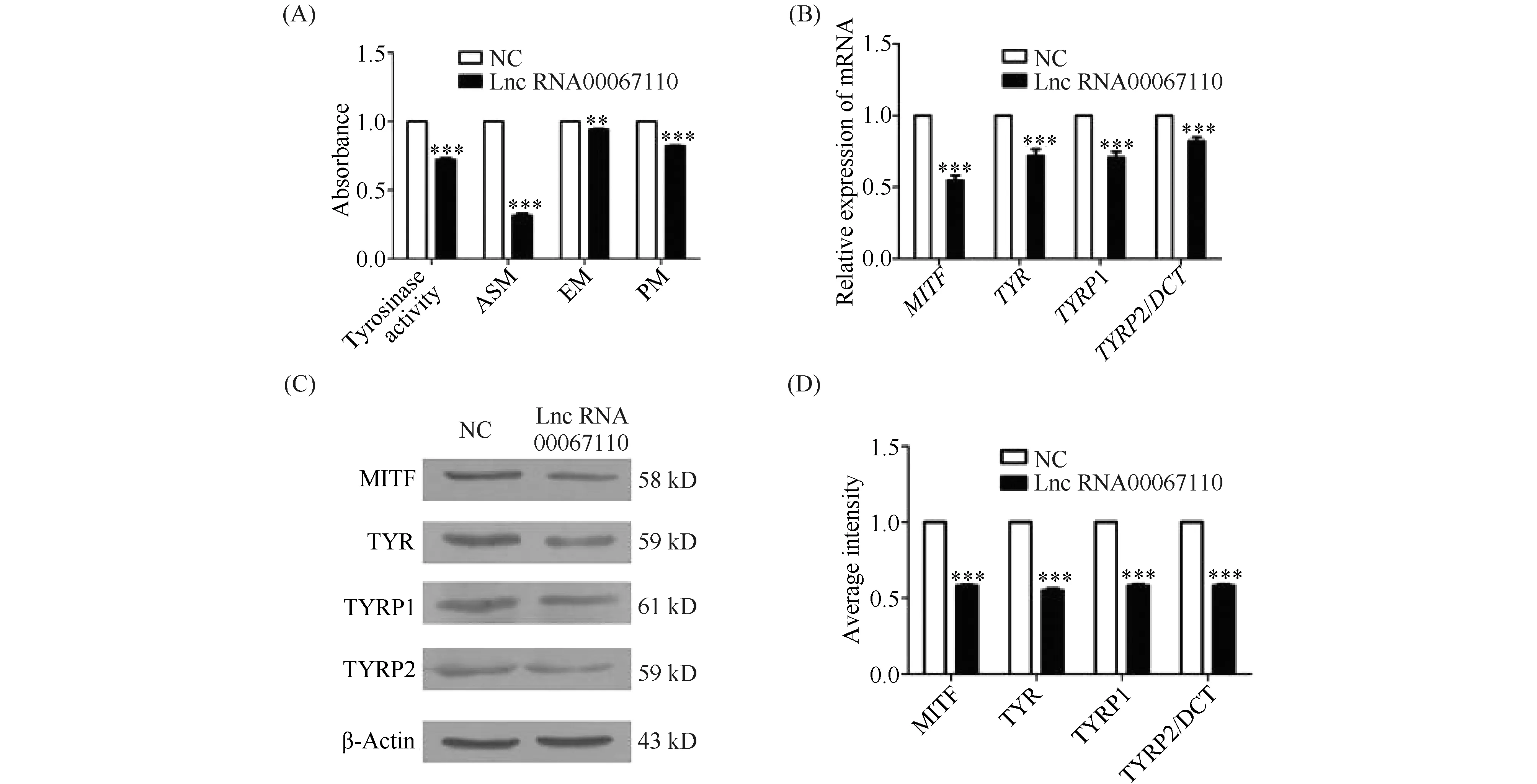
Fig.3 The effect of lncRNA0067110 overexpression on the tyrosinase activity and melanins production in B16-F10 cells (A) The effect of overexpression of lncRNA00067110 on tyrosinase activity, ASM, EM and PM in B16-F10 cells. The data were represented as the mean ± standard error (n = 3).**P <0.01,***P <0.001. (B) qPCR result showed the relative expression levels of MITF, TYR, TYRP1 and TYRP2 in B16-F10 cells were significantly reduced. The data were represented as the mean ± standard error (n = 4).***P <0.001. (C, D) Western blotting result showed that MITF, TYR, TYRP1 and TYRP2 protein expression in B16-F10 cells were significantly reduced. The data were represented as the mean ± standard error (n = 4).***P <0.001
2.4 LncRNA00067110 promoted B16-F10 cell apoptosis
The TUNEL result showed that the overexpression of lncRNA0067110 promoted the apoptosis of B16-F10 melanoma cells (Fig.4A).AKTplays an important role in cell survival and apoptosis, also known asPKBorRac. In order to investigate whether lncRNA00067110 was involved in the regulation ofAKTandBcl-2 in the apoptosis of melanoma cells, the mRNA expression and protein expression of AKT and Bcl-2 were tested and the results showed the down-regulation of the expression ofAKTandBcl-2 by the overexpression of lnc-RNA0067110 (Fig.4B, C, D, E).
Fig.4 LncRNA00067110 promoted the apoptosis of B16-F10 melanoma cells (A) TUNEL result showed the apoptosis of B16-F10 cells was promoted by the overexpression of lncRNA00067110. (B) qPCR result showed that the relative expression levels of AKT and Bcl-2 in B16-F10 cells were significantly reduced by the overexpression of lncRNA00067110. The data were represented as the mean ± standard error (n = 4).***P <0.001. (C, D, E) Western blotting was used to detect the relative expression of AKT, pAKT and Bcl-2 protein in B16-F10 cells with the overexpression of lncRNA00067110. The data were represented as the mean ± standard error(n = 4).**P <0.01,***P <0.001. This experiment was repeated three times
3 Discussion
Evidences have revealed that the etiology of melanoma involved environmental, endocrine and genetic factors, so a large number of researchers began to pay their attentions to the molecular mechanism, in order to find useful biomarkers (such as miRNA, protein, lnc-RNA) for non-invasive early detection or novel effective therapeutic targets in melanoma patients[13]. For instance, the expression of long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential in metastatic melanoma[14]. The long noncoding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation[15]. However, the mechanistic details of most lnc-RNA functions in melanoma remained unknown.
In previous research, lncRNA00067110 is differentially expressed in B16-F10 melanoma cells, which may be related to the biology of melanoma. In order to clarify the function and molecular mechanism of lnc-RNA00067110 in melanoma cells, RNA-seq technology was used to sequence the B16-F10 cells with the overexpression of lncRNA00067110. This study verified the expression of 5 genes with significant differences by qPCR and Western blotting. Among the 5 differentially expressed genes, lncRNA00067110 can bind to the predicted site ofCabyrand regulate the expression ofCabyr.
Cabyris a calcium binding tyrosine phosphorylation regulated fibrous sheath protein, which was initially reported to be testis specific[12]. Data have shown thatCabyr, as a new CT antigen, is widely distributed in various tumor tissues and cancer cells, such as liver cancer and esophageal cancer[11]. Considering thatCabyr contains an N-terminal dimerization/docking (D/D) domain, the similar domain of the R subunit of cAMP-dependent protein kinase (PKA) to the D/D domain was the breakthrough in the pathwayRap1/Raf1/MEK1/ERK/MNK1/CREB. Based on the the relationship of lncRNA00067110 targetingCabyr, the key genesMEK1/ERK/MNK1/CREBon the pathway were downregulated by the overexpression of lnc-RNA00067110 in B16 cells. The RAS/RAF/MEK/ERK pathway, one of the most well-known pathways involved in melanoma progression, is regulated by receptor tyrosine kinases, cytokines, and heterotrimeric G-protein-coupled receptors[16]. The small G protein RAS activates a downstream factor RAF followed by sequential activation ofMEKandERK, and this signal is finally transduced to regulation of transcription in the nucleus[17]. The downstream effects of inactivatedERKare extensive, including inhibition of proliferation. Due to the down-regulation of tumor suppressor and cyclin-dependent kinases (CDKs), the regulation ofMITFreduces the survival rate and up-regulates the apoptosis, invasion and metastasis caused by FAS-induced extracellular matrix remodeling, as well as angiogenesis. The cAMP responsive element (CRE) binding protein (CREB) can bind to CRE in theMITFpromoter[18].MITFis a communication center specific to melanocytes and melanoma cells, which mediate the significant differentiation effect of α-melanocyte stimulating hormone (α-MSH) through transcriptional regulation of enzymes essential for melanogenesis in differentiated melanocytes[19]. The down-regulation ofCREBaffected the activation of theMITFpromoter and down-regulated the activities of theTYR,TYRP1 andTYRP2 promoters.TYR,TYRP1, andTYRP2 catalyze the rate-limiting steps in melanins production and are functionally related to the expression of pigment phenotypes, whileTYRandMITFare more direct because they directly participate in melanin production. We speculate thatCabyrwas a target of LncRNA00067110, and its overexpression inhibited the proliferation of melanoma cells and the production of melanoma.
The expressions ofHoxb6,Zfp979,Dsn1 andRex2 are regulated by the overexpression of lnc-RNA00067110 in B16-F10 cells. Zfp protein is involved in the process of inflammation and apoptosis[20]. The overexpression ofDsn1 can maintain the stability of the genome[21]. Taking into account the functions of Zfp protein,Dsn1 andCabyr, through the detection of melanoma apoptosis markersAKT,Bcl-2 and TUNEL, we found that the up-regulation of lncRNA00067110 promoted melanoma apoptosis. The anti-apoptotic factorBcl-2 is a directMITFtarget gene. The apoptosis induced byMITFdepletion in primary melanocytes and melanoma can be partially rescued byBcl-2 overexpression[22]. The down-regulation ofBcl-2 andMITFfurther induces melanin apoptosis.
Thus, overexpression of lncRNA00067110 greatly inhibited the proliferation and melanins production of melanoma, and induced apoptosis of melanoma cells. Therefore, lncRNA00067110 might be a novel target for the treatment of melanoma.
