Assessment of the binding interactions of SARS-CoV-2 spike glycoprotein variants
2022-04-07DeepRghuPmelHmillArpithBnjiAmyMcLrenYuTingHsu
Deep Rghu,Pmel Hmill,Arpith Bnji,Amy McLren,Yu-Ting Hsu,c,1,*
aProduct Characterization,MilliporeSigma,Rockville,MD,20850,USA
bProduct Characterization,MilliporeSigma,Stirling,FK9 4NF,UK
cProcess and Analytical Development,MilliporeSigma,St.Louis,MO,63118,USA
Keywords:
Surface plasmon resonance
SARS-CoV-2
Monoclonal antibody
RBD
ACE2
A B S T R A C T
Severe acute respiratory syndrome-associated coronavirus 2 is a major global health issue and is driving the need for new therapeutics.The surface spike protein,which plays a central role in virus infection,is currently the target for vaccines and neutralizing treatments.The emergence of novel variants with multiple mutations in the spike protein may reduce the effectiveness of neutralizing antibodies by altering the binding activity of the protein with angiotensin-converting enzyme 2(ACE2).To understand the impact of spike protein mutations on the binding interactions required for virus infection and the effectiveness of neutralizing monoclonal antibody(mAb)therapies,the binding activities of the original spike protein receptor binding domain(RBD)sequence and the reported spike protein variants were investigated using surface plasmon resonance.In addition,the interactions of the ACE2 receptor,an antispike mAb(mAb1),a neutralizing mAb(mAb2),the original spike RBD sequence,and mutants D614G,N501Y,N439K,Y453F,and E484K were assessed.Compared to the original RBD,the Y453F and N501Y mutants displayed a significant increase in ACE2 binding affinity,whereas D614G had a substantial reduction in binding affinity.All mAb-RBD mutant proteins displayed a reduction in binding affinities relative to the original RBD,except for the E484K-mAb1 interaction.The potential neutralizing capability of mAb1 and mAb2 was investigated.Accordingly,mAb1 failed to inhibit the ACE2-RBD interaction while mAb2 inhibited the ACE2-RBD interactions for all RBD mutants,except mutant E484K,which only displayed partial blocking.
1.Introduction
Severe acute respiratory syndrome-associated coronavirus 2(SARS-CoV-2)was first identified in early January 2020[1-6].Since then,this virus has rapidly spread globally,causing a pandemic,as declared by the World Health Organization on March 11,2020.More than 160 million infections and 3.3 million deaths have been caused by SARS-CoV-2(as of May 13,2021)[5,7].As a result,through unprecedented efforts,the scientific community has been seeking to develop effective therapies and vaccines to prevent and treat SARS-CoV-2 infections.Within 4 months of its emergence,the complete genomic sequence of the virus was determined,which revealed that the virus belongs to the coronavirus group and is specifically a new member of the sub-genus,Sarbecovirus[4,8].Similar to other coronaviruses,SARS-CoV-2 expresses the spike(S),envelope,membrane,and nucleocapsid,with spikes playing a critical role in its life cycle by interacting with target cell receptors and enabling viral entry.Overall,the structure of the SARS-CoV-2 S protein closely resembles that of the SARS-CoV S protein[9,10].
The receptor binding domain(RBD)is a crucial component of the S protein subunit(S1),which binds to angiotensin-converting enzyme 2(ACE2),a recognized receptor for viral entry.SARSCoV-RBD binds to the cell surface receptor,ACE2,with an affinity in the low nanomolar range[9,11,12].As RBD-ACE2 binding occurs at the starting point of infection,RBD is a prime target for the development of therapeutic interventions.The mechanisms for therapies that aim to neutralize viral infection include either the inhibition of the interaction between the spike protein RBD and ACE2 or the disruption of the S protein S2 domain activity to prevent membrane fusion.Surface plasmon resonance(SPR)has become a leading methodology for analyzing the binding interactions between two molecules.Recent competitive assay studies using SPR revealed the ability of humanized single-domain antibodies to completely block the interaction between the S protein and ACE2[13].Such studies have thus demonstrated the utility of SPR for screening antibody-based therapies that can block the interaction with ACE2 and neutralize virus spike receptor binding interactions.
Attempts to use monoclonal antibodies(mAbs)targeting SARSCoV have achieved limited success in the cross-neutralization of SARS-CoV-2[14,15].This outcome may be explained by a recent study that described the features considered unique among class I viral fusion proteins,such as the presence of three hinge regions in the spike stalk region,leading to increased flexibility,which is speculated to improve virus fitness[16,17].The limited success of cross-neutralization may also be explained by the differences between SARS-CoV and SARS-CoV-2 RBD,such as an increase in electrostatic binding force,key residues forming salt bridges(Arg426-Glu329,Lys390-Glu37,Asp463-Lys26,and Lys465-Glu23),and differences in hydrogen bonds[18,19].Structural differences in the SARS-CoV-2 spike protein impact its interaction with anti-SARS-CoV antibodies,resulting in the limited inhibition of SARSCoV-2 RBD-ACE2 binding.Owing to its crucial role in the virus infection cycle and the importance of therapeutic and vaccine strategies,mutations in SARS-CoV-2 RBD are being closely monitored.
The rapid increase in coronavirus disease 2019(COVID-19)cases in the United Kingdom(UK)has enhanced epidemiological and virological investigations and monitoring.A SARS-CoV-2 variant,referred to as SARS-CoV-2 VOC 202012/01(variant of concern,year 2020,month 12,variant 01),was identified through viral genomic sequencing in the UK.This variant is defined as lineage B.1.1.7 and has multiple spike protein amino acid deletions and mutations,such as deletion 69-70,deletion 144,and mutations N501Y[20],A570D,D614G,P681H,T716I,S982A,and D1118H.This variant raised concerns as it had an unusual number of mutations on the spike protein.Some mutations(N501Yand E484K)have been found in multiple variants,such as the beta(B.1.351)[21]and gamma(P.1/B.1.1.28)variants[22].Mutations in the spike protein are of acute interest as they may impact the binding activity of the protein with ACE2,with possible implications for virus infectivity,and may result in a change in binding affinity with the antibodies raised through natural infection or spike-targeting vaccination.For example,mutation N501Y,which occurs in a position within the RBD,was identified as a key contributor to increase binding affinity with ACE2 associated with increased transmission of the virus[20,23].However,mutation D614G,which occurs outside the RBD,may cause a moderate increase in transmissibility[24].The previously identified high frequency variants,such as mutation N439K[25]and Y453F(mink mutation)[26],are also of significant concern as the antibodies raised after exposure to the original strain spike protein may not neutralize the virus containing these spike variants.E484K is known as an escape mutant in which antibodies targeting the original RBD protein sequence may fail to recognize this particular mutation,leading to consequences regarding the effectiveness of neutralizing antibodies as therapies or the efficacy of existing vaccines targeting the original sequence[27].
To understand the impact of RBD mutations on ACE2 and antispike mAb binding activities,the binding kinetics(association(kon),dissociation(koff)rates,and the equilibrium dissociation constant(KD))of the SARS-CoV-2 RBD(original isolated RBD sequence)protein and the selected RBD mutants(D614G,N501Y,N439K,Y453F,and E484K)against ACE2 and two different antibodies,anti-spike S1 antibody(mAb1)and anti-spike neutralizing antibody(mAb2),were investigated.The SPR technology was used to compare the binding affinity of ACE2-RBD,RBD-mAb1,and RBD-mAb2 interactions.The findings of this study revealed the feasibility of employing SPR to detect differences in the binding kinetics of different RBD mutants with ACE2 and anti-spike mAbs,and the capability of the antibodies to inhibit the ACE2-RBD interaction,which would be indicative of possible neutralizing activity.
2.Materials and methods
2.1.Equipment,software,and settings
A Biacore T200 system(Cytiva,Marlborough,MA,USA)with control software and evaluation software was used for the SPR binding studies and data analysis.PLA 3.0(Stegmann Systems GmbH,Rodgau,Germany)was used for the half maximal effective concentration(EC50)analysis,while GraphPad Prism(GraphPad Software,San Diego,CA,USA)/Minitab-19(Minitab LLC,State College,PA,USA)was used for graphical representations.The sample compartment and analysis temperatures of the Biacore T200 system were 4°C and 25°C,respectively.All analyses were performed at 25°C and pH 7.4.All binding studies were conducted with running buffer and dilution buffer consisting of 0.01 M HEPES,0.15 M NaCl,0.003 M EDTA,and 0.05%(V/V)surfactant P20.Double reference subtraction was performed for all experiments.
2.2.Sensor chips and reagents
The following Biacore sensor chips and reagents were purchased from Cytiva(Marlborough,MA,USA):Series S sensor chip CM5,Series S sensor chip protein A,Biotin CAPture Kit containing Sensor Chip CAP,Biotin CAPture Reagent,regeneration solution(8 M guanidine hydrochloride and 1 M sodium hydroxide),His-Capture Kit containing anti-histidine antibody,immobilization buffer(10 mM sodium acetate,pH 4.5),and regeneration solution(10 mM glycine,pH 1.5).SARS-CoV-2 spike S1 rabbit mAb1,SARS-CoV-2 spike neutralizing rabbit mAb2,and recombinant proteins corresponding to the original SARS-CoV-2 spike RBD;original S1;spike S1 variant D614G;and RBD protein variants N501Y,N439K,Y453F,and E84K were purchased from Sino Biological(Wayne,PA,USA).The recombinant human ACE2 receptor was purchased from Acro Biosystems(Cambridge,MA,USA).The specifications of the materials are presented in Table S1.
2.3.ACE2-spike RBD/S1 interactions
Biotinylated ACE2 protein was captured on the CAP sensor chip by injecting a biotin capture reagent for 120 s at a flow rate of 2μL/min,followed by a 35 s injection of biotinylated ACE2 receptor at aflow rate of 5 μL/min and concentration of 40 μg/mL.Serial dilutions of original and mutant RBD/S1 proteins were injected over a concentration range of 3.9-1000 nM at a flow rate of 30μL/min,with a contact time of 120 s and dissociation time of 600 s.The sensor chip surface was regenerated using 8 M guanidine hydrochloride and 1 M sodium hydroxide.Single-cycle kinetics(SCK)binding analyses were performed in this study.Two ACE2-original RBD SCK analyses were performed at the start and the end of the ACE2-mutant RBD SCK analysis,respectively,as assay controls to ensure the surface integrity and stability of the ligand and the analyte.
2.4.mAb-RBD/S1 interactions
Initially,two assay formats were developed to study the interaction between mAb1 and spike proteins.In the first approach,an anti-histidine(anti-His)antibody was immobilized on the CM5 sensor chip using a standard amine coupling approach.Histidinetagged spike protein S1 and RBD were captured separately on the anti-His surface as ligands and mAb was injected as an analyte.Serial dilutions(1-250 nM)of mAb1 were injected,followed by 10 mM glycine(pH 1.5)to regenerate the surface.In the alternative approach,a protein A sensor chip was used to capture mAb1.Thereafter,serial dilutions of S1 and RBD proteins(1-250 nM)were injected for a separate multicycle kinetic(MCK)analysis.The surface was regenerated using 10 mM glycine(pH 1.5).
All mAb-RBD/S1 interactions after the initial assessment were analyzed using the protein A capture approach.The mAbs were injected for 18 or 12 s(for mAb1/mAb2,respectively)at a flow rate of 10μL/min and concentration of 1μg/mL,for capture on a protein A sensor chip.Serial dilutions of the original and mutant RBD spike proteins were injected over a concentration range of 0.68-500 nM for mAb1 and 0.02-50 nM for mAb2 at a flow rate of 30μL/min,with contact time of 120 s and dissociation time of 300 s.The surface was regenerated using 10 mM glycine(pH 1.5)with a contact time of 60 s and a flow rate of 30μL/min.MCK was used to analyze mAb-RBD binding.
Two mAb-original RBD MCK analyses were performed(at the start and at the end of the MCK analysis of the mAb-mutant RBD interactions,respectively)as assay controls to ensure the surface integrity and stability of the ligand and the analyte.
2.5.Blocking(neutralizing)assay
An assay was performed to determine the ability of the mAb to block the binding of RBD to ACE2.The blocking assay was performed with the same format and settings used for the ACE2-RBD interaction analysis.In this assay,ACE2 was captured on the sensor surface;thereafter,injections of 1)mAb only,2)RBD only,and 3)pre-mixed mAb-RBD solution were carried out.For the blocking assessment,10 mM HBS-EP+buffer at pH 7.4(0.01 M HEPES,0.15 M NaCl,0.003 M EDTA and 0.05%(V/V)surfactant P20)was used as sample diluent.
The following four injections were performed in the initial blocking assessment of mAb1 on the ACE2 captured surface:1)60 nM RBD,2)100 nM mAb1,3)60 nM RBD+100 nM mAb1 preincubated for 45 min(ambient temperature),and 4)60 nM RBD+3.33μM mAb1 pre-incubated for 45 min(ambient temperature).The second assessment for mAb1 was performed on an ACE2 captured surface using the following injection sequence:1)60 nM RBD followed by the injection of 80 nM mAb1,and 2)60 nM RBD followed by the injection of 480 nM mAb1.
The mAb2 blocking assessment was performed with the following:1)60 nM RBD injection only,2)100 nM mAb2 injection only,and 3)injection of a mixture of 60 nM RBD+100 nM mAb2 pre-incubated for 45 min(ambient temperature).The assessment described above was applied to all RBD mutants.
2.6.Data analysis
The kinetic data were analyzed using the Biacore T200 Evaluation Software.The association and dissociation curves of the blank subtracted sensorgrams were fitted to a 1:1 binding model,and the kon,koff,and Rmax(saturation signal where all functional ligand molecules bound to analyte molecules)were determined as global fitting parameters.The analyte binding capacity Rmaxwas fitted globally.The KDwas calculated using the ratio of koffto kon.The relative binding affinity was determined by calculating the ratio of the mean KDof the original RBD(mean KDvalue for original RBD was calculated using the two values obtained within the same assay)to the KDof the mutant RBD interacting with mAb or ACE2.
3.Results and discussion
3.1.ACE2-RBD/S1 interactions
Biotinylated ACE2 was used to establish and optimize the binding interactions between the ACE2-RBD and ACE2-S1 proteins.The ACE2 capture level was optimized to obtain an optimal RBD/S1 binding response.A target Rmaxbetween 50 and 100 RU was used for this assay(Fig.S1 and Table S2).As the S1 subunit mainly consists of the RBD,which is responsible for recognizing cell surface receptors,the interactions observed between ACE2 and either recombinant S1 or RBD protein alone were hypothesized to be similar in this assay system.Based on the results,the binding affinity(KD)of ACE2 for both recombinant spike RBD protein(original sequence)and recombinant spike S1(original S1)protein was highly comparable(KDvalues of 10.12 nM and 10.51 nM,respectively)(Table S2).Hence,subsequent analysis was performed using the original RBD protein as a reference and control for the comparison with mutant RBD proteins.Such finding is also supported by literature data,where the binding affinity of RBD to human ACE2 was found to be comparable to that of the binding affinity of spike protein with human ACE2[11,28].
The sensorgram comparison tool was used to normalize the sensorgrams for the original and mutant RBD proteins,as shown in Fig.1.

Fig.1.Sensorgrams with normalized response(RU)showing the comparison of binding response for the original and receptor binding domain(RBD)/spike protein subunit 1(S1)mutants to the angiotensin-converting enzyme 2(ACE2).
Table 1 shows the kinetic parameters for the ACE2-RBD/S1 interaction for the original RBD and mutants.The konof mutant D614G to ACE2 was 4.8-fold slower than that of the original RBD,whereas the koffwas twice faster,resulting in a 10-fold decrease in the binding affinity[9].Such finding is consistent with that of recent studies where the increased infectivity of viruses with the D614G mutation was not found to be explained by a greater ACE2 binding activity[29].The konof N501Y and Y453F RBD mutants was comparable to that of the original RBD,whereas koffwas 3.4-fold slower for N501Y and 7.8-fold slower for Y453F,resulting in an overall 3.4-fold increase in the affinity for N501Y and a 6.7-fold increase for Y453F.Such finding correlates with various published data for these mutant spike proteins[26,30,31].The konand koffof N439K were found to be approximately 0.8-fold faster,resulting in a slightly higher affinity[11]than that of the original RBD.The konof the E484K mutant was 1.5-fold faster than the ACE2-original RBD interaction,whereas the koffwas twice faster than the original RBD,resulting in a 1.5-fold decrease in the affinity[32].Differences in the absolute KDvalues between the present study and other studiesmay arise due to different preparations of recombinant RBD/S1/ACE2 proteins,lot-to-lot variability,and the assay format utilized.
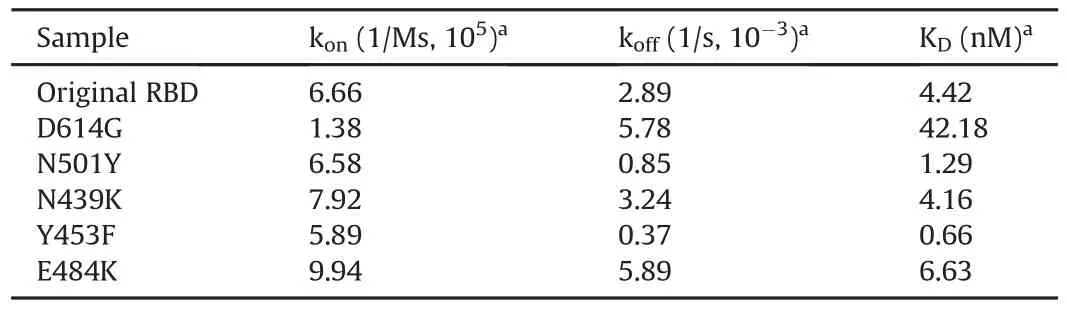
Table 1Average kinetic parameters for the angiotensin-converting enzyme 2(ACE2)-receptor binding domain(RBD)/S1 interaction(n=4).
In this study,kinetic analysis of the ACE2-RBD/S1 interactions could be used as a tool to compare the differences in kinetic parameters that could potentially impact virus infectivity.Aligning with recently reported data,the RBD proteins containing the Y453F[26]and N501Y[30,33]mutations were found to exhibit significantly enhanced binding to ACE2 relative to the original sequence RBD,which might be attributable to the slower kofffor these mutant RBDs.Consequently,these mutations can lead to stronger binding of the virus to the host cell receptor,resulting in higher transmission,which may impact the neutralization capability.
The relative kon,koff,and KDvalues for the ACE2-RBD/S1 variant interactions compared with those of the ACE2-original RBD interaction are presented in Table S3.The D614G and N501Y mutant RBD/S1 proteins displayed distinct kinetic profiles(Fig.1),which was also evident from their low similarity scores compared to those of the other mutants.
Fig.2 displays the mean KDfor the ACE2-original RBD and ACE2-mutant RBD/S1 interactions,as well as the variability of the data.The Y453F and N501Y mutant RBD proteins displayed a significant increase in ACE2-RBD binding affinity relative to the original RBD,and D614G had a substantial reduction in binding affinity.
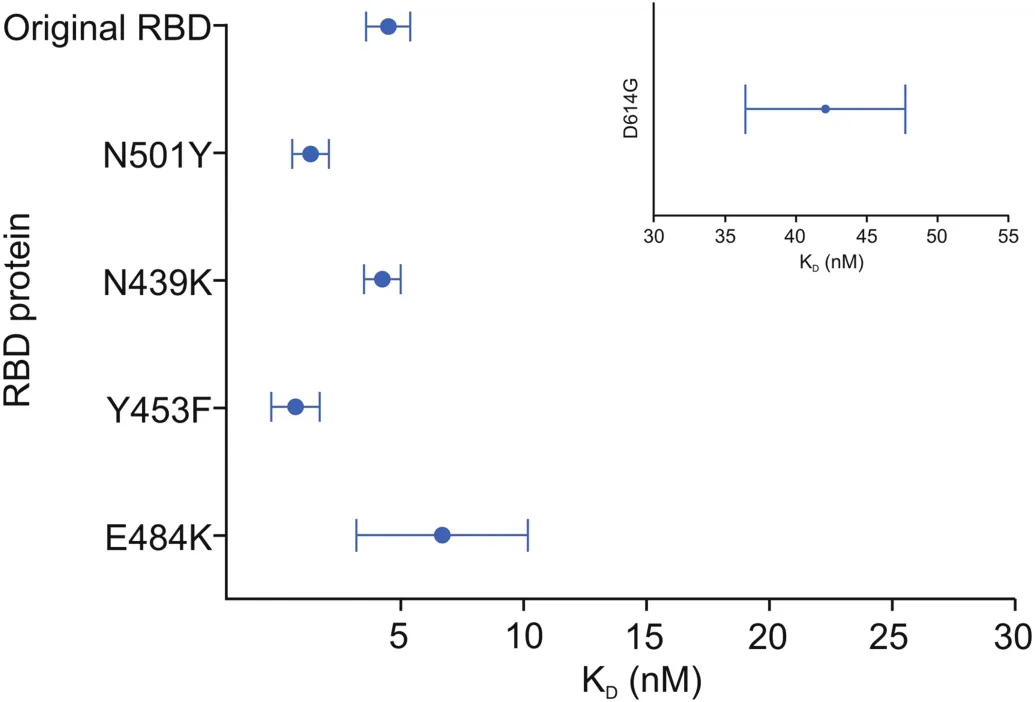
Fig.2.Graphical representation of the equilibrium dissociation constant(KD)values obtained for the ACE2-RBD/S1 and ACE2-S1(D614G)interactions;dot represents the mean KDvalue and the bar represents the 95% confidence interval.
3.2.mAb1-original RBD/S1 interactions
The initial investigation was performed to assess the comparability of the interaction between mAb-RBD and mAb-S1 proteins to characterize the spike protein and to carry out comparability studies of RBD/S1 mutants and the original RBD.The His-capture format was initially optimized for the analysis of the mAb1-RBD and mAb1-S1 interactions(Table S4).The capture level of the original RBD was first optimized to obtain an optimum response from the mAb1 injections,and a target Rmaxof 50 RU was used for this method.An alternative assay format was also optimized,where mAb1 was captured on a protein A sensor chip,followed by RBD protein injections.The kinetic binding results from His-capture format and protein A capture format were assessed(Table S4),and the KDvalues determined for both methods were found to be comparable.Hence,the protein A capture format was selected for subsequent analysis of mAb1-RBD/S1(Figs.S2 and S3).
3.3.Comparability studies of mAb and RBD/S1 interactions
To compare the binding properties of mAbs with the original RBD versus the mutant RBD/S1 proteins,the protein A capture format was selected,where mAb was immobilized as a ligand,ensuring constant capture levels of mAb and avoiding any variability due to potentially different capture levels of the RBD/S1 proteins.The binding profiles and affinities of the original RBD and mutant proteins were assessed.
Based on the results for the interactions between the original RBD and mutants with mAb1,all RBD/S1 mutants,except E484K,displayed a decrease in affinity(Table S5,Supplementary data).The KDvalues in this study were comparable to recently reported results for other mAbs[31,34].The exact KDvalues can vary depending on the analysis method and source of recombinant proteins.
The relative kinetic parameters for the mAb1-RBD/S1 interaction indicated that all mutant proteins had a reduced relative KDto the original RBD,except for E484K,where the affinity had increased by11.9%(Table S6,Supplementary data).The mAb-RBD/S1 comparability assay enables the characterization of the effect of RBD/S1 mutations on binding to mAbs,which allows the assessment of the potential impact on therapeutic properties.These findings suggest a potential reduction in the neutralization capability of mAb1 for each mutant(especially D614G)and a potential increase in the likelihood of the mutated virus escaping the immune response.The sensorgrams overlaid with a fitted 1:1 binding model for mAb1-original RBD and mAb1-mutant interactions revealed a significant difference in the kinetic profiles between D614G and the other RBD interactions(Fig.S4).The low similarity score(21.1%)of D614G relative to the original RBD,compared to the other mutants,also highlighted the impact of the mutation to the binding interaction(Table 2).
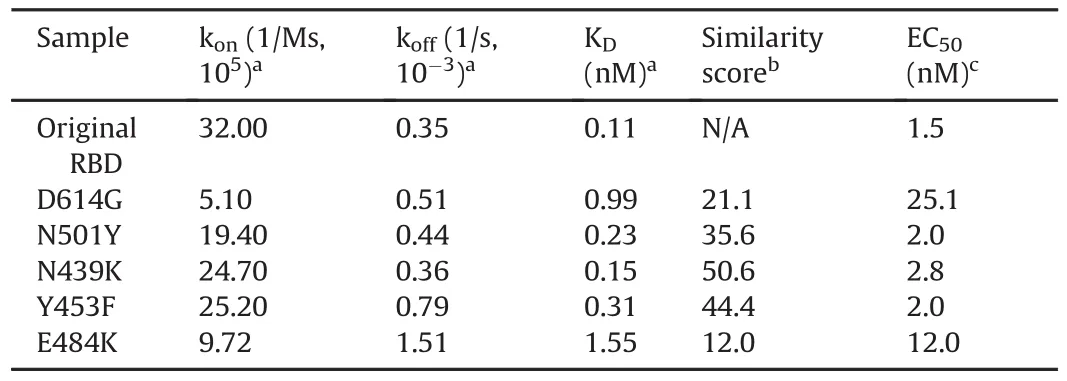
Table 2Kinetic parameters,similarity score,and EC50values for the spike neutralizing rabbit monoclonal antibody(mAb2)-RBD/S1 interaction(n=6).
The kinetic parameters for the interactions between mAb2 and the original RBD and mutants are listed in Table 2.All RBD/S1 mutants were found to display an increase in KDvalues,a decrease in kon,and an increase in koff.The surface responses(binding report point after analyte injection)were fitted using a 4-parameter logistic fit,and the EC50was calculated using the unrestricted model to compare the interaction of mAb2 with the mutants and the original RBD(Fig.S5).
For the interaction with mAb2,all mutant RBD/S1 proteins were found to have a reduced relative KDcompared to that of the original RBD(7.10%-73.33% relative to the original RBD,Table S7 and Fig.S6).The fitted sensorgrams(Fig.S7)indicated that mAb2-original RBD and mAb2-RBD/S1 mutants followed a 1:1 interaction,as the observed sensorgrams highly agreed with the selected model.
3.4.Blocking(neutralization)assay
Blocking assays were performed to assess the ability of mAb1 and mAb2 to prevent the ACE2-RBD interaction;this is because the ability of either antibody to bind to RBD and prevent the receptor/spike RBD interaction may be predictive of virus neutralizing ability.The mAb1/mAb2 concentrations selected for the blocking assessment were based on previously determined affinities(KD)for the ACE2-RBD and mAb-RBD interactions.To sufficiently inhibit the ACE2-RBD interaction,the affinity of the mAb-RBD interaction,as well as that of ACE2-RBD,will play a crucial role.Hence,several concentrations of mAb were assessed to determine the ability of the mAb to disrupt the ACE2-spike RBD interaction.The mAb concentrations used for the blocking assay were higher than the saturation concentration and were selected based on the KDvalue of the ACE2-RBD(in excess relative to the KDvalue).
3.4.1.mAb1-RBD blocking assay
The ACE2-original RBD interaction was assessed in the presence and absence of mAb1.The sensorgrams(Fig.S8)show the similarity of the binding response of RBD incubated with 3.3μM of mAb1 and RBD incubated with 100 nM of mAb1.The interaction with ACE2 was enhanced in the presence of mAb1.In the same study,mAb1 was not found to directly interact with ACE2.
To further investigate this phenomenon,an additional assay was performed using mAb1, where the binding response of mAb1 to RBD(60nM)at 480 and 80 nM was assessed on an ACE2 captured chip.An increase in the binding response between ACE2 and RBD in the presence of higher concentrations of mAb1 confirmed that mAb1 failed to inhibit the ACE2-RBD interaction(Fig.S9).This finding indicates that mAb1 and ACE2 bind to RBD at non-overlapping sites.The effectiveness of putative therapeutic neutralizing mAbs to prevent RBD binding to ACE2 is critically dependent on the location of the binding site.Such finding is consistent with that of recently published studies on alterative neutralizing mAb epitopes,which indicated that for certain mAbs,the binding site involved in the mAb-SARS-CoV-2 spike interaction does not overlap with the binding site for ACE2-RBD interaction[11,15,34].
Using the assay system,no inhibition of the ACE2-RBD interaction was observed with mAb1,a commercially available mAb that is generated for spike protein detection purposes and is raised against the entire S1 region as the immunogen.Hence,mAb1 may bind to an epitope outside of the RBD.Further,to our knowledge,the ability of this antibody to neutralize virus infectivity has not been observed[35].Further complementary techniques are required to understand the mechanism by which mAb1-RBD increases the binding between ACE2 and RBD.Additionally,these techniques provide insights into the region of S1 epitope dependence of spike-ACE2 binding.Analysis of the RBD mutant binding interactions with mAb1 was not performed as mAb1 failed to block the ACE2-original RBD interaction.
3.4.2.mAb2-RBD blocking assay
The binding of the original RBD to ACE2 was assessed in the presence and absence of mAb2.The sensorgrams show the binding response of RBD to ACE2 and the subsequent pre-incubation of different mAb2 concentrations with RBD(Fig.3).A dose-dependent response was observed,with 1 nM mAb2 having no impacton RBDACE2 binding,10 nM mAb2 resulting in limited inhibition,and 100 nM mAb2 resulting in complete blocking of the RBD-ACE2 interaction.The ability of mAb2 to block binding to ACE2 was also investigated for each of the mutant proteins and compared with the blocking effect exhibited by the original RBD.Preincubation of mAb2 resulted in the complete blocking of ACE2-RBD mutant binding interactions for all mutants,except for E484K,where only partial inhibition of the interaction was observed using 100 nM mAb2(Fig.4).A correlation was found between mAb2-RBD affinity and the extent of inhibition of the ACE2-RBD interactions;this is because the kon,koff,and KDvalues of the mAb2-E484K interaction were significantly lower than those of the original RBD(which had higher KDand EC50values,indicating lower binding affinity).Hence,a higher concentration of mAb2 would be required to block the interaction between ACE2 and E484K.The present study revealed the benefit of obtaining kinetic data for putative neutralizing mAbs at early stages as part of the screening process;such data may indicate the relative potency of the blocking activity.
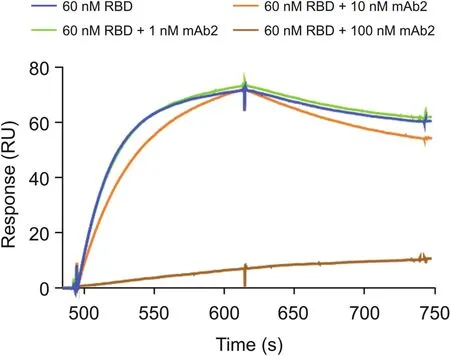
Fig.3.Spike neutralizing rabbit monoclonal antibody(mAb2)-RBD blocking assay.Response from the binding of RBD to ACE2 in the presence and absence of mAb2.ACE2 was captured on the chip;thereafter,the following was injected:60 nM RBD,60 nM RBD+1 nM mAb2,60 nM RBD+10 nM mAb2,and 60 nM RBD+100 nM mAb2(incubated for 45 min).
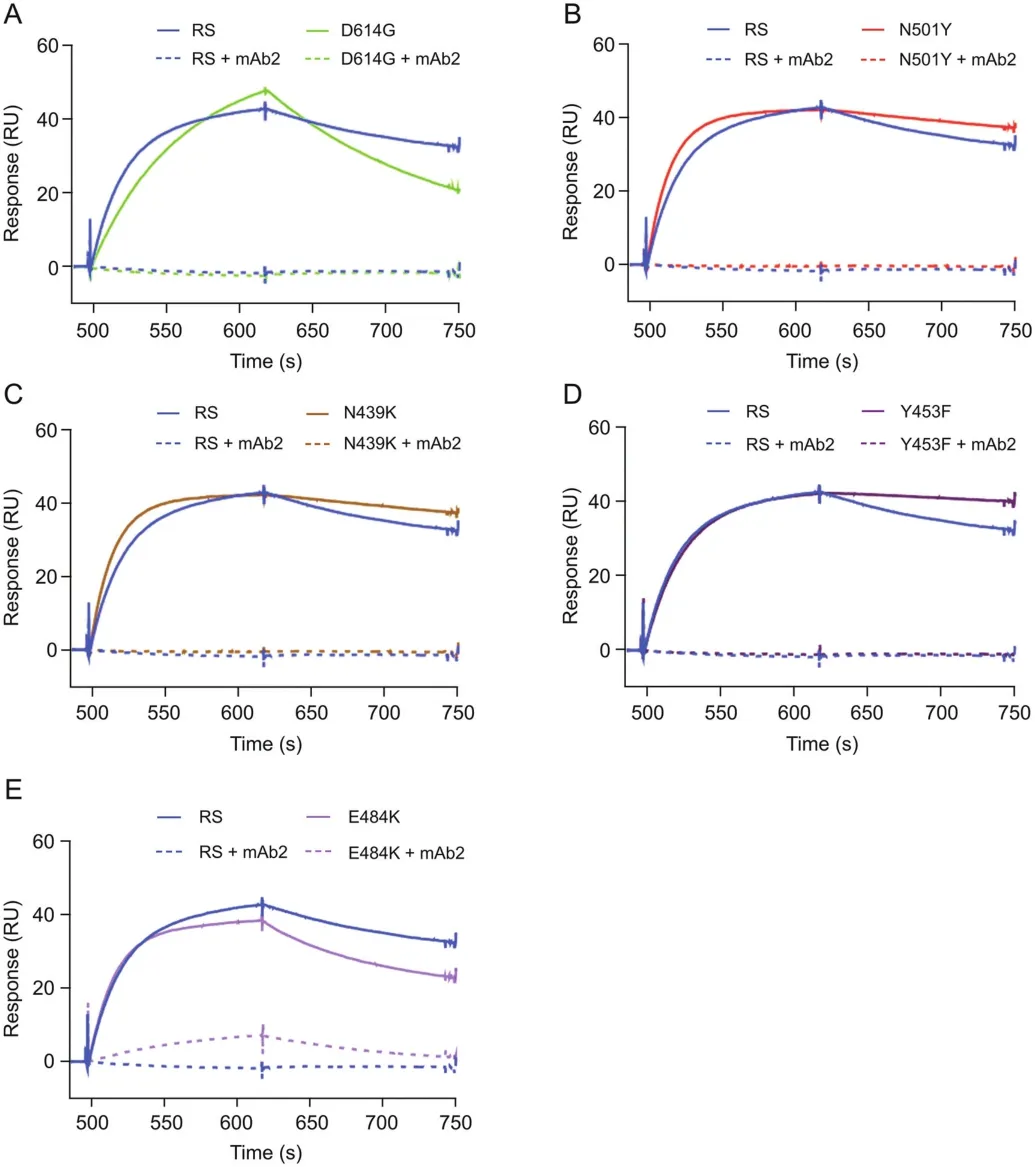
Fig.4.mAb2-RBD blocking assay.Responses from the binding of RBD and its variants to ACE2 in the presence and absence of mAb2.ACE2 was captured on the chip;thereafter,the following was injected:60 nM reference standard(RS/original RBD)/variant,60 nM RBD/variant+100 nM mAb2(incubated for 45 min).The variants used were(A)D614G,(B)N501Y,(C)N439K,(D)Y453F,and(E)E484K.
mAb2,a commercially available antibody purchased to mimic mAb therapeutic candidates,was employed to assess the suitability of the assays to screen for RBD-targeting mAbs and to demonstrate their potential interaction blocking ability.In contrast to mAb1,mAb2 has been utilized in a cell-based infectivity assay by the reagent manufacturer,using SARS-CoV-2 spike protein pseudotyped virus and human embryonic kidney 293T-ACE2 cells.The reported results indicate that mAb2 concentrations above 4 μg/mL(approximately 30 nM)achieved 99.9% inhibition,which implies that the mAb has virus-neutralizing activity[36].
The combination of data from both mAb-RBD interaction and ACE2 competition assays was required for potential mAb candidate assessment.Based on the data generated for mAb1(anti-spike S1 antibody)and mAb2(anti-spike neutralizing antibody),although mAb1 exhibited similar binding affinities to spike proteins(low nM range),it did not disrupt the binding between ACE2-spike protein and potentiated the interaction.Such finding indicates that mAb1 may interact with an epitope distinct from the ACE2 binding region,which may result in conformational changes in the spike-antibody complex,which might increase the binding to ACE2.In contrast,mAb2,a commercial mAb that has been demonstrated to possess virus-neutralizing activity,was found to block the ACE2-RBD interaction using the same assay system.Further studies analyzing the kinetic binding parameters of mAb2 with a panel of spike proteins revealed a correlation between the mAb-spike affinities and their blocking potency.
4.Conclusion
The SPR assays established in this study were successfully used to assess the binding interactions for ACE2-SARS-CoV-2 RBD and mAb-SARS-CoV-2 RBD.As the assays are sensitive to differences in binding behavior conferred by amino acid changes in the RBD,they can be used to screen therapeutic neutralizing mAbs against variants;this is because these changes provide a level of insight into the effect of the spike RBD mutations on ACE2 binding properties when they occur,ultimately forming the basis for candidate selection.
Although this in vitro system utilizes recombinant purified spike protein(monomeric)that is not present in the same conformation found in the native,virus envelope-associated form,it offers a rapid and simple approach for the characterization of RBD binding activities.The data generated in this study are consistent with those reported for the impact of mutations on increasing ACE2 binding and its association with enhanced transmission.However,this approach has limitations for gaining an understanding of the impact of mutations outside the RBD,in other regions of the spike,and how these mutations impact the binding directly with RBD through changes to the overall spike protein confirmation.A good example is the phenomenon of the D614G mutation,which is associated with increased virus transmissibility in the absence of enhanced binding to ACE2.The additional structural elements outside the RBD may explain the impact of viral entry into target cells.
Overall,this analysis system provides insights into the binding interactions of the minimal antigenic region targeted for antibody neutralization activity,which can also be complemented with approaches that enable the understanding of protein conformation,as well as infectivity assays,to broaden the understanding of the impact of spike protein mutations on virus transmissibility.In particular,the generation of kinetic binding data using SPR for the interactions between spike RBD,ACE2,and mAbs provides useful additional insights into complementary characterization techniques through endpoint analysis only(e.g.,enzyme-linked immunosorbent assay and cell-based infectivity assays).
CRediT author statement
Deepa Raghu:Methodology,Validation,Formal analysis,Investigation,Data curation,Writing-Original draft preparation,Visualization,Writing-Reviewing and Editing;Pamela Hamill:Conceptualization,Writing-Reviewing and Editing;Arpitha Banaji:Data curation;Amy McLaren:Data curation;Yu-Ting Hsu:Conceptualization,Methodology,Validation,Formal analysis,Investigation,Data curation,Writing-Original draft preparation,Visualization,Writing-Reviewing and Editing.
Declaration of competing interest
All authors are employed by Merck KGaA which provides products and services to the pharmaceutical industry.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2021.09.006.
杂志排行
Journal of Pharmaceutical Analysis的其它文章
- Screening strategies for quorum sensing inhibitors in combating bacterial infections
- Capillary electrophoresis methods for impurity profiling of drugs:A review of the past decade
- Antiviral,immunomodulatory,and anticoagulant effects of quercetin and its derivatives:Potential role in prevention and management of COVID-19
- Anti-inflammatory and analgesic properties of Moroccan medicinal plants:Phytochemistry,in vitro and in vivo investigations,mechanism insights,clinical evidences and perspectives
- Identifying the molecular basis of Jinhong tablets against chronic superficial gastritis via chemical profile identification and symptomguided network pharmacology analysis
- MTBSTFA derivatization-LC-MS/MS approach for the quantitative analysis of endogenous nucleotides in human colorectal carcinoma cells
