Association of the Expression of Glycolytic Pathway-related Genes with the Physiological Behavior of Lactobacillus plantarum
2021-12-03CHENXingGUOShuyuTAOYufeiDENGXiYANGShuhuiTUMingxiaWUAilingMASiyiRAOYuLIULei
CHEN Xing, GUO Shuyu, TAO Yufei, DENG Xi, YANG Shuhui, TU Mingxia, WU Ailing, MA Siyi, RAO Yu, LIU Lei
(School of Food and Biological Engineering, Xihua University, Chengdu 610039, China)
Abstract: This study aimed to investigate whether and how pgm, fba, and gap overexpression can affect the stress resistance,adhesiveness, and biofilm formation of Lactobacillus plantarum LR-1.Results showed that the overexpression of the pgm and fba genes enhanced the heat, acid and bile salt resistance, biofilm formation and adhesiveness of L.plantarum LR-1.The transcriptional level of the gap gene had no significant effect on the stress resistance, biofilm formation or adhesiveness.The up-regulation of pgm, fba, and gap increased the transcriptional levels of tuf and luxS in this strain with the first two being more effective than the last one.Furthermore, bioinformatic analyses suggested that pgm and fba could be associated with genes related to multiple pathways.These results provide a new understanding of the role of glycolytic pathway-related genes in regulating the stress resistance, adhesiveness, and biofilm formation of L.plantarum.
Keywords: Lactobacillus plantarum; glycolysis; stress resistance; adhesiveness; biofilm formation; bioinformatic analysis
Probiotics are orally administered live microorganisms,which, when taken in adequate amounts, confer a health benefit on the host[1].Lactobacillus paraplantarumis one of the most widely used species.Several studies suggest thatL.plantarumhas a positive impact on various diseases, such as irritable bowel syndrome, inflammatory bowel disease,diarrhoea and atopic dermatitis[2-4].Bacterial agents suffer adverse environments during the production and application process, and the survival activity of the cells needs to remain stable during storage and manufacturing process, as well as in the human gastrointestinal tract (GIT).Currently, the ability of bacteria to resist stress has been mainly improved via engineering methods, such as developing wall materials and additives[5-6].How to improve the properties of strains through intracellular gene modification remains unclear.Furthermore, the exact gene functions ofL.plantarumare not fully understood yet.
Phosphoglycerate mutase family protein (PGAM orPGM), fructose-bisphosphate aldolase (FBA), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH),encoded by thepgm,fbaandgapgenes, respectively,participate in the glycolytic pathway.This carbohydrate metabolism pathway is the primary energy source for the biological activity of lactic acid bacteria[7].Despite their roles in carbohydrate metabolism, they all have additional functions.Previous studies have demonstrated that they are regulated by the LuxS/autoinducer-2 (AI-2) quorum sensing system and that their expression affects the biofilm formation ofL.paraplantarum[8-9].In addition, dPGM is reportedly essential for the formation of symbiosis with the host plant,Burkholderia phymatum.Thefbaregulates transcription in a moonlighting role inFrancisellasp.[10-11].It also acts as an important adhesin, mediating binding to host cells via the fibronectin inMycoplasma hyopneumoniae[12].As for GAPDH, various functions have been found that are unrelated to glycolysis.It can interact with some types of DNA damage and proteins participating in DNA repair[13].Furthermore,GAPDH is one of the surface adhesins of many bacteria,such asStreptococcus agalactiae,Mycoplasma pneumoniae,Mycoplasma suisandClostridium perfringens[14-17].It also increase the adhesion of theLactobacillusstrains to human epithelial cells[18].
This study aims to construct several strains ofL.plantarumto upregulate either thepgm,fbaorgapgenes,and to investigate the impact of their overexpression on the physiological behaviour ofL.plantarum.Furthermore,this study determines whether the upregulation ofpgm,fbaandgapaffects the transcription levels oftufandluxSof this strain, andpgm,fba,gapare demonstrated to be multifunctional genes.Bioinformatic analysis is conducted to screen the potential interaction with these genes and the associated types.
1 Materials and Methods
1.1 Materials and reagents
1.1.1 Bacterial strains, cells, and culture conditions
L.plantarumLR-1, isolated initially from fermented pickles, was used as the parental strain.It was cultivated at 37 ℃ in de Man-Rogosa-Sharpe (MRS) broth (Beijing Land Bridge Technology Co.Ltd., Beijing) or aerobically in MRS-Agar, and stocks were kept in solidified MRS medium.Escherichia coliDH5α (Takara Biotechnology Co.Ltd.,Dalian) was grown in Lennox broth (LB) or LB agar at 37 ℃.Transformant strains ofE.coliDH5α were cultured in LB medium containing 200 μg/mL erythromycin.Transformant strains ofL.plantarumLR-1 were cultured in MRS broth or MRS-Agar containing 3 μg/mL erythromycin.
HT-29 cells were purchased from the American Type Culture Collection (ATCC, Manassas, WA, USA).These cells were cultured in Dulbecco’s Modified Eagle’s Medium(DMEM, Amresco) and supplemented with 10% (V/V) fetal calf serum (Zhejiang Tianhang Biological Technology Stock Co.Ltd., Zhejiang), penicillin (100 IU/mL, Gibco) and streptomycin (100 μg/mL, Gibco) at 37 ℃ in an atmosphere of 5% CO2/95% air at constant humidity.
1.1.2 Construction of plasmids and bacterial strains
The chromosomal DNA ofL.plantarumLR-1 was isolated using a TIANamp bacteria DNA Kit (Tiangen Biotech Co.Ltd., Beijing).Thepgm,fbaandgapgenes were amplified by polymerase chain reaction (PCR)using the primers listed in Table 1 containing restriction sites,XbaI andXhoI.The PCR product was cloned into a pMD18T vector (Takara Biotechnology Co.Ltd., Dalian)to construct vectorspgm-pMD18T,fba-pMD18T andgappMD18T.The recombinant vectors and pMG76e vector(kindly provided by Professor Shangwu Chen, China Agricultural University) were digested with FastDigest enzymes,XbaI andXhoI (Thermo Scientific), at 37 ℃ for 5 min to obtain the target gene fragments and dual-enzyme digested linearised pMG76e plasmid.Subsequently, the target gene fragments were separately inserted into the linearised pMG76e plasmid using a Rapid DNA Ligation Kit (Thermo Fisher Scientific, MA, USA).The ligation products weretransformed intoE.coliDH5α and verified by PCR using the primers 5’-TTCGGTCCTCGGGATATG-3’ (forward)and 5’-CTGTCTTGGCCGCTTCAA-3’ (reverse).The recombinant pMG76e plasmids containing either thepgm,fbaorgapgenes, as well as the empty pMG76e were electrotransformed intoL.plantarumLR-1 competence cells.The recombinant strains were selected with erythromycin(3 μg/mL) and verified by PCR using the primers mentioned above.

Table 1 Primer sequences used for PCR in this study
1.1.3 Other materials and reagents
TUREscript 1stStrand cDNA Synthesis Kit (Aidlab Biotechnologies Co.Ltd., Beijing); SYBR Green assay kit(Tiangen Biotech Co.Ltd., Beijing); 96-well plate (Corning,NY, USA).
1.2 Instruments and equipments
7500 Fast real-time PCR system (Applied Biosystems,USA); Synergy 2 microplate reader (Biotech, Winooski,VT, USA).
1.3 Methods
1.3.1 Real-time PCR
The recombinant strains and the wild type strain were cultured for 8 h until achieving the logarithmic phase.Total RNA was extracted from these cells using Invitrogen TRIzol reagent according to the manufacturer’s protocol.RNA quality was determined byA260nm/A280nm,A260nm/A230nmand electrophoresis.The isolated RNA was transcribed into single-stranded cDNA using a TUREscript 1stStrand cDNA Synthesis Kit.Real-time PCR was performed using an SYBR Green assay kit and the 7500 Fast real-time PCR system.Primers were designed using Primer 3 Input and listed in Table 2.16S rRNA was used as an internal reference.The relative expressions of thepgm,fbaandgapgenes were calculated using the 2−ΔΔCtmethod according to Livak et al.[19].Therefore, to determine the effect of the overexpression of thepgm,fbaandgapgenes on thetufandluxSgenes inL.planturamLR-1, the relative expressions of these genes were also calculated using the 2−ΔΔCtmethod, with the primers as shown in Table 2.
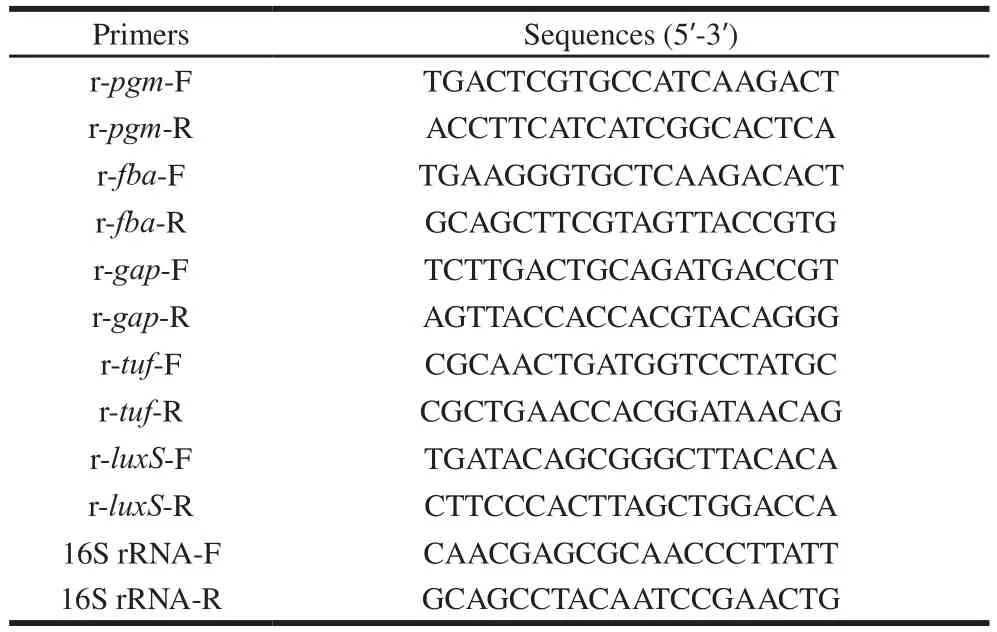
Table 2 Primer sequences used for real-time PCR in this study
1.3.2 Growth curves
The recombinant strainspgm-pMG76e-LR-1,fbapMG76e-LR-1,gap-pMG76e-LR-1, pMG76e-LR-1 and the wild type strain LR-1 were cultured in MRS without erythromycin.At 3, 6, 9, 12, 15, 18, 21, 24, 27, 30, 33 and 36 h, the cell densities were determined by measuring the OD600nmusing a UV-1800 spectrophotometer.Measurements were performed in triplicate.
1.3.3 Stress resistance assay
The recombinant strains and wild type strain were cultured in MRS without erythromycin for 8 h until reaching the logarithmic phase.Cells were centrifuged at 5 000 ×gat 4 ℃ for 5 min and washed with three times with phosphate buffered saline (PBS), after which they were treated at 80 ℃for 3 min.Living cells were determined via the flat colony counting method to assess the heat resistance.To compare acid- and bile- resistance, cells were centrifuged at 5 000 ×gfor 5 min and then resuspended in PBS (pH 2.0) or PBS containing bile salt (2 g/L).After 30 min, living cells were determined using flat colony counting method.The heat-,acid- and bile- resistance of cells were assessed by calculating cell survival rates using the following formula:

1.3.4 Biofilm formation
Crystal violet (CV) staining was used to quantify biofilm formation by the recombinant strains and wild type strain.Briefly, overnight cultures were diluted to an optical density of 0.1 at 600 nm.Then, 200 μL of diluted culture was transferred to a 96-well plate.After incubation at 37 ℃ for 36 h, the wells were gently washed three times with PBS, stained with 0.1% CV for 30 min at room temperature,rinsed with distilled water and air-dried.Next, 100 μL of 95% ethanol was added to each well to dissolve the CV.Theoptical density was determined at 595 nm using a Synergy 2 microplate reader.
The cells of thepgm-pMG76e-LR-1,fba-pMG76e-LR-1,gap-pMG76e-LR-1 and wild type strain LR-1 were cultured overnight in MRS to determine the biofilm formation of recombinant strains in acidic and bile salt environments.They were transferred to 96-well plate and treated and detected as described above.
1.3.5 Adhesion assay
HT-29 cells were cultured on 24-well plates, and a monolayer of cells (2.5 × 105cells/well) was used for adhesion assays after washing twice with PBS (pH 7.2).The wild type and recombinant strains of LR-1 were grown overnight in MRS broth.Then, the cells were collected(5 000 ×g, 4 ℃, 10 min), washed twice with PBS (pH 7.2),and resuspended in DMEM to a final concentration of 108CFU/mL.The suspensions (100 μL) were added to the wells containing HT-29 cells and incubated at 37 ℃ for 1.5 h.After the incubation period, the HT-29 cell cultures were washed twice with PBS to remove the non-adherent cells and then treated with Triton X100 (1%, 10 min) to release the adhered bacterial cells.The adhesion ratio of bacterial cells was calculated using the following formula:

1.3.6 Bioinformatics
To identify the potential genes in the bacteria interacting with either thepgmorfbagenes, bioinformatic analyses were conducted in iRefIndex or IntAct databases.Cytoscape(3.7.1) was applied to visualise the interaction networks between the genes.A KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis was performed online to characterise the properties of the associated genes.
1.4 Statistical analysis
The data were analysed by one-way analysis of variance using Graphpad prism 7.0.All data were shown as means for at least three independent experiments.Data points were presented as ±s.Pvalues of less than 0.05 were considered significant.
2 Results and Analysis
2.1 The transcription levels of the pgm, fba and gap genes in the recombinant strains
Recombinant strains were identified via PCR and agarose gel electrophoresis (data not shown).To determine the expression of thepgm,fbaandgapgenes in these recombinant strains, their transcription levels were detected using real-time PCR.The results are shown in Fig.1.The expression of these genes increased significantly in thepgm-LR-1,fba-LR-1 andgap-LR-1 recombinant strains,respectively.They were subject to a 79.76-, 72.35- and 54.33-fold increase in their corresponding recombinant strains compared to the wild type strain.
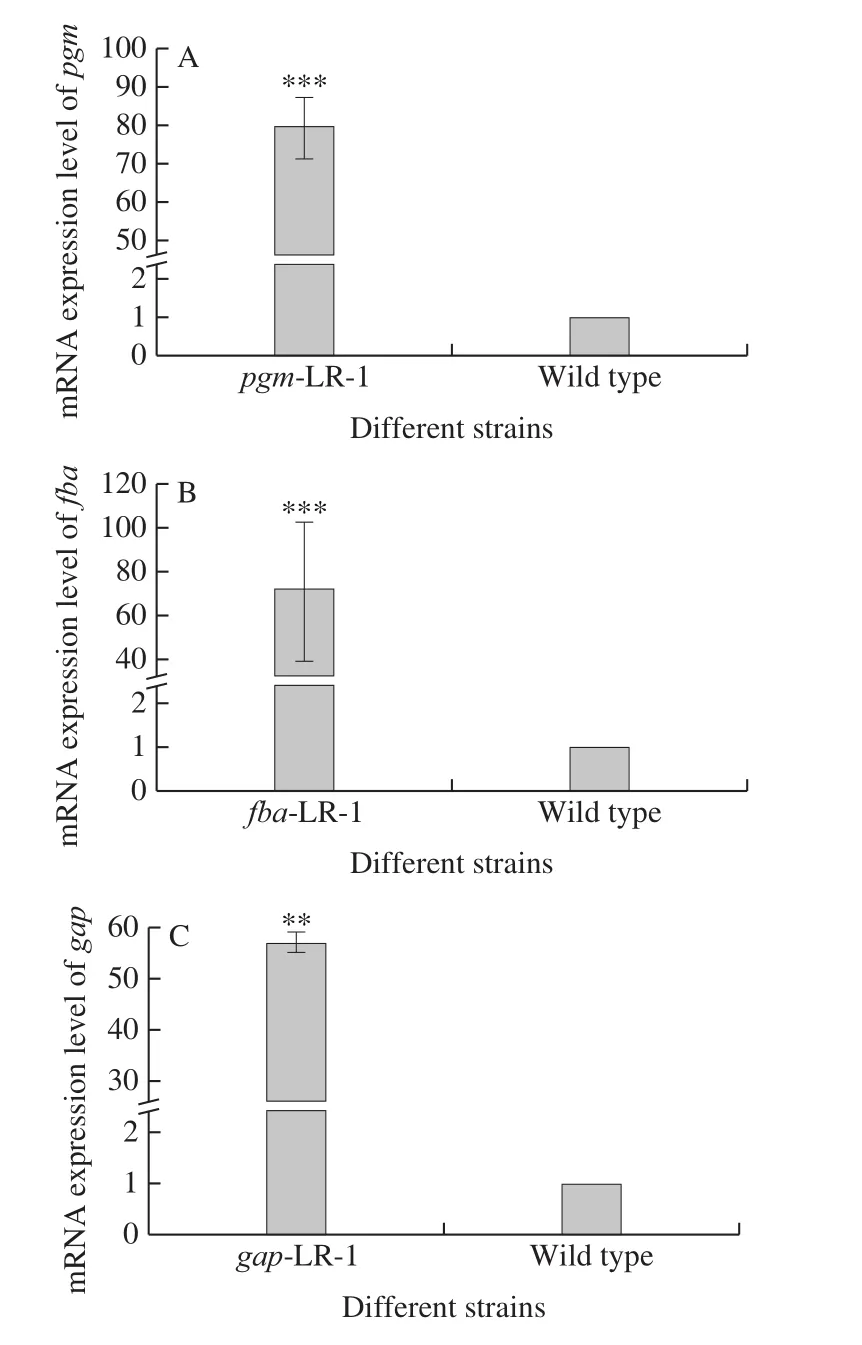
Fig.1 mRNA expression of pgm (A), fba (B) and gap (C) in recombinant strains overexpressing pgm-LR-1, fba-LR-1 and gap-LR-1 genes and wild type strain
2.2 The overexpression of the pgm, fba and gap genes affected the growth of LR-1
The growth curves of the strains are shown in Fig.2.The wild type strain entered the stationary phase after about 18 h.By contrast, the recombinant strains grew slightly slower than the wild type strain and reached the stationary phase after about 27 h.All these strains reached the maximum population of the OD600nmvalue 2.0.
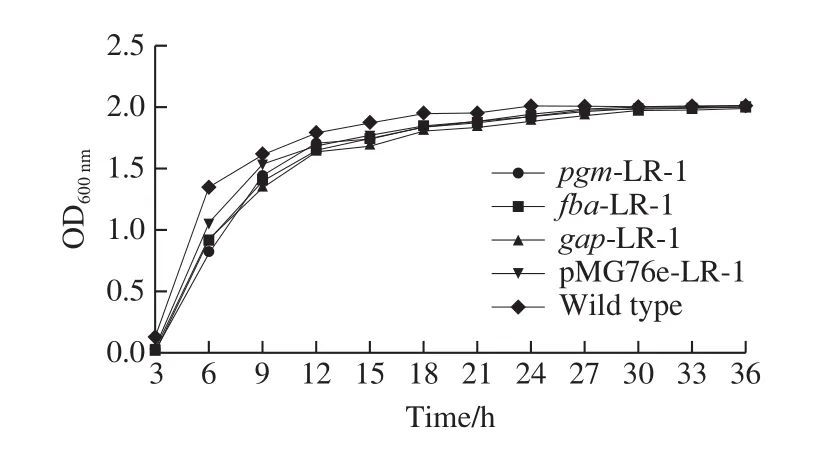
Fig.2 Growth curves of recombinant strains overexpressing pgm-LR-1,fba-LR-1, gap-LR-1 and pMG76e-LR-1 genes and wild type strain
The overexpression of the genes involved in glycolytic pathway may accelerate the carbon metabolism flux,however, the results here suggested that the recombinant strain LR-1 decreased the growth rate in the logarithmic phase.Possible reasons are as follows: 1) the operation of electrotransformation or/and the introduction of the plasmid pMG76e may resulted in the slower growth of the strain;2) metabolic acceleration may not influence the cell division of the strain; 3) glycolytic may compete with the cell growth of the strain[20]; 4) other intracelluar factors may repress cell growth and division of the strain.
2.3 The overexpression of the pgm, fba and gap genes affected the stress resistances of LR-1
As shown in Fig.3A, after exposure to 80 ℃ for 3 min, the survival rate of the wild type strain was 0.42‰.The introduction of pMG76e had no significant effect on the survival rate of LR-1 (data not shown).By contrast, the survival rates ofpgm-LR-1,fba-LR-1 andgap-LR-1 were 2.12‰, 1.40‰ and 0.92‰, respectively.They were subject to a 5.07-, 3.35- and a 2.21-fold increase compared to the wild type strain.There was no significant difference between thegap- LR-1 strain and the wild type strain.
The survival rates of the strains after exposure to an acidic environment are shown in Fig.3B.The survival rate of the wild type strain was 0.27‰.The introduction of pMG76e had no significant effect on the survival rate of LR-1 (data not shown).Contrarily, the survival rates ofpgm-LR-1,fba-LR-1 andgap-LR-1 were 0.87‰, 0.84‰ and 0.37‰, respectively.They were subject to a 3.38-, 3.28- and a 1.44-fold increase compared to the wild type strain.No significant differences were evident between thegap-LR-1 strain and the wild type strain.
The survival rates of the strains after exposure to a bile salt environment are shown in Fig.3C.The survival rate of the wild type strain was 0.27‰.The introduction of pMG76e had no significant effect on the survival rate of LR-1 (data not shown).By contrast, the survival rates ofpgm-LR-1,fba-LR-1 andgap-LR-1 were 1.17‰, 0.90‰ and 0.59‰,respectively.They were subject to a 4.35-, 3.33- and a 2.19-fold increase compared to the wild type strain.No significant differences were apparent between thegap-LR-1 strain and the wild type strain.
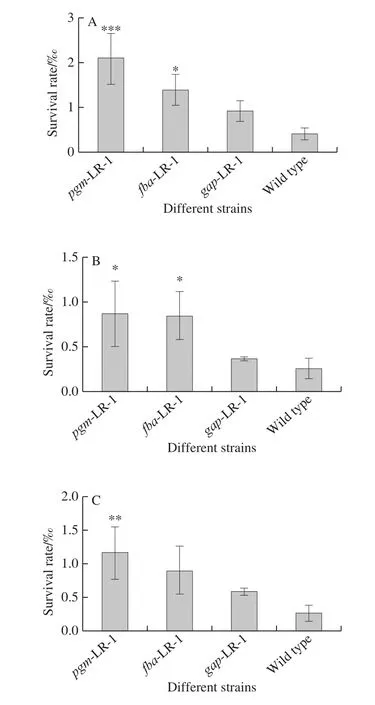
Fig.3 Heat (A), acid (B) and bile salt (C) resistance of recombinant strains containing pgm-LR-1, fba-LR-1, gap-LR-1 genes and wild type strain
2.4 The overexpression of the pgm, fba and gap genes affected the biofilm formation of LR-1
As shown in Fig.4A, the OD595nmvalue of the wild type strain is about 0.85.The introduction of pMG76e had no significant effect on the biofilm formation of LR-1(data not shown).Compared to the wild type strain, the biofilm formation increased 1.82-, 1.43- and 1.18-fold after overexpression of thepgm,fbaandgapgenes, with OD595nmvalues of 1.54, 1.22 and 1.00, respectively.There were no significant differences between thegap-LR-1 strain and the wild type strain.
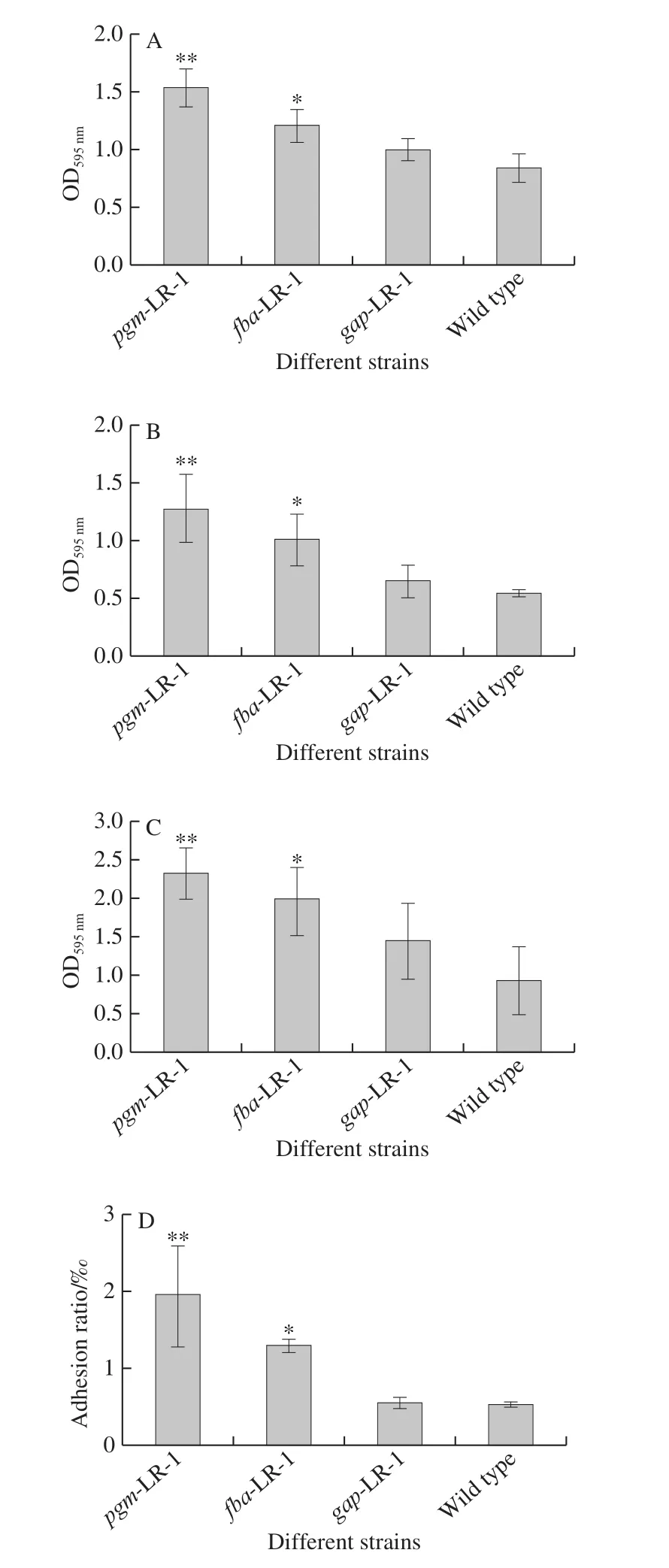
Fig.4 Biofilm formation (A), tolerance to acid (B) and bile salt (C),and adhesiveness (D) of recombination strains containing pgm-LR-1, fba-LR-1 and gap-LR-1 genes and wild type strain when cultured in MRS
The biofilm formation of these strains in acidic and bile salt environments are shown in Fig.4B, and Fig.4C.Compared to the wild type strain, the biofilm formation ability ofpgm-LR-1,fba-LR-1 andgap-LR-1 increased 2.32-,1.85- and 1.19-fold at pH 2.0, respectively.Moreover,the biofilm formation ability ofpgm-LR-1,fba-LR-1 andgap-LR-1 increased 2.49-, 2.13- and 1.55-fold at 0.2%bile salt, respectively.There was no significant difference between thegap-LR-1 strain and the wild type strain.
2.5 The overexpression of the pgm, fba and gap genes affected the adhesion ability of LR-1
The survival rate of these strains after adhesion to HT-29 cells are shown in Fig.4D.The adhesion ratio of the wild type strain was 0.53‰.The introduction of pMG76e had no significant effect on the adhesion ability of LR-1 (data not shown).The adhesion ratio ofpmg-LR-1,fba-LR-1 andgap-LR-1 increased to 1.96‰, 1.31‰ and 0.56‰, respectively.Compared to the wild type strain, the overexpression of thepgm,fbaandgapgenes enhanced the adhesion ability 3.66-, 2.43- and 1.04-fold on average.There was no significant difference between thegap-LR-1 strain and the wild type strain.
The metabolism of microbiology is a complex process including anabolism and catabolism of kinds of substances.Among them, carbohydrate metabolism is very important and widely studied.It was demonstrated that glycolysis play pivotal roles in the exopolysaccharides (EPS) yield[21],which is nearly the most important matrix in biofilms, and closely related with adhesion abilities.It is clear that the enzymePGMis required for the biosynthesis of EPS in other bacteria[22], and the lower EPS sythesis obtained was linked to a decrease inpgm[23].Thus, the promotion of the biofilm formation and adhesion by overexpression of glycolytic genes may result from the enhancement of EPS.Moreover, the carbon flux always couples with energy synthesis and release.ATP is rapidly regenerated mainly by glycolytic pathway in bacteria, which plays a critical role in all living beings as an energy source for various ATP-requiring enzymatic reactions[24].It is certain that the overexpression of glycolytic genes should influence the efficiency of glycolytic pathway,but the effect on the energy flex remain unclear which may plays animportant role in physiology of bacteria.
2.6 The overexpression of the pgm, fba and gap genes affected the transcription levels of other genes in LR-1
To determine the impact of overexpression of thepgm,fbaandgapgenes on the expression of thetufandluxSgenes,their transcription levels were detected using real-time PCR.As shown in Fig.5A, compared to the wild type strain, the expression oftufin recombinant strainspmg-LR-1,fba-LR-1 andgap-LR-1 increased 56.18-, 52.41- and 11.22-fold.Fig.5B showed that the expression ofluxSin recombinant strainspgm-LR-1,fba-LR-1 andgap- LR-1 respectively increased 96.94-, 54.83- and 8.28-fold on average compared to the wild type strain.
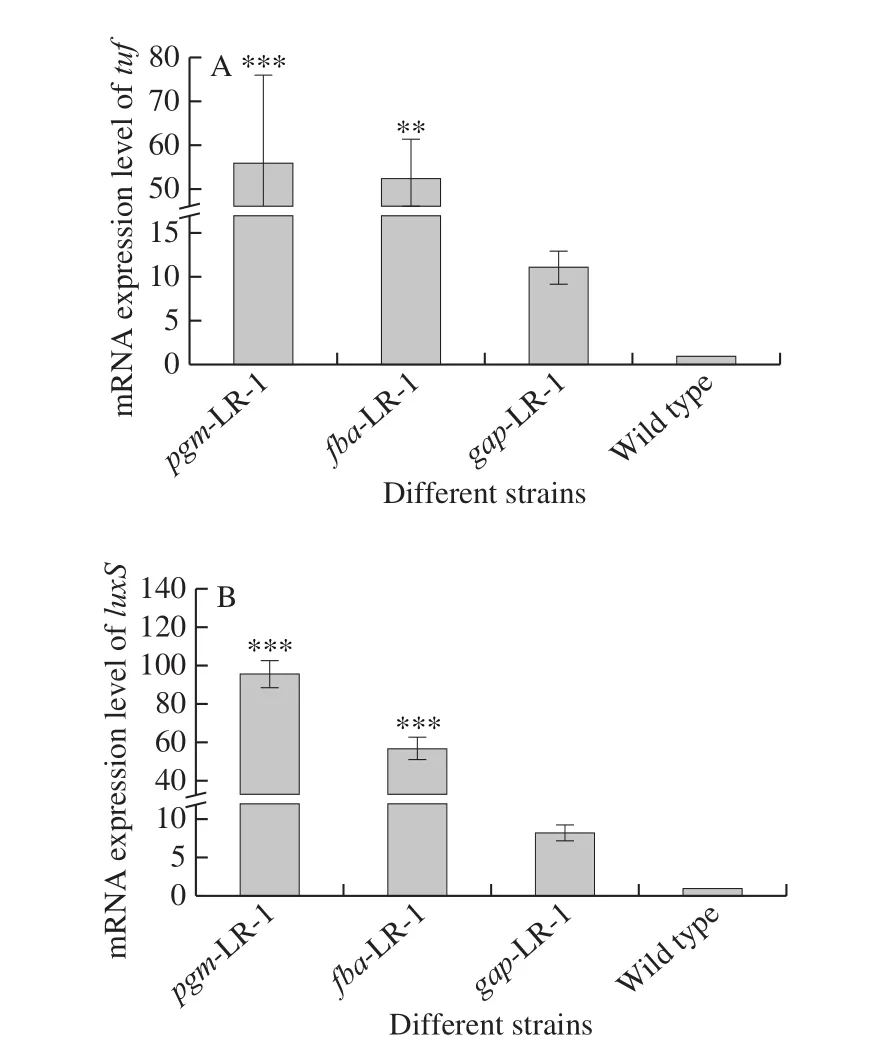
Fig.5 mRNA expression of tuf (A) and luxS (B) in recombinant strains pgm-LR-1, fba-LR-1 and gap-LR-1 compared with wild type strain
2.7 Bioinformatics analyses
The gene-gene interactions are shown in Fig.6.The network nodes represent genes, and the edges represent gene-gene associations.Fig.6A and Fig.6B show the potential genes and their association withpgminCampylobacter jejuniand inE.coliK12, respectively.Fig.6C and Fig.6D show the potential genes and their association withfbainC.jejuniand inE.coliK12,respectively.The detailed information is listed in Table 3 and Table 4.Thepgmgene was associated with 35 genes inC.jejuni, and 13 genes inE.coliK12, while thefbagene was associated with 9 genes inC.jejuni, and 7 genes inE.coliK12.Functionally, all these genes were classified into groups related to carbohydrate metabolism, metabolism of cofactors and vitamins, genetic information processing, signalling and cellular processes and unclassified metabolism.The functional classification of these genes suggested that thepgmandfbagenes can act via multiple pathways.

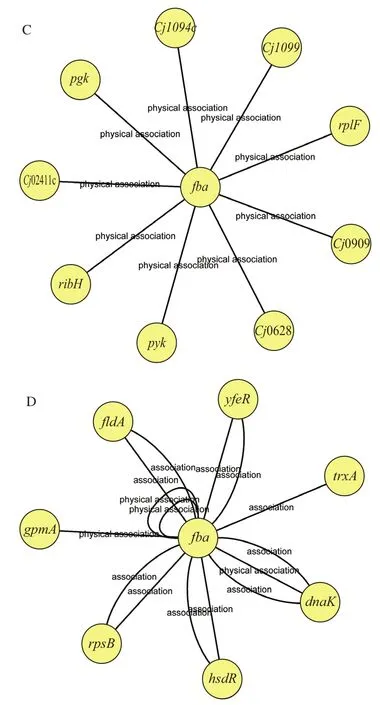
Fig.6 Association networks between genes and pgm in C.jejuni (A)and in E.coli K12 (B); and interaction networks between genes and fba in C.jejuni (C) and in E.coli K12 (D)

Table 3 Functional classification of genes interacting with pgm in C.jejuni and E.coli K12

Table 4 Functional classification of genes interacting with fba in C.jejuni and E.coli K12
According to the data of the bioinformatics analyses,several genes identified here have functions involved in biofilm formation, EPS synthesis, and stress bearing.Researches have indicated that genesgpsAandCj1325associated with biofilm formation in other bacteria[25-26].The expression of geneefpandpykpositively correlated with extracellular polysaccharide production[27-28].Similarly, the genehslVcould take part in the EPS production, and it is also involved in metabolic remodeling and in adapting to heat stress[29].GeneargHcould regulate production of the major biofilm-matrix EPS[30], which results in the biofilm formation of the bacteria.GenemrcAis associated with stress bearing and peptidolycan layer[31].All these genes combined withpgmandfbamay affect the physiological phenotype of the strain LR-1.
3 Discussion
The glycolytic pathway is one of the primary energy sources of lactic acid bacteria.In addition, glycolytic pathway genes have other functions besides being involved in carbohydrate catabolism.However, related research regardingL.plantarumis scarce.As far as is known, the impact of theoverexpression of these genes on physiological behaviour remains unknown.Because the proteins inside the cell form part of essential cellular processes, the creation of a knockout mutant is impossible.Consequently, few studies have provided direct experimental evidence for the role of these cell-surface proteins in the adhesion ofLactobacillusto host cells.In addition, although many studies have examined the effect of surface proteins on the adhesion of their originating strains, few studies have included their impact on other strains or on how to improve adhesion[18].Therefore, this paper examined the effect of the upregulation ofpgm,fbaandgapon important characteristics inL.plantarumLR-1, which was isolated from fermented vegetables.
By introducing the plasmid containingpgm,fba, andgapgenes into the LR-1 cells, their transcription levels increased significantly compared to the wild type strain.According to the growth curves, the recombinant strains includingpgm-LR-1,fba-LR-1,gap-LR-1 and pMG76e-LR-1 grew slower than the wild type strain, which could be attributed to the introduction of external pMG76e and the electrotransformation process.
L.plantarumis one of the most abundantLactobacillusspecies and is commonly found in starchy food, cereals,meat, dairy products, vegetables, fruits, and beverages[32-33].It is considered an important industrial microbe and has been widely utilised in food industries via food-related technologies[34-37].The bacteria will suffer adverse conditions during the production of food, such as high temperature.Therefore, the present study evaluated the thermal resistance of these recombinant strains.The results suggested that the overexpression of thepgmandfbagenes promoted thermal tolerance ofL.plantarumLR-1, but overexpression ofgaphad no significant effect.According to the WHO/FAO 2001[1],L.plantarumcan be utilised as probiotics because of their potential health-promoting effects.The primary criterion for probiotics is that they must be highly tolerant of the GIT environment[18].In previous studies involving lactobacilli,several glycolytic proteins, including PGM andFBAencoded by thepgmandfbagenes, respectively, are more abundant in response to acid stress conditions[38-41].Similarly, the present study found that the acid resistance of recombinant strains,pgm-LR-1 andfba-LR-1, increased significantly.These results indicate that not only can the acidic environment lead to the upregulation ofpgmandfba, but also that the transcription level of these two genes can influence the acid tolerance ofL.plantarum.However, the transcription level ofgaphad no significant effect on the acid resistance ofL.plantarum.Glycolytic proteins are reportedly upregulated under the bile stress response in probioticLactobacillus salivariusLI01[42].The present research showed that the overexpression ofpgmandfbacould enhance the bile resistance ofL.plantarum.All these results demonstrated that the expression ofpgmandfba, as well as their stress resistance abilities, could interact as both cause and effect inL.plantarum.
Biofilms constitute the predominant microbial style of life in natural and engineered ecosystems.The formation of biofilms, microbial communities embedded in self-produced polymeric matrices attached to a surface, is an ancient and universal trait that enables microorganisms to develop coordinated architectural and survival strategies[35-36,43-44].Biofilms are closely related to adhesion[45].This lifestyle allows microorganisms to survive in unfavourable environmental conditions[43].In addition,Lactobacillusbiofilms exhibit a higher tolerance than their planktonic counterparts[46].Glycolytic pathway genes plays a critical role in biofilm formation inSaccharomyces cerevisiae[47].As forListeria monocytogenes, the glycolytic pathway genes are expressed higher in biofilm cells than in planktonic cells[48].In accordance with this research, the results here demonstrated that the upregulation of thepgmandfbagenes could enhance the biofilm ability ofL.plantarum.
Adhesion to epithelial cells is considered important forLactobacillusto display probiotic effects.It has been found that glycolytic enzymes are related to the adhesion ability ofLactobacillusstrains.PGM encoded by thepgmgene was identified as a potential marker of adhesion inLactobacillus pentosus[49].Similarly, the current study indicated that the upregulation of thepgmgene promoted the adhesion ability ofL.plantarumto HT-29 cells.FBAencoded byfbahas been involved in additional functions[10-11,50].InM.hyopneumoniaeandMycoplasma bovis,fbamoonlights as an important adhesin, mediating binding to host cells via fibronectin[12,51].This study has also shown that the upregulation offbacan enhance the adhesion ofL.plantarum.GAPDH is a glycolytic enzyme found in a wide range of prokaryotes and eukaryotes.It has been identified as a surface protein related to adhesion[18].However, in this study, the upregulation of thegapgene had no significant effect on the adhesion ability ofL.plantarumLR-1.
Elongation factor (EF)-Tu encoded by thetufgene is a highly abundant protein participating in the translationprocess, in protein folding, and in stress resistance[52].Furthermore, EF-Tu can reportedly regulate the adhesion ability of bacteria[53-54].In several lactobacilli, EF-Tu has been described as a “moonlighting” protein and can be localised on the cell wall.Furthermore, a role in intestinal adhesion has been suggested for this protein[55].It is more abundant in the acid-adapted cells ofL.kefiranofaciensM1,L.caseiandL.delbrueckiisubsp.bulgaricus[56].This study showed that the transcription level oftufincreased significantly inpgm-LR-1,fba-LR-1 andgap-LR-1.It was higher inpgm-LR-1 andfba-LR-1 than ingap-LR-1, which might lead to an insignificant promotion of stress resistance, biofilm formation,and adhesion ability ingap-LR-1.LuxSencoded by theluxSgene catalyses the synthesis of the signalling molecule AI-2,which is proposed to be involved in interspecies bacterial communication.LuxSalso forms an essential part of the activated methyl cycle, where AI-2 is produced fromS-adenosylmethionine (SAM) in three enzymatic steps.SAM is used in various methylation reactions, and therefore, the SAM cycle plays a pivotal role in promoting the stability of macromolecules under stress conditions[55].Previous research revealed that the overexpression ofluxScould increase the transcription levels ofpgm,fbaandgap[9].This study found that the upregulation ofpgm,fbaandgapcould also increase the expression ofluxS.The underlying relationship between these genes remains unclear, and the molecular mechanism of how thepgm,fba, andgapgenes regulate this physiological behaviour inL.plantarumis worthy of further investigation.
The iRefIndex and IntAct databases provided an abundance of information related toC.jejuniandE.coliK12, but none was available forL.plantarum.Therefore, the potential genes and their association withpgmorfbain these bacteria were analysed and can provide references for further research involvingL.plantarum.These genes were classified into multiple pathways, suggesting that thepgmandfbagenes could play specific roles via various mechanisms.It has been demonstrated that some of these genes are associated with stress resistance, biofilm formation or adhesion.For example, the expression ofpsrAandgspAis associated with biofilm formation and adhesion inStaphylococcus aureusandCandidaspp.[57-59].trxAis essential for motility and contributes to the host infection ofL.monocytogenes[60].The genednaKcan control the heat-shock response[61](stressresponsive chaperone machine with the central metabolism),while the heterogeneous expression of thednaKgene fromAlicyclobacillus acidoterrestrisimproves the resistance ofE.coliagainst heat and acid stress[62].Proper control of theCaulobacter crescentuscell surface adhesion requires the general protein chaperone Dnak[63].gpmAhas been reported to be involved in biofilm formation and stress resistance inSalmonella enterica[64].Many other studies suggest that thesepgmorfbaassociative genes display functions related to stress resistance, biofilm formation or adhesion[65-67].All these findings provide useful references for further investigation into the underlying mechanism on how thepgmorfbagenes regulate the important physiological behaviour in bacteria.
4 Conclusions
In summary, this study indicate that the upregulation ofpgmandfbagenes inL.plantarumLR-1 enhances the thermal-, acid- and bile-salt-resistance of this strain.Furthermore, overexpression of these two genes promotes the biofilm formation and adhesion of this strain.The increased transcription level of thegapgene exhibits no significant effect on the stress resistance, biofilm formation and adhesion ofL.plantarumLR-1.The upregulation of thepgm,fbaandgapgenes increases the transcription levels oftufandluxSin this strain.This increase is higher in recombinant overexpressing gene strains,pgmandfba, than in those exhibiting overexpression of thegapgene.Furthermore,bioinformatic analysis suggests thatpgmandfbacan associate with genes that are related to multiple pathways.The underlying mechanism of how these genes regulate the physiological behaviour ofL.plantarumrequires further exploration.
