氯和紫外消毒过程中胞外抗性基因的产生特征
2021-10-26付树森王肖霖王尚杰张生博袁青彬
付树森,王 艺,王肖霖,王尚杰,程 远,卞 博,张生博,袁青彬
氯和紫外消毒过程中胞外抗性基因的产生特征
付树森,王 艺,王肖霖,王尚杰,程 远,卞 博,张生博,袁青彬*
(南京工业大学环境科学与工程学院,江苏 南京 211816)
考察了城市污水氯和紫外消毒过程中不同物理形态的胞外抗性基因的产生行为与及微生物群落的关联特征.结果表明,氯消毒尽管使胞内抗性基因丰度下降,但使结合型胞外抗性基因丰度明显上升(0.7±0.1) log,而游离型胞外抗性基因丰度下降(0.2±0.1)log.紫外消毒也使胞内抗性基因下降,但使游离型胞外抗性基因显著上升(0.4±0.2) log,而结合型胞外抗性基因丰度下降(0.3±0.1) log.氯消毒后, 结合型胞外DNA(a-eDNA)中变形菌门丰度下降而其他菌门的丰度上升,细菌多样性指数由4.2上升到4.7;而游离型胞外DNA(f-eDNA)中变形菌门上升了6.6%,多样性指数则从3.5降低到2.8.紫外消毒后,a-eDNA中变形菌门丰度下降了36.6%,多样性则上升到4.8,而f-eDNA中细菌丰度变化较小.分子生态网络分析揭示了抗性基因与细菌间广泛的寄存关系,、、和分别与17、15、15和5种菌属间存在共现性,表明抗性基因潜在宿主的变化是导致消毒后胞外抗性基因产生的关键原因.本研究表明氯和紫外消毒不能消除抗性基因风险,反而通过导致不同胞外抗性基因的大量产生,使风险的形式发生变化.
氯消毒;紫外消毒;胞外抗性基因;细菌群落;分子生态网络
全球范围抗生素的大量使用极大加剧了细菌抗药性的发展,已成为威胁公共健康的严峻问题.抗性基因被列为一种新兴污染物[1],在河流[2]、空气[3]和土壤[4]等各种环境中大量检出.其中污水处理厂作为各种废水的汇集地,已检出数百种抗生素抗性基因(ARG)[5-7],成为环境中抗性基因的主要储存库.作为生物污染的主要控制单元,以氯和紫外消毒为代表的污水消毒过程对ARG的去除效果也受到广泛关注.很多研究发现氯和紫外消毒不能有效地去除ARG[8-12],去除效果远低于其宿主耐药细菌[14-16],消毒后出水抗性基因仍高达1010~1013copies/L[17].即便抗性基因丰度有所降低,胞内抗性基因(iARG)也未被彻底破坏,而仅被释放到胞外,变成胞外抗性基因(eARG)[18-19].
eARG作为一种新形式抗性基因近年来受到关注.虽然位于细胞外,eARG并未失去生物活性,在适宜条件下仍可被吸收进入细胞表达抗性[20-21].相关研究将携带SHV基因的质粒成功转化到受体DH 5a,转移效率达(6.7±0.5)´102(CFU/ug DNA)[22].即使在自然环境中,eARG也可进入细胞转化.如枯草芽孢杆菌可以吸收水中eARG至体内表达,效率达3.0´10-3(transformation CFU/total CFU)[23].此外,eARG可以不同的物理形态存在,如游离于水中(游离型胞外抗性基因(f-eARG))或附着于颗粒物(结合型胞外抗性基因(a-eARG ))[24-25],两者的环境行为和风险可能存在显著差异.如a-eARG更能抵抗环境的降解[26-28],而f-eARG可能更易迁移和水平转移,研究表明f-eARG更容易与感受态细胞结合[29].同样,Kunito等研究发现含NDM-1抗性基因的质粒与土壤吸附结合后转化效率下降[30].因此,揭示不同形态eARG的归趋对于准确揭示抗性基因的风险至关重要.
本研究考察了城市污水氯和紫外消毒过程中结合态和游离态eARG的产生特征,选取广泛报道的两类抗性基因,四环素类(A,X)和磺胺类抗性基因(I,II)[31-32]作为代表,同时考察了该过程结合态胞外DNA(f-eDNA)和游离态胞外DNA(a- eDNA)中细菌群落的变化,并借助分子生态网络解析了两类eARG与细菌群落间的关联特征.研究结果将为揭示消毒过程中抗性基因的风险变化提供理论支撑.
1 材料与方法
1.1 消毒实验
水样取自南京市某城市污水处理厂二沉池出水,装于20L聚乙烯塑料桶中,1h内运送至实验室4℃存储,1周内分析完毕.对于氯消毒,采用1L灭菌烧杯作为反应容器,向500mL水样中投加次氯酸钠(NaClO)溶液(10mg/L),搅拌(300r/min)反应10min后加入硫代硫酸钠(Na2SO3)(5mg/L)终止.对于紫外消毒,将40mL水样置于一次性培养皿,以低压紫外平行光仪(30% UV-C, TL 120W/01,Philips)作为紫外消毒光源消毒,光源功率为120W,光束在样品表面中心处的照射强度为0.12mJ/cm2,通过控制紫外辐照时间[33]使最终消毒剂量为100mJ/cm2.
1.2 DNA的分离和提取
将10mL样品经滤膜(0.22μm)抽滤后,滤液用于f-eDNA提取,另外将抽滤后的滤膜浸入10mL磷酸盐缓冲溶液中,振荡(250r/min)10min后再次经滤膜(0.22μm)抽滤,得到的滤液用于a-eDNA提取,而两次抽滤的两张滤膜用于胞内DNA(iDNA)提取.胞外DNA(eDNA)的提取采用本实验室开发的磁珠法[25].即取2mL滤液加入15mL离心管,
随后加入4mLBuffer CL(含蛋白酶K(20mg/L)和盐酸胍(2mol/L))以及3mL异丙醇手摇混合,漩涡振荡2min.加入30μL悬浮磁珠,漩涡振荡4min使磁珠与样品中 DNA充分混合.将离心管放置于磁力架上静置70s后弃掉上清液.加入600μL Buffer CW (含7mol/L盐酸胍的异丙醇溶液),用移液枪打匀后振荡1min,继续放置到磁力架上静置70s,随后弃去液体.用75%乙醇清洗磁珠2次.随后将离心管于室温下静置10min,加入30μL提前预热(55℃)的Elution Buffer,振荡30s使其充分混合,随后每隔1min震荡30s,持续5min.将溶液与磁珠分离后即得到eDNA.对于iDNA,将滤膜剪碎到提取管中,按照土壤DNA提取试剂盒(FastDNATMSPIN kit for soil, MPbio,美国)操作说明书提取.
1.3 ARG丰度的测定

表1 各基因引物信息
应用荧光定量PCR(qPCR)测定样品中ARG的丰度.首先,将高浓度的标准品进行10倍梯度连续稀释5个梯度作为标准曲线,在qPCR仪(ABI7500,美国)上进行反应.qPCR的测定采用20μL反应体系,其中包括10μL 2×SuperReal PreMix Plus(Tiangen Biotech,中国),2μL 50×Rox染料(TiangenBiotech,中国),0.4μL上下引物、6.2μL ddH2O以及1μL标准品.反应条件为:95℃预热15min→40个扩增循环(95℃ 10s,退火20s,64℃保持30s).样品的测定步骤同标准曲线,并根据qPCR的扩增循环数Ct值计算ARG拷贝数.确保标准曲线的2>0.99.样品3组平行间偏差<5%,所有样品的扩增效率在90%~110%.各基因引物信息见表1.
1.4 细菌群落分析
将氯和紫外消毒前后的iDNA、f-eDNA和a-eDNA送至上海美吉生物医药科技有限公司Illumina miseq平台进行细菌16S rDNA高通量测序[35].即样品首先进行16S rDNA PCR扩增,引物为515F(5'-GTGCCAGCMGCCGCGG-3')和806R(5'- GGACTACHVGGGTWTCTAAT-3'),随后制备~ 400bp DNA片段的序列文库并测序.细菌群落组成,Shannon指数分析均在MajorBio I-Sanger云平台上进行(www.i-sanger.com).另外,采用分子生态网络考察ARG和菌属间的共现性,数据在公开网站Molecular Ecological Network Analysis Pipeline (http://ieg2.ou.edu/MENA)处理后,用Cytoscape软件作图[36-37].
2 结果与讨论
2.1 氯和紫外消毒过程中eARG的产生特征
图1展示了氯和UV消毒后3类ARG的丰度变化.可以发现,氯和UV消毒均使iARG的丰度下降.氯消毒之后,胞内A,X,I和II的丰度平均降低(0.2±0.1) log;而紫外消毒后四种抗性基因的丰度则平均降低(0.6±0.3)log.显然在常用剂量下,紫外对iARG的去除效率高于氯消毒,这可能和两种消毒作用机理不同有关.氯消毒并不能直接作用于DNA,而是破坏细胞膜和蛋白质等,消毒后部分细菌可能处于VBNC(存活但不可培养)状态[38-39]导致DNA仍位于胞内.相比之下,UV可直接作用于DNA[6]使ARG被破坏而无法被检出.
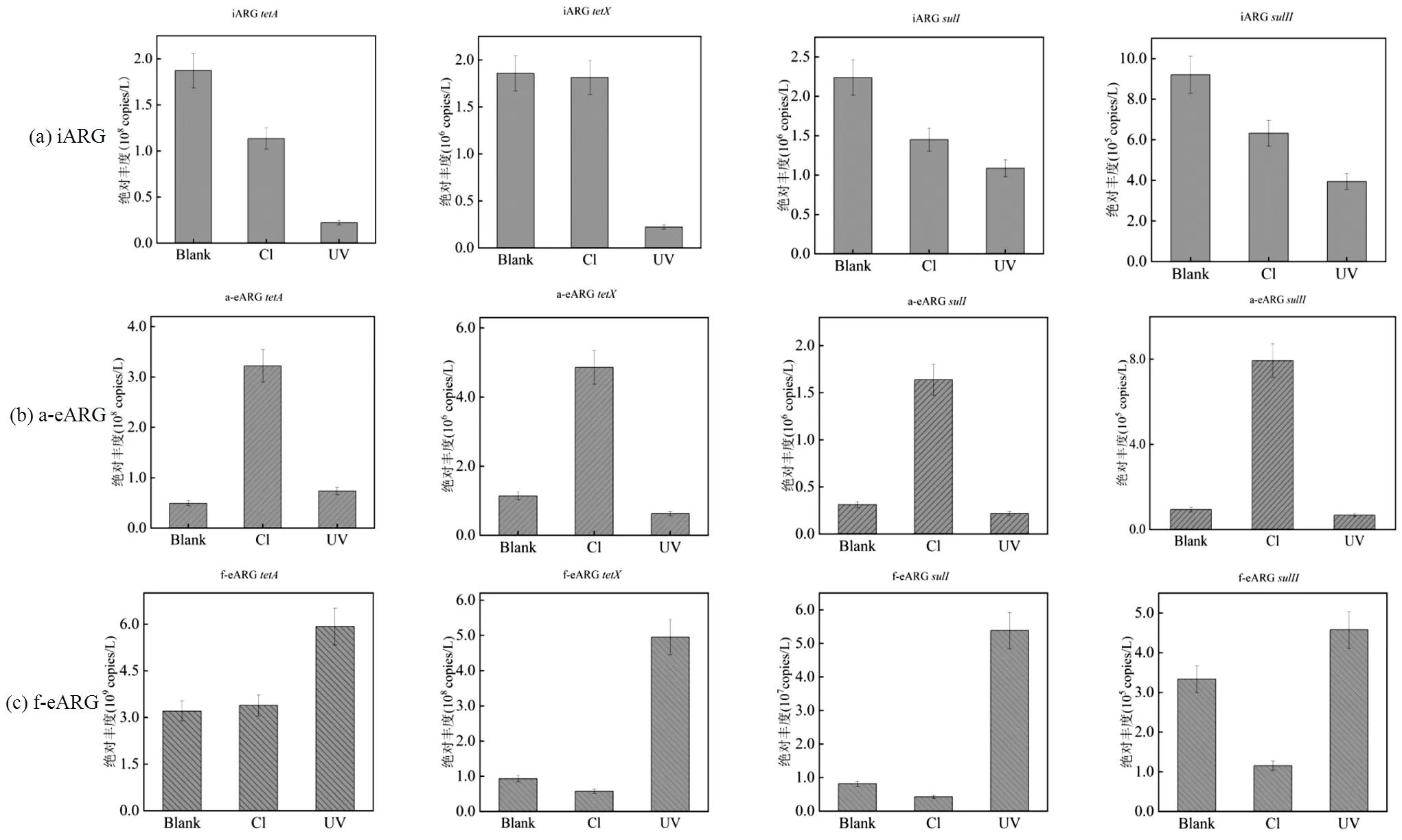
图1 氯和紫外消毒后iARG、a-eARG和f-eARG绝对丰度的变化
(Blank:空白,Cl:次氯酸钠(100mg Cl×min/L),UV:紫外(100mJ/cm2),Absolute Abundance:绝对丰度(copies/L))
两种eARG经氯和紫外消毒后变化趋势存在显著差异.对于a-eARG,氯消毒后4种ARG丰度显著提高了(0.7±0.1)log,同样a-eARG的相对占比(图2)分别由1.1%、1.7%、3.0%和7.0%提高至8.0%、 8.3%、22.0%和52.0%.而经UV处理之后,a-eARG丰度下降(0.1±0.2)log. a-eARG丰度的上升在前人研究中也被发现,如Liu等[18]报道氯消毒后,a-eARG的浓度上升了7.8倍.这表明,氯消毒后释放至胞外的抗性基因倾向于与水中物质结合形成a-eARG.这可能是因为DNA在水中类似于聚合电解质,具有较高的表面电荷和分子柔性,可被颗粒物吸附[40]而变为a-eARG.另外,细菌作为表面含生物大分子的胶体[41],经氯消毒氧化后可能破碎形成众多小颗粒胶体,从而提供更多吸附位点使ARG形成a-eARG.
与a-eARG不同,f-eARG经氯消毒之后,丰度下降(0.3±0.1)log;而经UV处理之后,丰度上升(0.4±0.2)log,其相对占比(图2)同样上升3.0%~25.0%.表明UV消毒更有利于f-eARG的形成.这可能仍与UV消毒机制有关.UV消毒是通过作用于胸腺嘧啶键形成二聚体,破坏DNA的结构[6],阻止遗传物质的复制而引起细菌死亡.死亡的细菌悬浮在水体中,可能阻碍UV对DNA的破坏,从而使得f-eARG更容易存活下来.同时,与氯消毒相比,UV特定的DNA靶向作用机理使细菌较难破碎形成小分子胶体,导致DNA缺少吸附位点而以游离型为主,使f-eARG丰度升高.
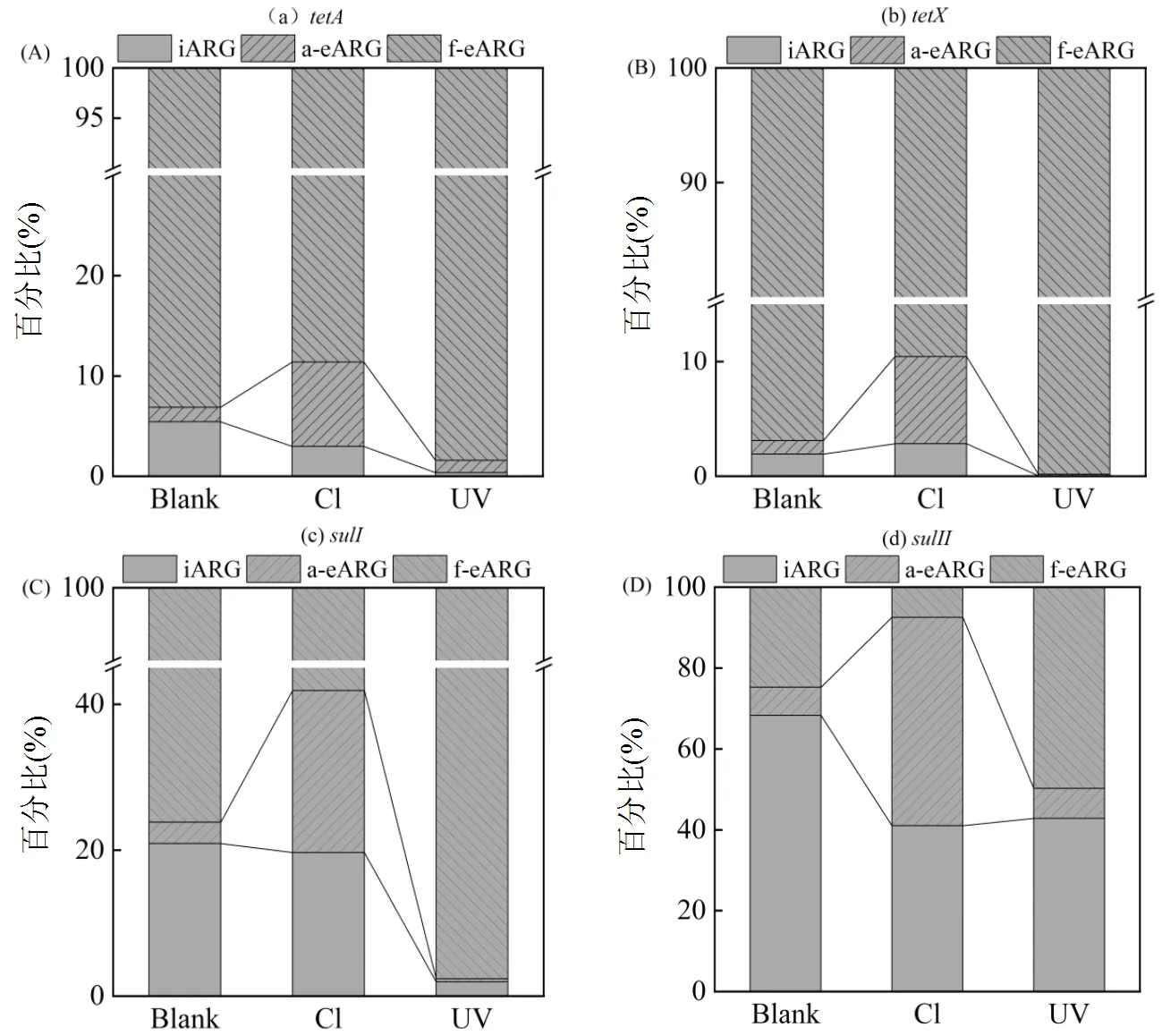
图2 氯和紫外消毒前后三种类型ARG (iARG、f-eARG和a-eARG)的相对百分比
Blank:空白, Cl:次氯酸钠(100mg Cl min/L)UV:紫外(100mJ/cm2)
2.2 氯和紫外消毒后细菌群落在3种DNA变化
变形菌门(Proteobacteria)、蓝菌门(Cyanobacteria)、Patescibacter超门和拟杆菌门(Bacteroidetes)是消毒前iDNA的主要菌门,其相对丰度分别为45.0%、17.5%、16.9%和3.2%(图3(a)).消毒之后,除变形菌门外,其他门相对丰度均降低,群落多样性指数的降低也证实了这一点.氯和紫外消毒之后,iDNA香农指数从5.3分别下降到4.2和4.9.a-eDNA和f-eDNA中的变形菌门相对丰度在消毒之后分别升高93.1%和86.7%.此结果与本文之前报道的结果相符[25]:变形菌门在污水消毒过程中更容易释放DNA成为eDNA.对a-eDNA和f-eDNA中细菌种属(图3(b))的进一步研究发现等菌属的增加导致了变形菌门丰度的提高.
氯消毒使iDNA中的变形菌门和栖热菌门(Deinococcus-Thermus)的丰度升高,这表明与其他菌门相比,这两种菌门更难释放DNA到环境当中.在a-eDNA中除变形菌门外绝大多数菌门丰度均上升,如Cyanobacteria的丰度由0.5%上升到5.0%,Bacteroidetes的丰度则上升了11.0%,而细菌浓度的上升可能会引起结合型ARG丰度的升高,而在f-eDNA中,变形菌门上升了6.6%,而其他菌门的丰度变化较小.与此对应,氯消毒之后f-eARG中细菌的多样性指数从3.5降低到2.8,而a-eDNA中细菌多样性指数则由4.2上升到4.7.值得注意的是,氯消毒降低了iDNA中一些致病菌的丰度,例如和.紫外消毒使iDNA中拟杆菌门()和放线菌门()丰度上升,表明这两种菌门比其它菌门更能耐受UV.
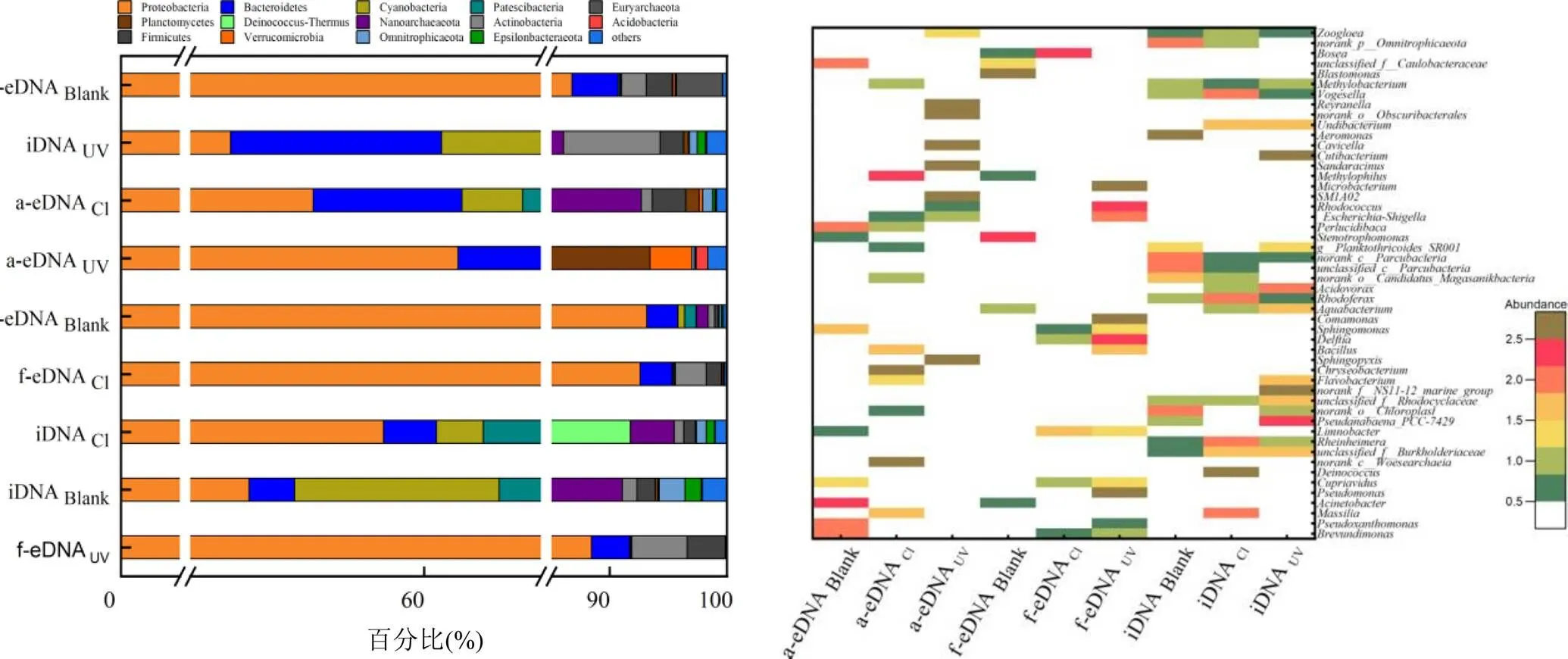
图3 氯和UV消毒前后三种类型DNA中细菌群落在门和属水平的丰度变化
(Blank:空白,Cl:次氯酸钠(100mg Cl min/L),UV:紫外(100mJ/cm2)
与之相比,UV消毒后a-eDNA中变形菌门的丰度下降了36.6 %,其他菌门的丰度反而上升.而在f-eDNA中大多数的菌门丰度变化较小.与氯消毒相似的是,UV消毒之后f-eDNA的香农系数下降至2.9,而a-eDNA的则上升到4.8.同时,UV消毒降低了iDNA中人体致病菌的丰度,包括等.
2.3 ARG与菌属间的共现性分析
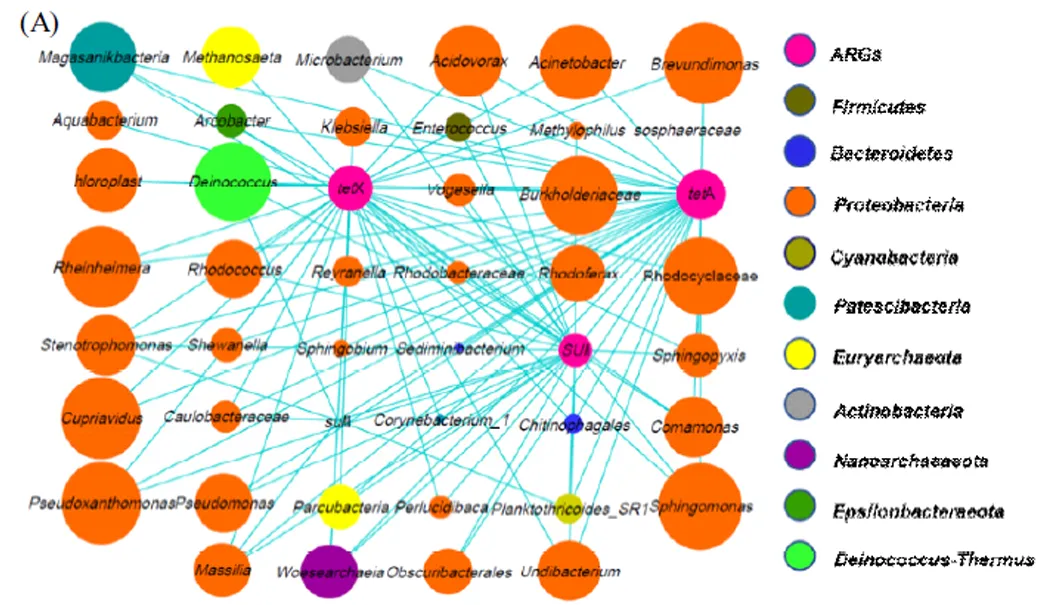
(A) 为ARG和菌属之间的共现性.(B) 为每种ARG和其相关菌属的共现性,其中只统计了每个样品中OTUs丰度大于1 %的菌属,节点的大小表示丰度的大小
利用分子生态网络图考察了ARG和细菌菌属之间的共线性,从而揭示可能存在的宿主关系[42].结果表明,A、X、I和II分别与17、15、15和5种菌属存在共现性,这表明A,X均可能寄存于和等菌属,而I和II可能分别在和等菌属中.上述多种ARG与细菌间的寄存关系也被前人报道,如A存在于[41]、[42]和[45],X存在于[46]、I存在于[47]和等.这种寄存关系意味着潜在宿主的类型及数量变化可能是消毒过程中eARG丰度变化的关键原因.如A与lI由于潜在宿主较多,消毒时可能有更多的iARG转变会eARG.而A和X的宿主有[46]和[49]等耐氯菌,I的宿主有耐UV菌属[50],可能使iARG在消毒过程中较难被释放.此外,研究还发现ARG存在于部分人类致病菌,如A、X与[51],I、II与[50]及人畜共患病原菌[52]间存在寄存关系.病原菌中ARG的存在可能使抗生素治疗失效,给公共健康带来严重威胁.
3 结论
3.1 氯和紫外消毒后两种eARG的变化存在显著差异:氯消毒(100mg Cl×min/L)后a-eARG丰度上升(0.7±0.1) log,而f-eARG丰度下降(0.2±0.1) log;紫外消毒(100mJ/cm2)后a-eARG丰度下降0.3±0.1log,而f-eARG丰度上升(0.4±0.2 )log.
3.2 氯消毒后a-eDNA中除变形菌门外大多数菌门的丰度均提高,细菌多样性指数由4.2上升到4.7;f-eDNA中变形菌门提高6.6 %,多样性指数则从3.5降至2.8.紫外消毒后a-eDNA中变形菌门丰度下降36.6 %,多样性则上升到4.8,而f-eDNA中菌门丰度变化较小.
3.3A、X、I和II分别与17, 15, 15和5种菌属之间存在共现性关系,表明ARG-细菌间广泛的寄存关系.消毒过程中潜在宿主的变化是导致eARG丰度变化的关键原因.此外,ARG 还可寄存于和等病原菌,给公共健康带来严重威胁.
[1] Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance-the need for global solutions [J]. Lancet Infect Dis, 2013,13(12):1057-98.
[2] Nnadozie C F, Odume O N. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes [J]. Environmental Pollution, 2019,254: 113067.
[3] Trudel M V, Vincent A T, Attéré S A, et al. Diversity of antibiotic- resistance genes in Canadian isolates of Aeromonas salmonicida subsp. salmonicida: dominance of pSN254b and discovery of pAsa8 [J]. Entific Reports, 2016,6:35617.
[4] Burch T R, Sadowsky M J, Lapara T M. Fate of Antibiotic Resistance Genes and Class 1Integrons in Soil Microcosms Following the Application of Treated Residual Municipal Wastewater Solids [J]. Environmental Science & Technology, 2014,48(10):5620-5626.
[5] Su Z, Li A, Chen J,et al. Wastewater discharge drives ARGs spread in the coastal area: A case study in Hangzhou Bay, China [J]. Marine Pollution Bulletin, 2020,151:110856.
[6] Dodd M C. Potential impacts of disinfection processes on elimination and deactivation of antibiotic resistance genes during water and wastewater treatment [J]. Journal of Environmental Monitoring, 2012, 14(7):1754-1771.
[7] Pietramellara G, Ascher-Jenull J, Borgogni F, et al. Extracellular DNA in soil and sediment: Fate and ecological relevance [J]. Biol Fertil Soils, 2008,45:219-235.
[8] Childress H, Sullivan B, Kaur J, et al.Effects of ultraviolet light disinfection on tetracycline-resistant bacteria in wastewater effluents [J]. Journal of Water and Health, 2013,12(3):404-409.
[9] Huang J J, Hu H Y, Tang F, et al.Inactivation and Reactivation of Antibiotic-resistant Bacteria by Chlorination in Secondary Effluents of a Municipal Wastewater Treatment Plant [J]. Water Research, 2011,45:2775-81.
[10] Miller J H. Fate of Antibiotic Resistance Genes During Anaerobic Digestion of Wastewater Solids [D]. Dissertation submitted to the faculty of the Virginia Polytechnic Institute and State University, 2014.
[11] Childress H, Sullivan B, Kaur J, et al. Effects of ultraviolet light disinfection on tetracycline-resistant bacteria in wastewater effluents [J]. Journal of water and health, 2014,12:404-409.
[12] 张崇淼,牛治瑶,王 真,等.氯消毒对产ESBLs菌β-内酰胺酶类抗性基因接合转移的抑制及作用机制研究[J/OL]. 中国环境科学: 1-9 [2021-04-15]. https://doi.org/10.19674/j.cnki.issn1000-6923.20210414.002.
Zhang C M, Liu Z Y, Wang Z, et al. Inhibition and associated mechanism of conjugative transfer of β-lactamase resistance genes in ESBLs producing bacteria by chlorine disinfection [J/OL]. China Environmental Science, 1-9 [2021-04-15]. https://doi.org/10.19674/ j.cnki.issn1000-6923.20210414.002.
[13] Gao P, Munir M, Xagoraraki I, Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant [J]. Science of The Total Environment, 2012,(421-422):173-183.
[14] McKinney C W, Pruden A, Ultraviolet Disinfection of Antibiotic Resistant Bacteria and Their Antibiotic Resistance Genes in Water and Wastewater [J]. Environmental Science & Technology, 2012,46(24): 13393-13400.
[15] Mckinney C W, Pruden A. Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater [J]. Environmental Science & Technology, 2012,46(24):13393.
[16] Van Aken B, Lin L S. Effect of the disinfection agents chlorine, UV irradiation, silver ions, and TiO2nanoparticles/near-UV on DNA molecules [J]. Water Science & Technology, 2011,64(6):1226-1232.
[17] Kim S, Park H, Chandran K. Propensity of activated sludge to amplify or attenuate tetracycline resistance genes and tetracycline resistant bacteria: A mathematical modeling approach [J]. Chemosphere, 2010, 78(9):1071-1077.
[18] Liu S S, Qu H M, Yang D, et al.Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant [J]. Water Research, 2018,136:131-136.
[19] 李树铭,王 锦,王海潮.UV、O3及UV/O3削减耐药菌和抗性基因性能[J]. 中国环境科学, 2019,39(12):5145-5153.
Li S M, Wang M, Wang H C. Reduction of ARB and ARGs by ultraviolet, ozone and combined disinfection technology [J]. China Environmental Science, 2019,39(12):5145-5153.
[20] Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure [J]. Current Opinion in Biotechnology, 2003,14(3):255-261.
[21] Wang D N, Liu L, Qiu Z G, et al. A new adsorption-elution technique for the concentration of aquatic extracellular antibiotic resistance genes from large volumes of water [J]. Water Research, 2016,92:188-198.
[22] Dong P, Wang H, Fang T, et al. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG [J]. Environ. Int., 2019,125:90-96.
[23] 李美菊,陈向东,谢志雄,等.通过DNA释放及感受态建立进行的枯草杆菌细胞间自然转化[J]. 武汉大学学报(理学版), 2003,(4):514-518.
Li M J, Chen X D, Xie Z X. Bacillus subtilis undergo Cell-to-Cell natural transformation via DNA releasing and competence acquiring [J]. Journal of Wuhan University (Natural Science Edition), 2003,(4):514-518.
[24] Lorenz M G, Wackernagel W. Natural genetic transformation of Pseudomonas stutzeri by sand-adsorbed DNA [J]. Archives of Microbiology, 1990,154(4):380-385.
[25] Yuan Q B, Huang Y M, Wu W B, et al. Redistribution of intracellular and extracellular free &adsorbed antibiotic resistance genes through a wastewater treatment plant by an enhanced extracellular DNA extraction method with magnetic beads [J]. Environ. Int., 2019,131:104986.
[26] Chandrasekaran S, Venkatesh B, Lalithakumari D. Transfer and expression of a multiple antibiotic resistance plasmid in marine bacteria [J]. Curr. Microbiol., 1998,37(5):347-51.
[27] Paget E, Simonet P, On the track of natural transformation in soil [J]. FEMS Microbiology Ecology, 1994,15(1):109-117.
[28] Hendrickx L, Hausner M, Wuertz S. Natural genetic transformation in monoculturesp. Strain BD413 Biofilms [J]. Applied & Environmental Microbiology, 2003,69(3):1721-1727.
[29] Palmen R, Hellingwerf K J. Uptake and processing of DNA by Acinetobacter calcoaceticus – a review 1Presented at the Workshop on `Type-4pili – biogenesis, adhesins, protein export and DNA import', Schloss Ringberg, Germany, 26–29 November 1995.1 [J]. Fuel & Energy Abstracts, 1997,192(1):179-190.
[30] Kunito T, Ihyo Y, Miyahara H, et al. Soil properties affecting adsorption of plasmid DNA and its transformation efficiency in Escherichia coli [J]. Biology and Fertility of Soils, 2016,52(2):223-231.
[31] Wang J, Chu L,Wojnárovits L, et al. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: An overview [J]. Science of the Total Environment, 2020,744:140997.
[32] Phattarapattamawong S, Chareewan N, Polprasert C. Comparative removal of two antibiotic resistant bacteria and genes by the simultaneous use of chlorine and UV irradiation (UV/chlorine): Influence of free radicals on gene degradation [J]. Science of the Total Environment, 2021,755:142696.
[33] Guo M T, Yuan Q B, Yang J. Ultraviolet reduction of erythromycin and tetracycline resistant heterotrophic bacteria and their resistance genes in municipal wastewater [J]. Chemosphere, 2013,93(11):2864- 2868.
[34] Yuan Q B, Guo M T, Yang J. Monitoring and assessing the impact of wastewater treatment on release of both antibiotic-resistant bacteria and their typical genes in a Chinese municipal wastewater treatment plant [J]. Environ Sci Process Impacts, 2014,16(8):1930-1937.
[35] Guo M T, Yuan Q B, Yang J. Insights into the amplification of bacterial resistance to erythromycin in activated sludge [J]. Chemosphere, 2015,136(oct.):79-85.
[36] Díaz Montaña J J, Gómez Vela F, Díaz Díaz N. GNC–app: A new Cytoscape app to rate gene networks biological coherence using gene–gene indirect relationships [J]. Biosystems, 2018,166:61-65.
[37] Wang R N, Zhang Y, Cao Z-H, et al. Occurrence of super antibiotic resistance genes in the downstream of the Yangtze River in China: Prevalence and antibiotic resistance profiles [J]. Science of the Total Environment, 2019,651:1946-1957.
[38] Chen S, Li X, Wang Y, et al. Induction ofinto a VBNC state through chlorination/chloramination and differences in characteristics of the bacterium between states [J]. Water Res, 2018,142:279-288.
[39] Ma X, Bibby K. Free chlorine and monochloramine inactivation kinetics of Aspergillus and Penicillium in drinking water [J]. Water Research, 2017,120:265-271.
[40] Poly F, Chenu C, Simonet P, et al. Differences between linear chromosomal and supercoiled plasmid DNA in their mechanisms and extent of adsorption on clay minerals [J]. Langmuir, 2000,16(3): 1233-1238.
[41] 王文生,魏德洲,郑龙熙.微生物在矿物表面吸附的意义及研究方法[J]. 国外金属矿选矿, 1998,(3):37-40.
Wang W S, Wei D Z, Zheng L X. Significance and research methods of microbe adsorption on mineral surface [J]. Metallic Ore Dressing Abroad, 1998,(3):37-40.
[42] Ju F, Li B, Ma L. et al. Antibiotic resistance genes and human bacterial pathogens: Co-occurrence, removal, and enrichment in municipal sewage sludge digesters [J]. Water Research, 2016,91(Mar.15):1-10.
[43] Nigro S J, Brown M H,Hall R M.AbGRI1-5, a novel AbGRI1variant in an Acinetobacter baumannii GC2 isolate from Adelaide, Australia [J]. Journal of Antimicrobial Chemotherapy, 2018,74(3):821-823.
[44] Du X, He F, Shi Q. et al. The rapid emergence of tigecycline resistance in blaKPC–2Harboring, as Mediated in Vivo by Mutation in tetA During Tigecycline Treatment [J]. Frontiers in microbiology, 2018,9:648.
[45] Shi L, Liang Q, Feng J, et al. Coexistence of two novel resistance plasmids, blaKPC-2-carrying p14057A and tetA(A) -carrying p14057B, in Pseudomonas aeruginosa [J]. Virulence, 2018,9(1):306-311.
[46] Deng M, Zhu M H, Li J J,et al. Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital [J]. Antimicrobial Agents & Chemotherapy, 2014,58(1):297-303.
[47] Bonomo R A, Szabo D.Mechanisms of Multidrug Resistance in Acinetobacter Species and Pseudomonas aeruginosa [J]. Clin. Infect. Dis., 2006,43(Supplement_2):S49-S56.
[48] Shakhawat, Chowdhury. Heterotrophic bacteria in drinking water distribution system: a review [J]. Environmental Monitoring and Assessment, 2011,184(10):6087.
[49] Liu G, Verberk J, Dijk J. Bacteriology of drinking water distribution systems: an integral and multidimensional review [J]. Applied Microbiology and Biotechnology, 2013,97(21):9265-9276.
[50] InÈs Mehri, Turki Y, HanÈne ChÉrif, et al. Influence of biological treatment and ultraviolet disinfection system onspp. diversity in wastewater as assessed by denaturing gradient gel electrophoresis [J]. CLEAN – Soil, Air, Water, 2014,42(5):1-8.
[51] 周春爽.紫外活化过硫酸盐去除污水中抗性菌和抗性基因效能研究[D]. 哈尔滨:哈尔滨工业大学, 2020.
Zhou C S. Removal efficiency of resistant bacterial and resistant genes from wastewater by UV activated persulfate [D]. Harbin: Harbin Institute of Techndogy, 2020.
[52] 吴春笃,许小红,宁德刚,等.城市污水细菌多样性及其生物安全性研究[J]. 中国安全科学学报, 2008,(1):119-122,180.
Wu C D, Xu X H, Ning D G, et al. Study on bacterial diversity and biological safety of municipal sewage [J]. China Safety Science Journal, 2008,(1):119-122,180.
Generation of extracellular antibiotic resistance genes during municipal wastewater chlorination and UV disinfection.
FU Shu-sen, WANG Yi, WANG Xiao-lin, WANG Shang-jie, CHENG Yuan, BIAN Bo, ZHANG Sheng-bo, YUAN Qing-bin*
(College of Environmental Science and Engineering, Nanjing Tech University, Nanjing 211816, China)., 2021,41(10):4756~4762
In this study, the generation of eARGs in different physical forms during wastewater chlorination and ultraviolet (UV) disinfection was investigated, and the correlation with microbial communities was explored. Results indicated that though chlorination decreased the abundance of intracellular ARGs, absorbed eARGs was significantly amplified by (0.7±0.1)log, while the abundance of free eARGs decreased by (0.2±0.1)log. UV disinfection also decreased the abundance of intracellular ARGs, but caused a significant increase of free eARGs ((0.4±0.2)log) and a decrease of absorbed eARGs ((0.3±0.1)log). Post chlorination, the abundance of most phyla increased whiledecreased in the absorbed extracellular DNA (a-eDNA), resulting in a increase of the bacterial diversity index from 4.2 to 4.7. Whereas,increased by 6.6% in the free extracellular DNA (f-eDNA) after chlorination, causing a decrease of the bacterial diversity index from 3.5 to 2.8. Post UV disinfection, the abundance ofin a-eDNA decreased by 36.6%, while the bacterial diversity index increased to 4.8; the abundance of bacteria in f-eDNA changed slightly. The molecular ecological network analysis indicated a wide hosting relationship between ARGs and bacteria genera.,,andwere correlated with 17, 15, 15 and 5genera respectively, suggesting changes in potential hosts post disinfection were essential mechanisms of the eARGs generation. This study shows that chlorination and UV disinfection can’t eliminate the risk of antibiotic resistance but only change patterns of the risk by inducing the generation of adsorbed and free eARGs.
chlorination;ultraviolet disinfection;extracellular antibiotic resistance genes;bacterial community;molecular ecological network
X172
A
1000-6923(2021)10-4756-07
付树森(1996-),男,江苏徐州人,南京工业大学硕士研究生,研究方向为水环境中抗性基因的检出、分布和去除技术.
2021-02-19
江苏省自然科学基金资助项目(BK20201367);国家自然科学基金资助项目(51608260)
* 责任作者, 副教授, yuanqb@njtech.edu.cn
