Transcriptome-wide Effect of Salix SmSPR1 in etiolated seedling of Arabidopsis
2021-04-30XiaoxiaLiuJianguoZhangLiXueGuodongRao
Xiaoxia Liu · Jianguo Zhang ,2 · Li Xue ·Guodong Rao
Abstract Microtubules and their regulatory proteins are involved in the regulation of plant cell morphology. SPIRAL1 (SPR1), a plant-specif ic microtubule-binding protein, is critical in regulating the anisotropic growth of plant cells. Our previous study showed that overexpressed Salix SmSPR1 genes in Arabidopsis thaliana caused right-handed spiral elongation in etiolated seedlings, but there were no morphological diff erences between wild-type and transgenic seedlings under varied light conditions. We then studied the transcriptional regulation patterns in transgenic plants engineered with the SmSPR1 gene. Transcriptomic results showed that a large number of diff erentially expressed genes were involved in plant light signal reception, chlorophyll synthesis and photosystem structure. Eleven gene families with 42 photosynthesis-related genes and 6 light-responsive genes were involved in regulation of cell morphology. Our results showed that these genes in the SmSPR1-ox line were particularly down-regulated under dark conditions. In addition, 33 TFs showed diff erences between SmSPR1-ox and wild-type lines. Taken together, the transcriptome analysis provides new insight into investigating the molecular mechanisms of light-induced cell morphological changes mediated by the microtubule binding protein SPR1.
Keywords Arabidopsis thaliana · Cell anisotropism ·SmSPR1 · Transcription factor · Transcriptome analysis
Introduction
Microtubules (MTs), components of the cytoskeleton,are important in maintaining cell morphology, promoting intracellular material transport, signaling, and regulating cell mitosis and diff erentiation (Gardiner et al. 2011). In the cell cycle, plant cells at diff erent stages have diff erent microtubule arrays, each of which performs special functions in the corresponding period, e.g., cell wall formation,cell morphology, and polar growth. The microtubule arrays of each stage are highly dynamic (Hashimoto 2015). Light is one of the most important environmental factors that regulates plant growth and development. Plant tissues and cells exhibit diff erent morphological patterns under light versus darkness. These patterns include a short hypocotyl and rapid root growth in light as well as termination of root elongation and a rapidly elongating hypocotyl in darkness. Thus,both microtubules and light are related to the morphology of plant cells.
Numerous studies have shown that hypocotyl elongation in light and darkness is regulated via internal signals. These signals can be prorogated via plant hormones, TFs, and plant photoreceptor systems (Zhang et al. 2017; Ling et al. 2017).Microtubule-associated proteins (MAPs) aff ect plant cell growth and diff erentiation by changing the array, stability,and dynamics of microtubules. Many MAPs have been identif ied, and these have diff erent functions in plant growth and development (Gardiner 2013). InArabidopsis, AtEB1 regulates the dynamic change of the plus end of the microtubule by inhibiting its contraction. AtEB1 also responds to touch and gravity signals in roots, and the AtEB1 mutant exhibits leftward deviations in root growth (Bisgrove et al. 2008).WDL3 participates in the physiological process of photoinhibition of hypocotyl growth by regulating the arrangement of periplasmic microtubules under light conditions. In dark conditions, WDL3 is degraded via the ubiquitin proteasome pathway, thus relieving the inhibitory Effect of WDL3 on the growth of the hypocotyl (Liu et al. 2013).
Willow is an important deciduous tree that grows rapidly.It has a wide distribution and exhibits high genetic variation.Salix matsudanavar. f.tortuosa(Vilm.) Rehd. is a variant ofS. matsudanaKoidz. with beautifully curved branches. This variant provides highly suitable natural mutant material for studying the diff erences in woody plant traits. For genetic improvement of trees, it is important to study the function of microtubules and their binding proteins. SPIRAL1 (SPR1) is a plant-specif ic protein, which belongs to a six-member family inArabidopsis(Nakajima et al. 2006). Mutation ofspr1inArabidopsiscaused isotropic growth of plant endoderm and cortical cells, and resulted in the rotation of the root axis. The petiole ofatspr1lines exhibited a typical righthanded spiral growth phenotype, and the roots tilted to the right when cultured vertically on hard medium (Sedbrook et al. 2004). The abnormal arrangement of cortical microtubules occurred in the somatic cells of thespr1mutant(Furutani et al. 2000). However, SPR1 has been intensively studied only inArabidopsis. Few studies have investigated SPR1 in woody plants. The spiral phenotype caused byspr1is very similar to the curved phenotype ofS. matsudanavar.f.tortuosa(Vilm.) Rehd., which prompted our investigation into the function ofSalix SPR1. We transferred theSalix SPR1intoArabidopsisand obtained the transgenic plants.
In this study, we established that overexpression ofSmSPR1inArabidopsiscan promote etiolated seedlings exhibiting right-handed helical elongation. However, it remained unclear how the SPR1 protein coordinates these downstream regulators to mediate hypocotyl growth in darkness. To address this question, we selected four RNA samples of transgenic and wild-typeArabidopsisseedlings that were grown in light and dark conditions, respectively.We sequenced the samples to elucidate dark-induced helical growth in transgenic lines by using Illumina deep sequencing technique. We aimed to develop a system to explore the function ofSmSPR1by inducing intracellular changes in theArabidopsistranscriptome via exposure to light and darkness. Our goal was to establish a genetic basis for molecular breeding and the improvement of horticultural forest morphology.
Materials and methods
Plant materials, RNA extraction, transcriptome sequencing
Experimental plant material was taken fromS. matsudanaindividuals. Branches and buds of adult willows were surface sterilized and then frozen rapidly in liquid nitrogen.TheArabidopsisseeds (Col-0) of this study were sterilized with 70% (v/v) ethanol for 1 min following 15% (v/v)sodium hypochlorite (~ 10%) for 10 min. The seeds were then washed f ive times in sterilized water and sown on Murashige and Skoog (MS) medium for transgenic selection with 50 mg·L −1 kanamycin. The germination assays were conducted at 22 °C after being stratif ied at 4 °C for 48 h.
We designed four RNASeq libraries to obtain a general overview of the light-responsive hypocotyl helix transcriptome. These libraries used wild-type seedlings with a 7-day light / dark culture andSmSPR1-oxseedlings with a 7-day light / dark culture. The total RNA of 7-day-old seedlings was extracted by a plant total RNA extraction kit (Aidlab,Beijing, China). Each library was sequenced in the Illumina HiSeq 4000 system by using single-ended mode. Finally, a paired end read of 125 bp / 150 bp length was generated via the Illumina Hiseq platform (Novogene Technology, Beijing) and we then calculated the clean readings for Q20,Q30, and GC and used high-quality clean readings for further analysis. All diff erent pure reads were mapped to theArabidopsisreference genome database (TAIR10).
Functional classif ication of diff erentially expressed genes (DEGs) and quantitative-PCR analysis
DEGs were identif ied between wild-type and transgenic lines. Functional analysis of DEGs was performed by Gene Ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes) through > 2 or < − twofold changes and correctedPvalues of < 0.005 to analyze functional classif ication of gene expression diff erences. Quantitative real-time PCR (qRT-PCR) was used to determine the expression of selected genes and GoTaq qPCR Master Mix (Promega) was used according to the manufacturer’s instructions. The thermal cycling conditions were 95 °C / 3 min, then 95 °C / 15 s,60 °C / 30 s, 72 °C / 30 s for 40 cycles. Relative expression levels were calculated using the 2-ΔCt method. AtActin2(At3G18780) was selected and verif ied as an internal standard. Three biological replicates were used in this experiment.Table S1 lists all the primers used in this analysis.
Statistical analyses
Quantitative data were analyzed using the one-way analysis of variance (ANOVA) with SPSS. The signif icance analysis was performed atP≤ 0.05.
Accession numbers
RNA Sequence data presented here can be accessed in the GenBank libraries using accession numbers: CK_D7, SRR10321777; CK_L7, SRR10321776; OE_D7,SRR10321775; OE_L7, SRR10321774.
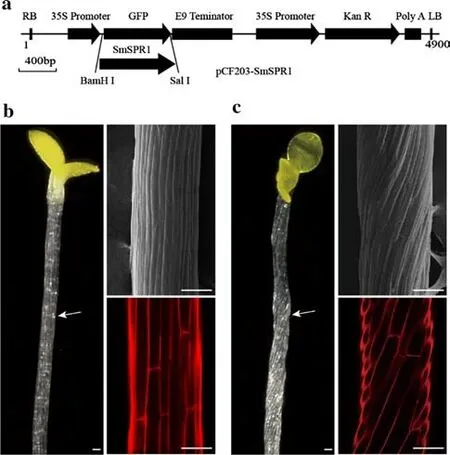
Fig. 1 Phenotypes of the wild-type and SmSPR1-ox lines. a Construction of the pCF203- SmSPR1 vector. b, c The phenotypes and overview of wild-type ( b) and SmSPR1-ox ( c) after 7 days of darkness treatments (Liu et al. 2019), Bar = 100 μm
Results
Overexpression of SmSPR1 caused hypocotyl helix in etiolated seedlings of Arabidopsis
The cDNA ofSmSPR1was cloned and inserted into the binary vector pCF203 under the control of 35S promoter(Fig. 1 a). The recombinant plasmid pCF203-SmSPR1was transformed intoAgrobacteriumstrain GV3101. Fourteen resistant lines were selected, and one homozygous overexpression line with a distinct phenotype was selected for further research. The results showed that the seedlings of wildtype andSmSPR1-oxlines exhibited no obvious diff erences when cultured under light. When seeds were cultured in darkness for 7 d, we found that the hypocotyls ofSmSPR1-oxexhibited spiral growth that was absent in wild-type plants(Fig. 1 b and c). These results indicate that overexpression ofSmSPR1is involved in skotomorphogenesis of seedlings,and this process may be suppressed by light signals. In order to gain insights into this mechanism, we conducted global mRNA sequencing of four treatments: wild-type seedlings cultured for 7 d under light or dark conditions,SmSPR1-oxseedlings cultured for 7 d under light or dark conditions.
Transcriptome sequencing and identif ication of SmSPR1-ox-regulated genes
Four libraries were constructed from the diff erent lines ofArabidopsisseedlings after 7 days of light and darkness treatments. These generated 50-68 million clean reads from RNA-seq, with 8.85G nucleotides of WT_L7, 7.64G of OE_L7, 10.23G of WT_D7 and 8.3G of OE_D7. The GC content of all four samples was around 45% (Table 1). An overview of the raw reads and clean reads is shown in Fig. S1.
To explain the impact ofSmSPR1overexpression in the spiral growth of hypocotyls, diff erences in gene expression between the dark and light treatments were compared for the wild-type andSmSPR1-oxseedlings. In the wild-type,1,510 genes were up-regulated and 1,786 genes were downregulated respectively in the dark treatment compared to the light treatment.SmSPR1-oxexhibited a greater number of changes when compared with the wild-type, with 2,129 up-regulated and 2,235 down-regulated genes (Fig. 2 a, b).This indicates that overexpression ofSmSPR1aff ects the transcription of more downstream genes during light anddarkness conversion. We also compared the DEGs of wildtype andSmSPR1-oxseedlings in the same condition (dark or light). There were no up-regulated genes (two-fold or greater changes) in theSmSPR1-oxline when compared with the wild-type line, and only 20 unigenes were down-regulated under the light growth conditions (OE_L7/WT_L7)(Fig. 2 c, Table S2). After 7-day darkness treatment, there were 118 (up-regulated) and 116 (down-regulated) unigenes that exhibited changes (≥ twofold changes) inSmSPR1-oxlines when compared to the wild-type line (OE_D7/WT_D7)(Fig. 2 c, Table S3). The results showed that overexpression ofSmSPR1inArabidopsisexhibited a weaker Effect under light growth conditions. However, hundreds of genes were aff ected by overexpression ofSmSPR1when transgenic lines were cultured in darkness, indicating that expression of these genes may be related to the spiral growth of hypocotyls.
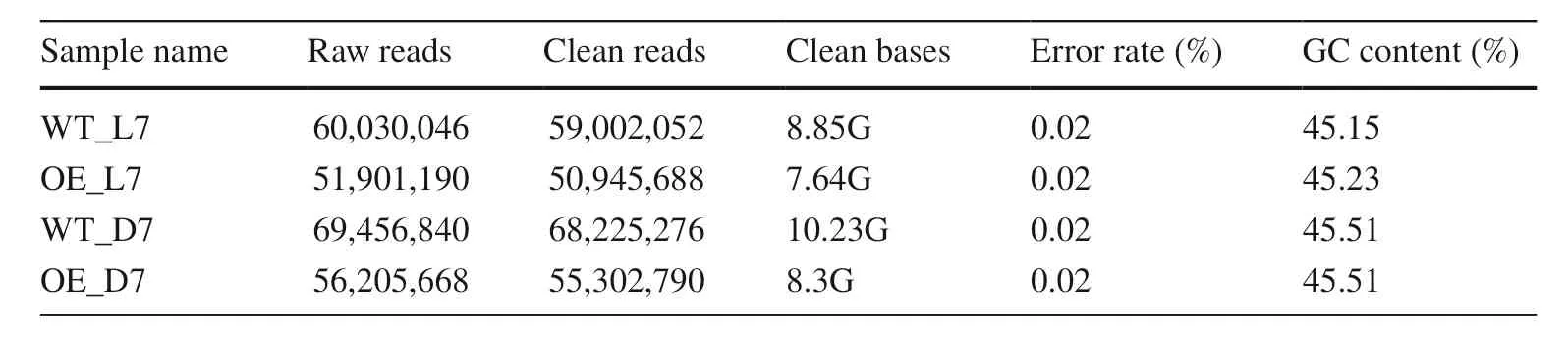
Table 1 Summary statistics of RNA sequencing data

Fig. 2 Changes in gene expression prof iles of the wild-type line and SmSPR1-ox line during light induction. a Venn diagram of upregulated genes between light and darkness, respectively, in wild-type(WT_L7 VS WT_D7) and SmSPR1-ox line (OE_L7 VS OE_D7). b Venn diagram of down-regulated genes between light and darkness,respectively, in wild-type (WT_L7 VS WT_D7) and S mSPR1-ox line(OE_L7 VS OE_D7). c The number of DEGs between wild-type and SmSPR1-ox seedlings after 7-days darkness (OE_D7/WT_D7) or light (OE_L7/WT_L7)
Functional analysis of SmSPR1-ox-regulated genes
To identify DEGs associated withSmSPR1and to understand the metabolic or signal transduction pathways between them, we compared the DEGs between the wild-type andSmSPR1-oxsubjected to light and darkness treatments. All DEGs were assigned to functional categories via GO and KEGG analysis. Notably, theSmSPR1-oxin the dark treatment exhibited more changes in gene expression than did the wild-type. These genes were classif ied on the basis of“biological process (BP)”, “cellular component (CC),” and“molecular function (MF)” (Fig. 3). The up-regulated genes were assigned to BP, with wide-ranging sub-categories. The down-regulated genes were mainly assigned in the CC. The“metabolic process,” “cellular component,” “molecular function,” “cell part,” “biological process,” and “cell” categories were the most common groups for both up-regulated or down-regulated genes. Changes in the “biological process” classif ication were most signif icant in GO analysis(Fig. 3). KEGG pathway mapping facilitates understanding of biological pathways for specif ied functions and networks.A total of 234 (118 up-regulated and 116 down-regulated)genes were mapped into 49 KEGG pathways. These results showed that the most represented pathways were “Metabolic pathways” and “Biosynthesis of secondary metabolites.”Many of the genes are related to “photosynthesis”, such as“Photosynthesis-antenna proteins” and “porphyrin and chlorophyll metabolism,” indicating that overexpression ofSmSPR1aff ects photoreceptor-related pathways (Table 2).
ForSmSPR1-oxplants, the GO and KEGG analyses indicated that the up-regulated genes were mostly related to response to light stimulus. The down-regulated genes were signif icantly related to photosynthesis and chlorophyll synthesis.
Screening of crucial transcription factors in SmSPR1-ox lines compared to wild-type after dark treatment
Transcription factors (TFs) are proteins that regulate gene transcription by binding to a specif ic domain of the target gene promoter. Among 234 DEGs, 33 TFs showed signif icant diff erences betweenSmSPR1-oxlines and wild-type lines (Table 3). These genes belong to diverse categories,including zinc-f inger-related TFs, MYB-like TFs, transcription activators, and ethylene-responsive TFs. Moreover, 25 of the TFs were up-regulated, while only 8 were down-regulated (Fig. S2). In this study, we identif ied 7 MYB and these genes were up-regulated inSmSPR1-oxlines. Furthermore,3 B-box-type zinc f inger family proteins were observed.The expressions of these three TFs were up-regulated in transgenic lines subjected to the dark treatment. It is worth noting that among the diff erentially expressed genes, we discovered an important light-responsive TF, which we named HY5. However, in our study, the expression of HY5 was up-regulated inSmSPR1-oxlines in the dark treatment when compared with the wild-type. This indicates that HY5 may have other functions.

Fig. 3 GO enrichment of DEGs in SmSPR1-ox plants. a Signaling pathway enrichment of up-regulated genes in SmSPR1-ox transcriptome. b Signaling pathway enrichment of downregulated genes in SmSPR1-ox transcriptome
Plant light signal and photosynthesis -related genes
We identif ied 6 DEGs that participate in light signal transduction including SPA1, SPA3, BBX24, BBX25, BBX28,and HY5 (Table S4). The six genes were up-regulated inSmSPR1-oxseedlings. In this study, we identif ied 4 DEGs that were associated with plant hormone modulation. InSmSPR1-oxplants, 2 genes related to IAA were up-regulated and 2 genes related to ethylene were down-regulated when compared with the wild-type.
Sixty-seven DEGs were identif ied as participants in plant photosynthesis between wild-type andSmSPR1-oxplants in the dark treatment. All these genes have been reported to play direct or indirect roles in chlorophyll synthesis and photosystem structure. Among them, 64 were down-regulated inSmSPR1-oxplants compared with wild-type. These downregulated genes are mainly related to photosystem reaction center complex (PSAD2, PSAG, PSAO, PSAE1, PSAK,PSAH2, PSBW, PSBT, and PSBX), photosystem light harvesting complex (LHCA1, LHCA3, LHCA4, LHCA6,LHCB1.1, LHCB1.2, LHCB1.3, LHCB2.1, LHCB2.2,LHCB2.4, LHCB3, LHCB4.2, LHCB5, and LHCB6),ribosomal protein (RPL5, RPL15, RPL18, and RPL19),and the Rubisco small subunit (RBCS) multigene family(RBCS-1A, RBCS-1B, RBCS-2B, and RBCS-3B).Arabidopsiscontains two PSBQ genes of photosystem II (PS II). In our experiment, the expression level of PSBQ genes inSmSPR1-oxlines was lower than in wild-type lines. InArabidopsis, the genome encodes three structurally related but diff erentially regulated NADPH: protochlorophyllide oxidoreductase (POR) genes: PORA, PORB, and PORC.These genes catalyze the reduction of protochlorophyllide to chlorophyllide (Paddock et al. 2010). In our study, these three genes were down-regulated in theSmSPR1-oxlines subjected to the darkness treatment. PORA was particularly down-regulated. The above results indicate thatSmSPR1-oxwidely aff ects plant photosynthesis and photosystem structure genes (Fig. 4, Table S5). To f ind the regulatory photosynthesis genes related to anisotropic growth of cells in the dark, we selected down-regulated genes and constructed a co-expression network in which the hub genes showed the tightest connections (Fig. S3). The central genes with the highest number of edges are PSA and PSB.

Table 2 KEGG analysis of DEGs involved in SmSPR1-ox Arabidopsis

Table 3 Transcription factors in DEGs under darkness
To further verify the accuracy of RNA-seq results, we screened 15 representative genes for qRT-PCR analysis. Theresults showed that the diff erence of gene expression levels between wild lines andSmSPR1-oxlines was highly correlated with RNA-seq data (Fig. 5).

Fig. 4 Diff erentially expressed photosynthesis-related genes after 7 days of dark culture. a The number and percentage of chlorophyll synthesis genes and photosystem structure genes regulated by SmSPR1-ox. b Heat-map showing the expression fold change of gene families associated with chlorophyll synthesis and photosystem structure. The color scale at the right represents the log-transformed Fragments Per Kilobase Million (FPKM) value
Discussion
SmSPR1was identif ied and transferred intoArabidopsis,and there were no phenotypic diff erences in transgenic lines under normal growth conditions. However, when the seedlings were cultured in darkness for 7 d, the hypocotyl of theSmSPR1-oxline showed spiral growth (Fig. 1 b and c), as reported earlier (Liu et al. 2019). This suggested that the function ofSPR1was inhibited by culture in daylight. Diff erences in gene expression in wild-type andSmSPR1-oxlines were functionally assigned to predict the genes that may participate in the spiral growth of hypocotyl. KEGG analysis of DEGs also revealed that photosynthesis, porphyrin, and chlorophyll metabolism were severely changed. This indicated that the hypocotyl helix phenotype inSmSPR1-oxlines are associated with photosynthesis and might interact with other TFs. Overexpression ofSmSPR1caused hypocotyl helix only in the dark treatment, illustrating that light signals are also a key factor leading to this phenotype. Therefore, we focused our analysis on (1) the light signal response pathway that causes plant skotomorphogenesis and (2) genes that may lead to anisotropic growth of plant hypocotyl cells.
Numerous studies have shown that the Effects of light and darkness on hypocotyl elongation are regulated by external and internal factors, including TFs, plant photoreceptors and plant hormones (Ma et al. 2018; Liu et al. 2013; Luo et al.2010). Several studies have shown thatSPR1aff ects cellular morphology (Galva et al. 2014; Nakajima et al. 2004;Sedbrook et al. 2004). In this study, we generated transgenicArabidopsisthat overexpressedSmSPR1. In the dark, these transgenic lines exhibited a right-handed spiral and tilted at a skewed angle. In the comparison of signif icant diff erences between OE_L7 and WT_L7, we found only 20 down-regulated DEGs and no up-regulated DEGs. Under dark conditions, OE_D7 and WT_D7 exhibited 118 up-regulated DEGs and 116 down-regulated DEGs when compared to OE_L7 and WT_L7, respectively. Diff erential expression of these genes led to helical growth of hypocotyls in overexpressing lines (Fig. 2 b), indicating thatSmSPR1may cause changes of cell morphology in the dark by participating in the elongation of hypocotyl cells. These DEGs may deliver interesting insights into what processes, mechanisms, and pathways are aff ected bySmSPR1.
Among these DEGs, 33 TFs showed significant differences betweenSmSPR1-oxlines and wild-type lines(Table 3). These include TFs that are associated with changes in cell morphology observed in hypocotyls grown under dark conditions. Among these TFs, four proved to be light-induced TFs (BBX24, BBX25, BBX29, and HY5)and four were hormone-responsive TFs (RAP2-3, RAP2-13, IAA19, and IAA29). Previous studies have reported that plant hormones are widely involved in cell extension, division and diff erentiation (Alabadi et al. 2009, 2004). This indicates that light response and hormone signaling pathways may play important roles in etiolated spiral growth.
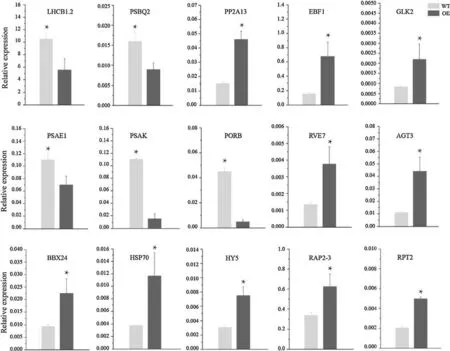
Fig. 5 qRT-PCR analysis of DEGs in wild-type and SmSPR1-ox line during darkness. P values were determined by one-way ANOVA and Tukey HSD test. * P < 0.05
Chloroplasts are important plant organelles involved in photosynthesis and the production of amino acids, lipids,and plant hormones (Ericson et al. 1986; Okazaki et al.2010). In higher plants, the chloroplasts contain photosynthetic thylakoid membranes and four multi-subunit protein complexes (i.e., photosystem I [PSI], photosystem II [PSII],ATP synthase, and cytochrome b6f complex). These four complexes comprise at least 70 diff erent proteins (Friso et al. 2004). Chloroplasts begin as proplastid in small meristematic cells and then change from a simple proplastid to a photosynthetic chloroplast (Jiang et al. 2012; Gang et al.2019). Xu et al. ( 2016) reported that chloroplast signaling counteracts light signaling in terms of inhibition of hypocotyl elongation. The chloroplasts in the dark plant cells are in the form of etioplasts. Etioplasts can develop into chloroplasts after exposure to light. These etioplasts are also an essential organelle for plants and are responsible for many basic intermediate metabolic reactions.
A plastid is produced by the bipartite division of preexisting plastids, so plastid division represents an important process for maintaining the appropriate number of plastids in plant cells. In order to maintain proper proplastid separation during cell division, proplastids must be split prior to cytokinesis (Aldridge et al. 2005). In this study, we identif ied 64 DEGs signif icantly enriched in the terms related to photosynthesis and chlorophyll synthesis (Fig. 4). Interestingly,transcriptome results showed that the expression levels of all of these genes were down-regulated in overexpressing plants relative to wild-type plants. There is no obvious cell division in the cortex and epidermis. The post-embryonic growth ofArabidopsishypocotyls is the result of cell elongation. Therefore, it is speculated that the down-regulation of plastid-related genes caused by overexpression ofSmSPR1may be associated with the elongation of cells in the dark,thereby aff ecting the anisotropic growth of cells.
The development of plant seedlings is divided into light morphogenesis under light conditions and dark morphogenesis in darkness, which was characterized by elongated hypocotyls and closed cotyledons. It is known that plant seedling hypocotyls exhibit rapid elongation in darkness.During this process, COP1 acts as a functional E3 ligase and plays an important role which degrades the positive factors, but it stabilizes negative factors to mainly inhibit photomorphogenesis in the dark (Chen et al. 2015; Seo et al.2004). Previous studies have reported that suppressor of phytochrome A (SPA1-SPA4) proteins interact with COP1 to inhibit seedling photomorphogenic growth (Balcerowicz et al. 2017; Ling et al. 2017; Saijo et al. 2003). In our study,two members of theSPAgene family (SPA1andSPA3)were up-regulated under darkness in theSmSPR1-oxlines(Table S3). The B-box proteins, as a family of zinc f inger TFs, play diff erent roles in regulatory networks that control growth and developmental processes. It has been reported that two B-box proteins of BBX21 and BBX22 could interact with HY5 and enhance its activity. Interestingly, BBX24 and BBX25 inhibit the function of HY5 by combining with it to form inactive heterodimers (Gangappa and Botto 2014;2016). In our study, the expressions of BBX24 and BBX25 were up-regulated in transgenic lines. All these results demonstrate thatSmSPR1may aff ect plant cell morphology by regulating skotomorphogenesis-related genes. HY5 was a TF with the function of promoting photomorphogenesis. It plays a regulatory role under light conditions and is degraded by COP1 in dark conditions (Gangappa and Botto 2016;Srivastava et al. 2015). However, in the present study, the expression of HY5 was up-regulated inSmSPR1-oxlines in the dark compared with the wild-type. This indicates that it may be involved in other biological signaling pathways in dark conditions. Previous studies have reported that HY5 is not completely degraded in the dark and exists in phosphorylated form. HY5 is then rapidly unphosphorylated during the transition to light during seedling photomorphogenesis(Gangappa and Botto 2016). It is speculated that the function of HY5 after phosphorylation is similar to that of SPR1 and indirectly promotes the stability of MT. On the basis of the above studies, we generated a simplif ied model for the role of HY5 in the hypocotyl helix caused by overexpression ofSmSPR1(Fig. 6).
Conclusions
Microtubules and their regulatory proteins are involved in the regulation of plant cell morphology. SPIRAL1 (SPR1)is a plant-specific microtubule-binding protein that has attracted attention in the direction of microtubule elongation and plant growth. In this study, we found that the DEGs betweenSmSPR1-oxlines and wild-type lines inA. thalianaare concentrated in light-induced TFs, hormone signaling,and photosynthetic energy metabolism. It is indicated that light response and hormone signaling pathways and chlorophyll synthesis genes and photosystem structural genes may play important roles in etiolated spiral growth. This study provides a new basis to explore the function ofSmSPR1by inducing intracellular changes in theArabidopsistranscriptome during light and darkness.

Fig. 6 Phosphorylation- and dephosphorylation-mediated HY5 promotion of cell morphology through regulation of microtubules and chloroplastid during light-dark transformation
Publisher’s NoteSpringer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affi liations.
杂志排行
Journal of Forestry Research的其它文章
- Sacred groves of India: repositories of a rich heritage and tools for biodiversity conservation
- Relationship between H 2 O 2 accumulation and NO synthesis during osmotic stress: promoted somatic embryogenesis of Fraxinus mandshurica
- Changes in leaf stomatal traits of diff erent aged temperate forest stands
- Somatic embryogenesis and plant regeneration in Betula platyphalla
- Hydrogen peroxide as a systemic messenger in the photosynthetic induction of mulberry leaves
- Production and quality of eucalyptus mini-cuttings using kaolin-based particle f ilms
