Molecular Cloning and Bioinformatics Analysis of araC Gene of Vibrio alginolyticus
2021-04-08FanglingMOGyamfuaAFRIYIEJialingHUJunlingWANGShihuiZHOUZhongduoWANGKUEBUTORNYEChuanhaoPANHuanyingPANG
Fangling MO, Gyamfua AFRIYIE, Jialing HU, Junling WANG, Shihui ZHOU, Zhongduo WANG, F.K.A. KUEBUTORNYE, Chuanhao PAN, Huanying PANG*
1. Fisheries College of Guangdong Ocean University, Zhanjiang 524088, China; 2.Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals, Key Laboratory of Diseases Controlling for Aquatic Economic Animals of Guangdong Higher Education Institutions, Zhanjiang 524088, China
Abstract [Objective] To clone araC gene of Vibrio alginolyticus HY9901 strain, and analyze bioinformatics. [Methods] the whole genome sequence of Vibrio alginolyticus on GenBank was used to design specific primers. According to the principle of PCR amplification sequence, the target gene araC was amplified, and then the sequence was further analyzed by bioinformatics method to establish the phylogenetic tree of araC gene and its corresponding subunit three-dimensional structure model. [Results] Sequence analysis revealed araC gene is 711 bp and encodes a putative protein of 236 amino acids. The predicted molecular mass of AraC was 26.92 ku. Using Signal P 4.0 and TMHMM Server 2.0 software for analysis, it was predicted that the AraC protein did not contain a signal peptide or a transmembranous region. The AraC protein had two cAMP and cGMP dependent protein kinase phosphorylation site, five protein kinase C phosphorylation sites, three casein kinase II phosphorylation sites, one prenyl group binding site (CAAX box) and five microbodies C-terminal targeting signal. The predicted results of protein subcellular localization showed that AraC was located in the mitochondria, nucleus and cytoplasm. Its protein is unstable and hydrophilic. The AraC protein is a transcriptional regulatory protein which belongs to HTH_18 superfamily. According to the prediction, secondary structure: a-helix (Alpha helix) accounted for 52.12%, random coil (31.78%), extended strand (11.02%), b-fold (Beta turn) accounted for 5.08%. V. alginolyticus, Vibrio parahaemolyticus and Vibrio palustris were clustered together, which implies that the genetic relationship between these three species was the closest.
Key words Vibrio alginolyticus, araC gene, Gene cloning, Bioinformatics analysis
1 Introduction
Vibrio
alginolyticus
is a gram-negative halophilic bacterium that is commonly found as part of the normal microbial flora in marine environments. It has the shape of a short rod and contains no spores and capsule.V
.alginolyticus
has a large geographic distribution in marine and estuarine waters especially in bathing areas.Studies have shown thatV
.alginolyticus
is considered the most frequent species living freely in water and sedimentsand can survive in sea water even under conditions of nutrient stress while maintaining their virulence.Due to the halophilic nature ofV
.alginolyticus
, it is associated with several diseases of marine animals such as molluscs, fin fish and crustaceans.V
.alginolyticus
could cause infectiousVibriosis
in finfish, shrimp and shellfish in a larger scalethat can lead to immense mortalities and economic loss for the aquaculture industry. Additionally, in humans, studies have proven thatV
.alginolyticus
is responsible for gastroenteritisand peritonitis. It quickly adapts to changing environmental conditions. This reflects both the increasing pressure of human-induced coastal areas and the genetic and phenotypic plasticity ofVibrio
. As global warming increases ocean temperatures, incidences of out-of-season infections caused byV
.alginolyticus
are escalating and drawing more attention.On a global scale, the aquaculture industry has experienced high growth over the last two decades. This can be attributed to increased fish demand which is associated to high population growth and urbanization. Aquatic organisms such as fish, shrimps,etc
. are sources of high-protein animal and are digestible with high biological value as well. Finfish, shellfish and other aquatic organisms had been cultured and provide about two-thirds of the total fish supply, globally. Aquaculture with its resource-efficient method of producing aquatic organisms provide one-third of aquatic animals to the total supply of fish.However, the progress of aquaculture is challenged, such as disease outbreaks wherebyV
.alginolyticus
and otherVibrio
species are the principal agents responsible. TheseVibrio
species causes severe economic losses globally that obstructs the development of the aquaculture sector. The World Bank in 1997 reported that, 3 billion USD per year is the global estimation of disease losses.V
.alginolyticus
affects a varied range of marine vertebrates, invertebrates and marine teleost including penaeids, sea horse, bivalves and cephalopods.Nevertheless, the virulence factors ofV
.alginolyticus
are not completely understood.In this study,araC
gene ofV
.alginolyticus
was cloned sequenced and bioinformatically analyzed to provide a theoretical basis for further study of the function of AraC protein. ThearaC
gene together with other genes such asaraB
,araA
andAraD
(collectively known asaraBAD) exist in L-arabinose operon. L-arabinose operon directs the catabolism of arabinose inEscherichia
coli
.araBAD
encodes for three metabolic enzymes that are required for the metabolism of L-arabinose. AraC, on the other hand controls the expression of the L-arabinose operon as a single unit together with the catabolite activator protein (CAP)-cAMP complex. L-arabinose is a type of Arabinose found in component of biopolymers such as hemicellulose and pectin.2 Materials and methods
2.1 Strain and vector
The strain ofV
.alginolyticus
, as a highly virulent strain, was isolated from a diseasedLutjanus
erythopterus
in Zhanjiang watershed in the Guangdong Province and was stored at Fisheries College, Guangdong Ocean University. The cloning vector pmd18T was purchased from TaKaRa Biotechnology (Dalian) Company Ltd.2.2 Reagents
Ampicillin stock solution (100 mg/mL) was prepared with sterile water and stored at -20 ℃. The working concentration was 100 μL/100 mL in medium. Chloramphenicol stock solution was prepared with anhydrous ethanol, and the working concentration was 25 μg/mL. Calcium chloride solution (0.1 mol/L) used to prepare competent cells was prepared by dissolving 1.47 g of CaCl·2HO in 100 mL of sterile water, filtered through 0.22 μm filter and stored in a refrigerator at 4 ℃.UNIQ-10 Column Bacterial Genomic DNA Extraction Kit was purchased from Sangon Biotech (Shanghai) Company Ltd. EasyPure Plasmid MiniPrep Kit and EasyPureTM Quick Gel Extraction Kit were purchased from Beijing Transgen Biotech Co., Ltd. TIANamp Bacteria DNA Kit was purchased from Tiangen Biotech (Beijing) Co., Ltd. TaKaRa Ex Taq DNA Polymerase and T4 DNA ligase were purchased from TaKaRa Biotechnology (Dalian) Co., Ltd. Easy Pfu DNA polymerase was purchased from Tiangen Biotech (Beijing) Company Ltd. Primer synthesis and sequence analysis were done by Sangon Biotech (Shanghai) Company Ltd. The media used in this experiment include TSA medium (pH 7.3±0.2), TSB medium (pH 7.3±0.2), LB liquid medium and LB solid medium (containing 1.5% agar).
2.3 DNA extraction
Single colonies ofV
.alginolyticus
were cultured with TSB 28 ℃ for 18 h. Then, 1 mL of bacteria medium was taken and subjected to DNA extraction using UNIQ-10 Column Bacterial Genomic DNA Extraction Kit according to the instruction of DNA purification kit. And 1 mL of bacterial solution was centrifuged at 5 000 r/min for 5 min. The supernatant was discarded. The solution was washed twice with PBS and re-suspended in 200 μL of double distilled water (ddw). It was boiled for 5 min at 100 ℃ then centrifuged at 5 000 r/min for 5 min. The resulting supernatant was transferred into sterile EP tube and centrifuged at 12 000 r/min for 5 min and the clear supernatant containing the bacterial genomic DNA template was collected and preserved at -20 ℃ until use.2.4 Cloning of AraC gene from the
.
The primers were designed using Primer Premier Version 5.0 software according to the whole AraC sequence obtained from the GenBank (ACCESSION:NZ_AAPS00000000). The primers were 5′ ATGAAATCTAGCTTAAGCA 3′ and 5′ CTATCGGGAAAACTCTCTA 3′ as forward and reverse primers respectively. Polymerase Chain Reaction (PCR) was carried out on DNA sample fromV
.alginolyticus
for the purpose of amplification. The reaction system contained 1 μL of each primer, 1 μL of extracted DNA sample, 12.5 μL r-taq buffer, 9.5 μL of double distilled water. The PCR reaction was started with a pre denaturation step at 96 ℃ for 5 min followed by 33 thermal cycles of denaturation step at 96 ℃ for 30 s, annealing/renaturation at 60 ℃ for 45 s and extension at 72 ℃ for 1 min, terminated with a final extension of 10 min at 72 ℃. Finally, the resulting products of PCR were ran in 1.0% agarosed gel, electrophoresis.2.5 Ligation, transformation and sequencing of target gene
The PCR product was incubated with the vector pmd18T at 16 ℃ overnight. The ligation product was sequenced by Sangon Biotech (Shanghai) Company Ltd.
2.6 Bioinformatics analysis
The physical and chemical properties of the protein, signal peptide prediction, N-glycosylation sites prediction and protein structure domain were analyzed using ExPASy software with reference to literature. The N-terminal signal peptide sequence of AraC protein was predicted using SignalP4.1 Server software. Transmembrane domains were predicted using TMHMM Server 2.0 software. Subcellular localization was performed using PSORT. The potential phosphorylation sites and glycosylation sites were predicted using Soft Berry-Psite software. The amino acid sequences of otherVibrio
strains similar to the studied protein were searched in NCBI using the provided BLAST online searching software. These amino acid sequences were analyzed using MEGA7.0 software by Neighbour-joining method. The SWISS-MODEL software was also used to predict the 3-D structure ofV
.alginolyticus
AraC protein, performing the homologous modeling.3 Results
3.1 Cloning and amplification of
gene
ThearaC
gene was amplified via PCR. Agarosegelelectrophoresis analysis was carried out on the PCR products. Through the amplification of PCR, the specific bands of the PCR products of about 711bp was obtained as shown in Fig.1. The product was ligated with the cloning vector pmd18T, and the ligation product was sequenced by Sangon Biotech (Shanghai) Company Ltd. The sequence result revealed that thearaC
gene contains a 711bp open reading frame (ORF), and could encode 236 amino acids. The gene was submitted to GenBank, with an accession number of MN328349.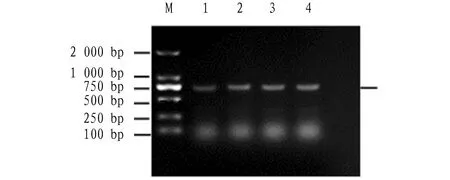
Note: Column M: DNA marker DL2000; Columns 1-4: AraC PCR product.
3.2 Physicochemical properties and sequence analysis
The physicochemical properties ofV
.aginolyticus
AraC protein were analyzed using ExPASy (http://web.expasy.org/protparam/) software. The molecular weight , theoretical isoelectric point, instability index, grand average of hydropathicy, number of negatively charged residues (Asp + Glu), and number of positively charged residues (Arg + Lys) of AraC protein was 26.92, 8.94, 58.18, -0.152, 24, and 28, respectively. The molecular structural formula of AraC was CHNOSwith a total of 3 801 atoms. AraC is an unstable hydrophilic protein with instability index (II) of 58.18 and a theoretical pI value of 8.94.The AraC protein was subjected to signal peptide prediction using SignalP 4.1 Server online software. The results showed that the protein contains no significant signal peptide cleavage sites (http://www.cbs.dtu.dk/services/SignalP/). PORST (https://wolfpsort.hgc.jp/) subcellular localization prediction showed that AraC exists in mitochondria, nucleus and cytoplasm. The prediction using TMHMM Server (http://www.cbs.dtu.dk/) online software showed that AraC contains no transmembrane domain.
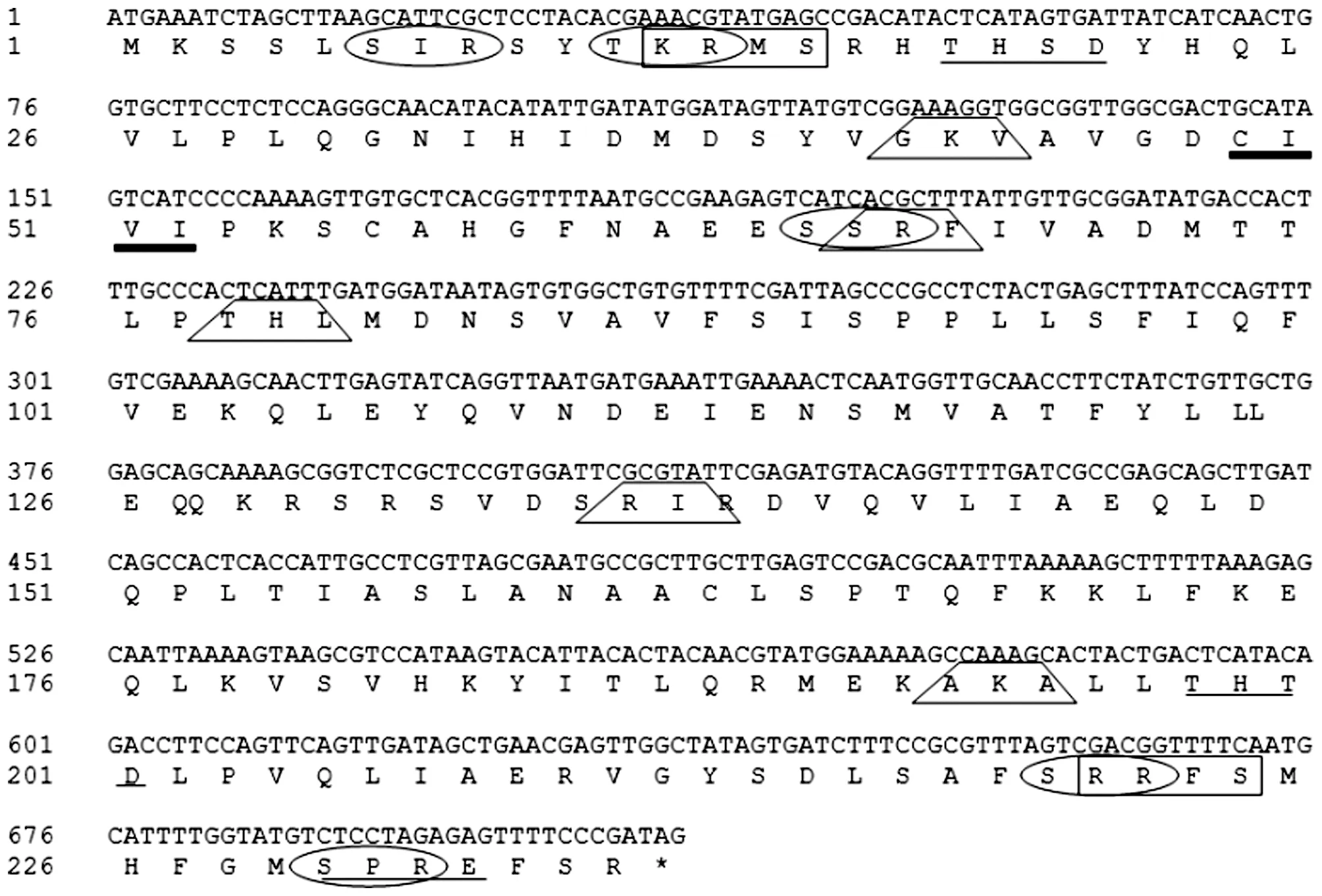
Note: : cAMP- and cGMP-dependent protein kinase phosphorylation site; : Casein kinase II phosphorylation site; : Protein kinase C phosphorylation site; : Prenyl group binding site (CAAX box); : Microbodies C-terminal targeting signal; *: Terminator.
Using SoftBerry-Psite software, the amino acid sequence of AraC protein potentially contains two cAMP- and cGMP-dependent protein kinase phosphorylation sites, five protein kinase C phosphorylation sites, three casein kinase II phosphorylation sites, one prenyl group binding site(CAAX box) and five microbodies C-terminal targeting signal (http://linux1.softberry.com/cgi-bin/programs/proloc/psite.pl) as shown in Fig.2.
3.3 Homology analysis with similar protein sequences of other
strains
The GeneDoc software was used for the homology analysis. AraC protein from otherVibrio
strains includingVibrio
vulnificus
,Vibrio
splendidus
,Vibrio
tasmaniensis
andVibrio
lentus
were compared to AraC protein sequence ofV
.alginolyticus
and the result is shown in Fig.3.
Fig.3 Homology comparison of amino acid sequence of AraC protein from Vibrio alginolyticus with Vibrio AraC protein sequence
A phylogenetic tree was built with AraC protein sequence ofV
.alginolyticus
and similar protein sequences of otherVibrio
strains using MEGA 7.0 system software. Based on the results,V
.alginolyticus
,V
.parahaemolyticus
andV
.palustris
were clustered together, indicating that the genetic relationship between the three species was closest (Fig.4). This implies that these species have common evolutionary ancestor.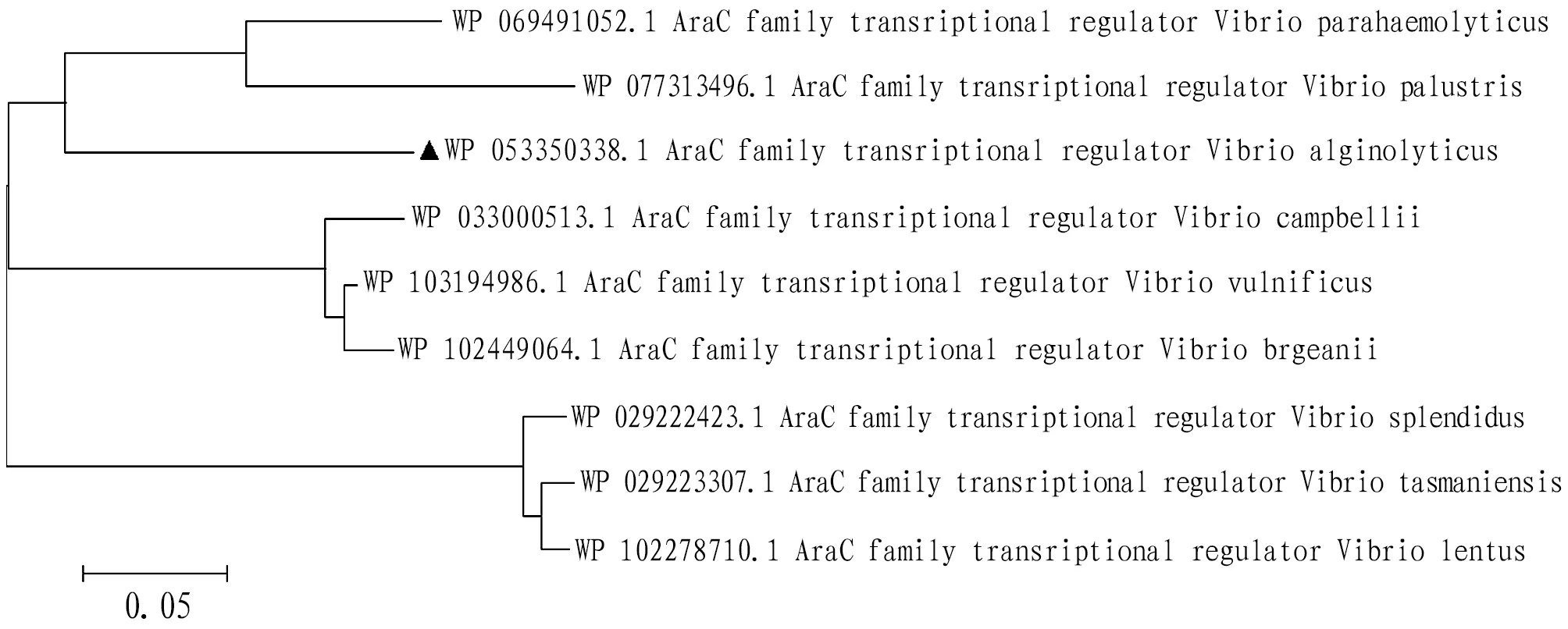
Fig.4 Phylogenetic tree of Vibrio alginolyticus AraC protein constructed by Neighbour-joining method
3.4 Structure domain analysis of AraC protein
The amino acid sequence of AraC protein was committed to NCBI online and performed protein Blast analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the conserved domain was obtained as shown in Fig.5. The result showed that AraC protein belonged to HTH_18 superfamily.
Fig.5 Analysis of functional domain of AraC protein
3.5 Prediction of secondary structure of AraC protein
The SOPMA secondary structure prediction method was carried out on the AraC protein (https://npsa-prabi.ibcp.fr/). The protein’s secondary structure consisted of 52.12% alpha helix (Hh), 11.02% extended strand (Ee), 5.08% beta turn (Tt) and 31.78% random coil (Cc) as shown in Fig.6.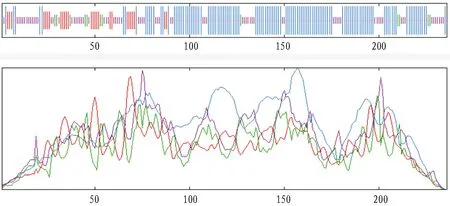
Fig.6 Secondary structure of AraC protein in Vibrio alginolyticus
3.6 Prediction of tertiary structure
The homologous modeling ofV
.alginolyticus
AraC protein was performed using SWISS-MODEL online software (http://swissmodel.expasy.org/) to predict the tertiary structure of AraC gene. The 3-D structure model of AraC protein subunit was obtained (Fig.7). The result indicated that AraC protein is a transcriptional regulatory protein.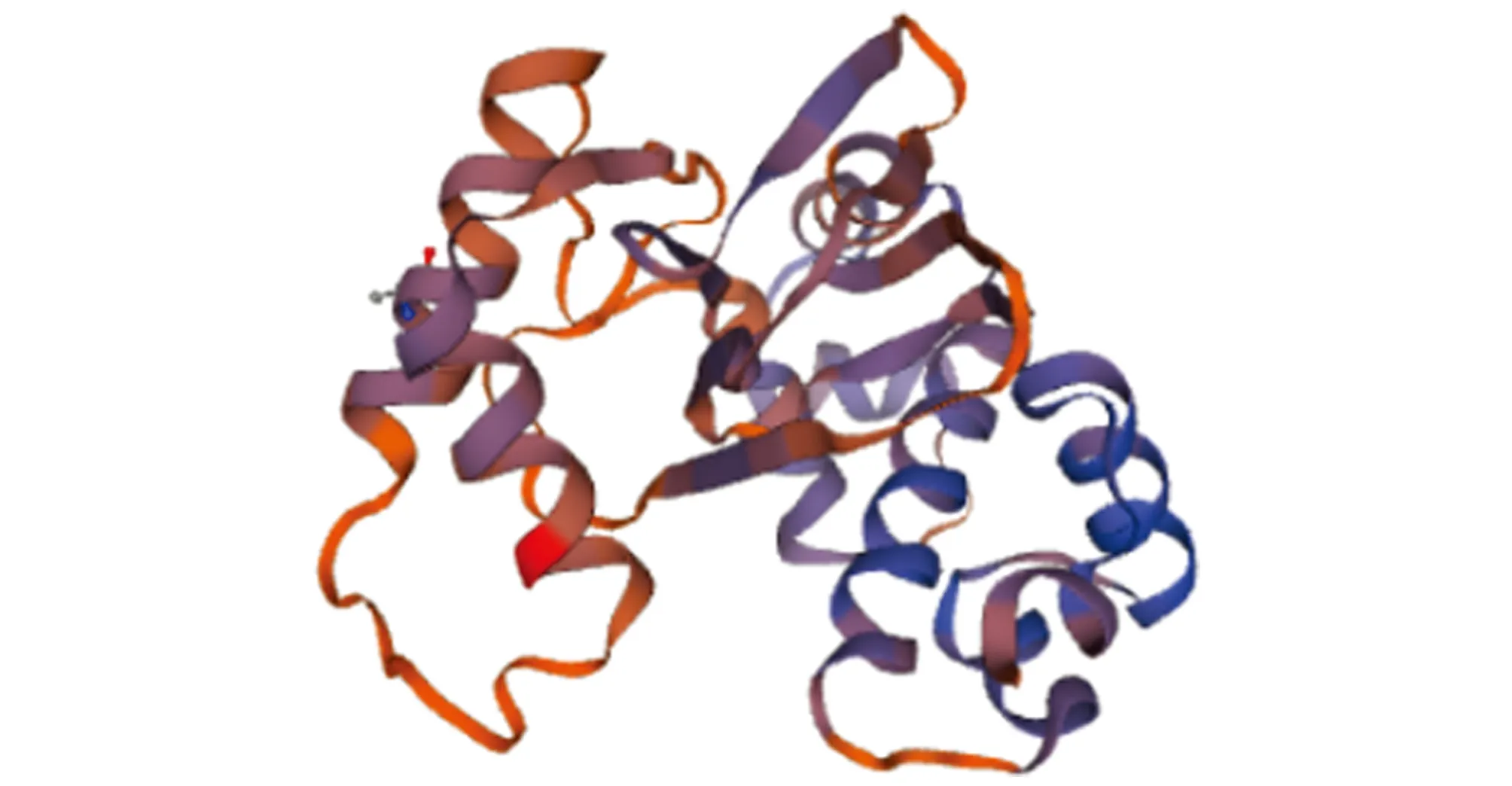
Fig.7 Three-dimensional structure of AraC protein in Vibrio alginolyticus
4 Discussion
AraC protein had small amino acids content when compared with other secreted proteins with a theoretical molecular mass of 26.92 kDa and has aliphatic index of 92.92. Also, its instability index was 58.18 and the GRAVY value of the protein was -0.152.This implies that the AraC protein can be activatedin
vivo
. The protein consists of multiple binding sites such as two cAMP- and cGMP-dependent protein kinase phosphorylation sites, five protein kinase C phosphorylation sites, three casein kinase II phosphorylation sites, one prenyl group binding site and five microbodies C-terminal targeting signal. These sites facilitate the protein to enter into the host cells and anchor in intracellular structures.The NCBI Blast analysis of AraC protein show that the protein contains aHTH_18 superfamily conserved domain. In proteins, the helix-turn-helix (HTH) is a major structural motif capable of binding DNA. The recognition and binding to DNA by helix-turn-helix proteins is done by the two α helices, one occupying the N-terminal end of the motif, the other at the C-terminus. AraC protein belongs to HTH-type transcriptional regulatory superfamily and the domain is correlated with transcriptional regulator. It has been demonstrated that the AraC protein causes an auto inducer-dependent activation of transcription of L-arabinose operon. The result shows that AraC protein ofV
.alginolyticus
has no signal peptide, meaning, after the synthesis of AraC, the protein was not transported hence it was not a secretory protein or some cytoplasmic matrix synthesis proteins. The 3-D structure model of AraC protein subunit was obtained using SWISS-MODEL software. The AraC regulatory protein is sensitive to the level of arabinose and plays a dual role as both an activator in the presence of arabinose and a repressor in the absence of arabinose to regulate the expression of AraBAD which encodes for three metabolic enzymes that are required for the metabolism of L-arabinose in theEscherichia
coli
. Additionally, AraC protein auto-regulates its own expression at high AraC levels.5 Conclusions
In summary,V
.alginolyticus
was isolated from a diseasedE
.coioides
and obtainedAraC
protein from this bacterium. A full-lengthAraC
gene of the bacteria was cloned and was subjected to bioinformatics analysis. The length ofAraC
gene sequence was 711bp and coded 236 amino acids. This protein was unstable hydrophilic with an instability index (II) of 58.18 and a theoretical pI value of 8.94. Structural analysis showed that the protein belonged to the HTH_18 superfamily. The 3D structure and conserved domains were predicted and analyzed. Also, AraC function as an activator, repressor and can control gene expression directly and indirectly. This study lays a foundation for further research and better understanding of the mechanism of transcriptional regulation of AraC protein.About
KIT
The Royal Tropical Institute (KIT) in Amsterdam is an independent centre of knowledge and expertise in the areas of international and intercultural cooperation, operating at the interface between theory and practice and between policy and implementation. The Institute contributes to sustainable development, poverty alleviation and cultural preservation and exchange.
杂志排行
Asian Agricultural Research的其它文章
- Ecological Investigation and Ecological Security Assessment of Xizhijiang River Basin in Huizhou City of Guangdong Province
- Occurrence Regularity and Integrated Control Technology of Canker in Citrus
- Activation Effect of Hydrochemical Energy in Regenerative Agriculture on Nutrients of Arsenic Sandstone
- IS-LM Model Based Analysis on the Role of China’s Financial Policy for Supporting Agriculture
- Prediction and Protection of Cultivated Land in China
- Emergy Estimation of the Main Forest Biomass in Shangri-La, Tibetan Area of Yunnan Province
