小麦Glu-3位点基因拷贝数的变异分析
2021-03-25陈璨韩南南刘洋史晓维司红起马传喜
陈璨,韩南南,刘洋,史晓维,司红起,马传喜
小麦位点基因拷贝数的变异分析
陈璨,韩南南,刘洋,史晓维,司红起,马传喜
安徽农业大学农学院/农业农村部黄淮南部小麦生物学与遗传育种重点实验室,合肥 230036
【】基因拷贝数变异是一种常见又重要的基因结构变异,往往影响个体表型。低分子量麦谷蛋白(low-molecular-weight glutenin subunit,LMW-GS)是小麦贮藏蛋白的主要组成部分,位于位点。小麦作为异源六倍体,其庞大且复杂的基因组结构导致难以利用传统方法检测目的基因的拷贝数,针对小麦基因组,筛选可靠稳定的内参基因和体系,探索适合复杂基因组的拷贝数变异测定技术,测定位点LWM-GS基因拷贝数。以为内参基因,根据基因序列设计内参引物和探针,通过定性和定量PCR测定内参基因在12个普通小麦品种中的拷贝数,分析该基因拷贝数在不同品种间的稳定性;又以小麦品种篙优2018的5个稀释浓度的基因组DNA为模板,利用qRT-PCR验证内参系统的重复性和准确性;根据位点LMW-GS基因序列设计特异性引物及探针,利用qRT-PCR和ddPCR 2种方法检测8个小麦品种位点基因拷贝数,比较后选择更优的高通量基因拷贝数检测方法;再根据和位点LMW-GS基因序列设计相应的特异性引物及探针,并利用ddPCR技术检测和分析了231份小麦品种的、和位点上LMW-GS基因拷贝数。在12个普通小麦品种间、同一品种5个DNA稀释浓度间的拷贝数测定结果一致,技术重复间的变异系数仅为0.07%—0.77%,所构建的内参系统稳定;比较qRT-PCR和ddPCR 2种拷贝数检测方法,8个品种所测的位点拷贝数结果一致,分别为3、5、3、4、3、3、3和3;且ddPCR检测重复间的变异系数为0.30%—1.67%,远低于qRT-PCR的3.14%—12.72%,更加可靠;利用ddPCR对231份普通小麦品种的、和位点上LMW-GS基因拷贝检测后分析发现,大多数小麦品种在3个位点上的拷贝数为4,所占频率分别为51.95%、32.03%和28.57%,位点总拷贝数变异范围为10—21,变异系数为16.12%。内参系统具有良好的稳定性和重复性,可以用作小麦位点和其他目的基因拷贝数检测的内参;qRT-PCR和ddPCR均可用于小麦基因拷贝数的检测,但后者更稳定、可靠,且操作简单、检测通量高。
小麦;低分子量麦谷蛋白;微滴式数字PCR;拷贝数变异
0 引言
【研究意义】小麦低分子量麦谷蛋白(low molecular weight glutenin subunit,LMW-GS)约占贮藏蛋白的1/3[1],其组成和结构对小麦面粉品质具有重要影响,尤其是面团面筋强度,并决定面团的粘弹性[2-3]。研究表明,LMW-GS的编码基因大部分定位于小麦第一同源群染色体1A、1B和1D短臂近端粒处,依次被命名为、和位点,每个位点都包含着多个LMW-GS基因,构成了非常复杂的LMW-GS基因家族[4-6]。LMW-GS基因的组成、拷贝数及其表达情况都影响着小麦品质性状[7-9]。但由于LMW-GS基因组成复杂,目前缺少有效的分离方法,很大程度上限制了对其拷贝数的研究。因此,探索和构建高通量检测拷贝数变异的方法尤为重要。【前人研究进展】基因的拷贝数变异(copy number variation,CNV)是一种常见而又重要的基因结构变异,一般认为是由染色体重排等染色体结构变异引起的,会导致基因组遗传不稳定,对个体性状会产生一定的影响[10-11]。对于拷贝数的检测,常用的技术主要包括实时荧光定量PCR(quantitative real-time PCR,qRT-PCR)、荧光原位杂交(fluorescence in situ hybridization,FISH)、Sothern印迹杂交(southern blot)、短片段多重定量PCR(quantitative multiplex PCR of short fragments,QMPSF)以及数字PCR(digital PCR,dPCR)技术。20世纪末,Vogelstein等[12]提出了dPCR的概念,即将有限稀释法、泊松分布和PCR技术联合使用的新方法[13-14]。一种新的dPCR分析系统——微滴式数字PCR(droplet digital PCR,ddPCR),用于检测目的基因的拷贝数,可以使用荧光探针对2个靶基因同时进行检测,先使用微滴生产仪将一个样本分成几万份(>10 000),分配到不同的微滴单元,每个单元包含一个或多个拷贝数的目标分子(DNA模板),在每个反应单元中分别对目标分子进行PCR扩增,扩增结束后利用微滴分析仪对各个反应单元的荧光信号进行统计学分析,最后根据泊松分布原理及阳性微滴的个数与比例得出靶分子的起始拷贝数或浓度[15-16]。在利用ddPCR进行基因拷贝数检测时需要一个已知的单拷贝或低拷贝基因作为内参基因,乙酰辅酶A羧化酶(acetyl-CoA carboxylase,ACCase)是植物脂肪酸生物合成中的一种限速酶[17-19]。在禾本科作物基因组中,同时含有编码质体ACCase()和胞质ACCase()的基因[20-21],其中,被定位于2A、2B和2D染色体靠近端粒的短臂上,且Southern印迹杂交试验表明普通小麦中在每个染色体组都是单拷贝[22]。因此,可以作为检测小麦中其他基因拷贝数的内参基因。【本研究切入点】由于六倍体小麦为异源多倍体作物,其基因组庞大且复杂,利用传统的Sothern印迹杂交等技术检测小麦基因拷贝数费时费力,并且无法达到高通量测定要求。【拟解决的关键问题】本研究拟选择适当内参基因,通过ddPCR技术构建一种高通量、快速且稳定的小麦基因拷贝数测定方法;并通过构建的内参体系和方法测定普通小麦位点LMW-GS基因拷贝数。
1 材料与方法
1.1 材料
231份不同品种六倍体小麦秋播于合肥高新技术农业园试验站,每个材料种植2行,行长2 m,行距25 cm,田间管理同大田管理。
1.2 内参的选择
构建可靠的CNV检测方法需要选择特异性的DNA序列作为内参基因,并且需要满足3个要求:具有物种特异性,具有单个或稳定的低拷贝数,以及不同品种之间拷贝数相同[23-24]。通过查阅文献,选择作为内参基因,从NCBI网站上下载已知序列,在保守区域设计引物和探针,构建内参系统。
1.3 定性PCR条件
采用全式金生物公司的TransTaq®Hifi酶在T100TMThermal Cycler (BIO-RAD Laboratories,USA)热循环仪进行定性PCR分析,反应体系为2.0 μl样本DNA、0.5 μl上游引物(10 μmol·L-1)、0.5 μl下游引物(10 μmol·L-1)、2.0 μl dNTPs(2.5 mmol·L-1)、2.5 μl 10×Buffer、0.25 μl DNA酶,补加ddH2O至25 μl。PCR扩增程序为95℃ 5 min;95℃ 50 s,50—65℃ 30 s,72℃ 30s,40个循环;72℃ 10min;4℃保存。扩增产物用1.2%琼脂糖凝胶电泳进行检测。
1.4 定量PCR条件
qRT-PCR采用愚公生命科技有限公司的Taq-HS Probe qPCR Premix,在CF×96TMReal-Time System(BIO-RAD Laboratories,USA)中进行,反应体系为2.0 μl样本DNA、1.0 μl上游引物(10 μmol·L-1)、1.0 μl下游引物(10 μmol·L-1)、1.0 μl探针(10 μmol·L-1)和1.25 μl Taq-HS Probe qPCR Premix(10×),补加ddH2O至25 μl。qPCR扩增程序为95℃ 3 min;95℃ 30 s,50—65℃ 30 s,72℃ 30 s,40个循环;在每个循环的延伸阶段采集荧光信号。参考Weng等[25]报道的绝对定量法来计算不同品种中目的基因拷贝数。
1.5 基因组DNA的酶切处理
采用NEB公司的HⅠ限制性内切酶对基因组DNA进行酶切处理,50 μl的反应体系包含1 μg基因组DNA、1 μlHⅠ限制性内切酶和5 μl 10×NE Buffer,补加ddH2O至50 μl。在37℃恒温条件下反应5 h,于4℃保存备用。
1.6 微滴式数字PCR分析基因拷贝数
先配制20×目的基因(或内参基因)引物/探针混合液,包含目的基因(或内参基因)上、下游引物(100 μmol·L-1)各18 μL和目的基因(或内参基因)探针(100 μmol·L-1)5 μL,补加ddH2O至100 μL。再采用BIO-RAD公司的ddPCR Supermix for Probes(No dUTP)制备ddPCR反应液,反应体系为10 μL 2×ddPCR Supermix for Probes(No dUTP)、1 μL 20×目的基因引物/探针混合液、1 μL 20×内参基因引物/探针混合液和30 ng酶切后基因组DNA,补加ddH2O至20 μL。
随后将反应液通过QX200TM微滴发生器(QX200TMDroplet Generator, BIO-RAD Laboratories,USA)生成纳升级别的油包水的液滴(理论上可形成20 000个微滴),紧接着转移到96孔PCR板(Eppendorf, Germany)中,并利用热封仪(PX1TMPCR Plate Sealer, BIO-RAD Laboratories,USA)进行封膜。然后转移到T100TMThermal Cycler(BIO-RAD Laboratories,USA)上进行PCR扩增,扩增程序为95℃ 10 min;94℃ 30 s;50—65℃ 1 min,40个循环;98℃ 10 min;4℃保存待用,升降温速度为2.0 ℃·s-1。
PCR扩增完成后,将其转移到QX200TM微滴分析仪(QX200TMDroplet Reader, BIO-RAD Laboratories,USA)对所有样品孔进行荧光检测。最后利用QuantaSoft软件(V1.3.10)对ddPCR数据进行分析,计算每个样品目的基因拷贝数。
2 结果
2.1 内参基因的选择及其引物与探针的设计
通过查阅文献搜索到低拷贝数的候选基因质体乙酰辅酶A羧化酶基因(),在普通小麦中具有3个拷贝。从NCBI网站上下载已公布的序列,运用DNAMAN生物软件对下载的序列进行序列对比,运用Primer express3.0软件在的保守区域设计引物(Acc1F和Acc1R)和探针(T-Acc1)(表1)。
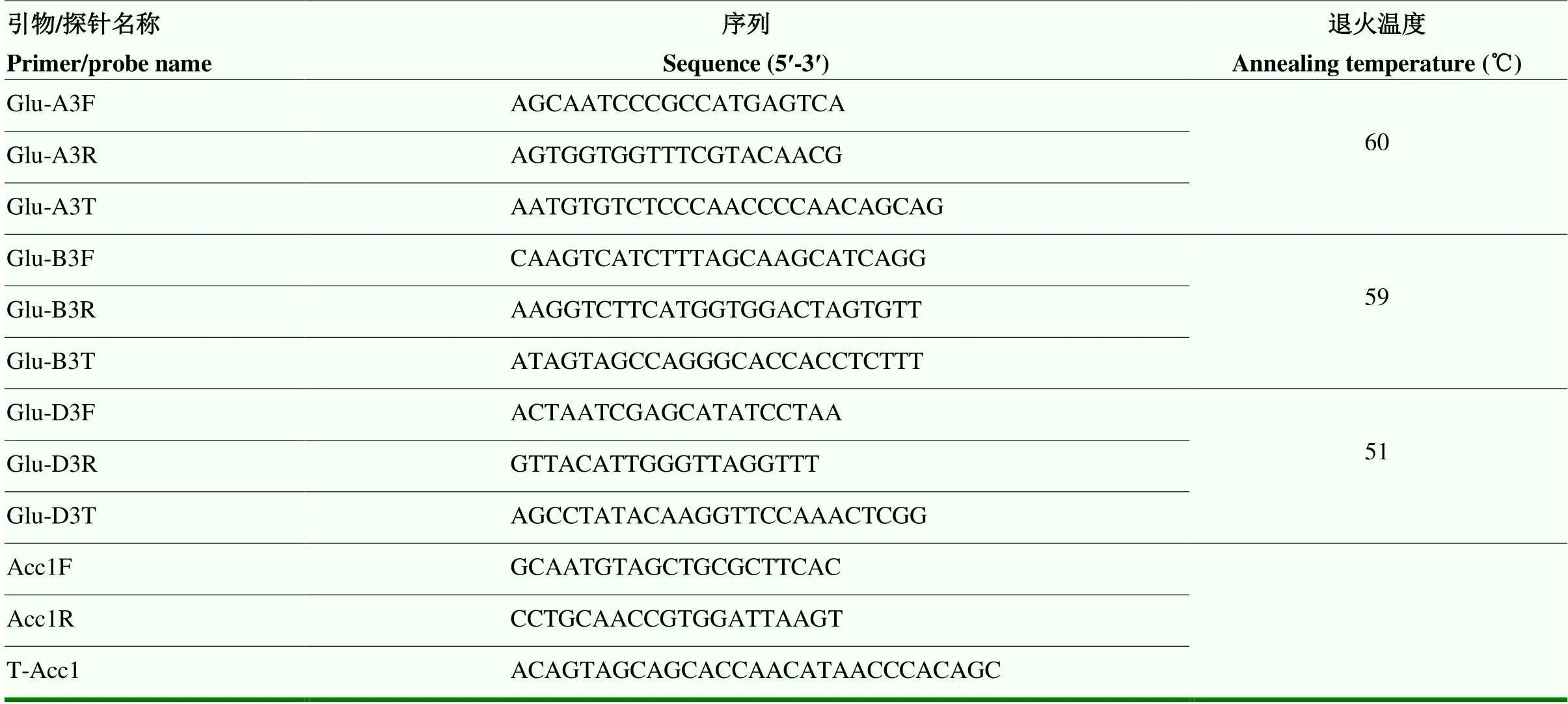
表1 特异性引物和探针
2.2 定性和定量PCR检测Acc1在不同六倍体小麦中的稳定性
合适的内参基因应在同一物种的不同品种之间具有相同的拷贝数,为了检测不同普通小麦品种之间拷贝数的种内稳定性,选取12个品种的普通小麦提取基因组DNA,每个品种各取30 ng DNA,以Acc1F、Acc1R为引物进行定性PCR扩增,结果表明,不同品种之间获得相同大小和等效强度的PCR产物,并且没有显示出非特异性的条带(图1),表明所设计的引物能够稳定地扩增出序列,特异性较高,并且普通小麦品种间拷贝数没有明显差异。

1:黔11240-2;2:郑麦583;3:中麦14;4:德宏福麦2号;5:瑞泉麦168;6:中合-75;7:益科麦5号;8:内麦836;9:阳光838;10:安农1020;11:新麦31;12:黔090304-4
选取12个不同品种小麦50 ng基因组DNA,以Acc1F、Acc1R为引物,T-Acc1为探针进行实时荧光定量PCR,定量PCR的结果显示不同品种的小麦扩增曲线极为相似,并且获得的Ct值只有轻微的变异性(图2),范围为29.18—30.00,变异系数为1.03%,拷贝数在供试品种之间被认为是相同的。
2.3 定量PCR检测Acc1定量系统的重复性
为了进一步验证定量系统的准确性,选用篙优2018的5个稀释度的基因组DNA为模板,以Acc1F、Acc1R为引物,T-Acc1为探针进行定量PCR(表2),结果表明,复孔间变异系数为0.07%—0.77%,SD值为0.0208—0.2100,由于这些值相对较小,因此,定量系统被认为是稳定且可靠的,可作为内参基因来测定其他基因的拷贝数。
2.4 Glu-3位点LMW-GS基因的引物与探针设计及其特异性分析
从NCBI网站上下载已知的分别位于、和位点上的LMW-GS基因,通过对LMW-GS基因序列进行比对,并设计相应的特异性引物与探针(表1),其中,Glu-A3T、Glu-B3T和Glu-D3T探针的5′端、3′端所连的荧光集团分别为5′-FAM和3′-BHQ1;T-Acc1探针的5′端、3′端所连的荧光集团分别为5′-HEX和3′-BHQ1。
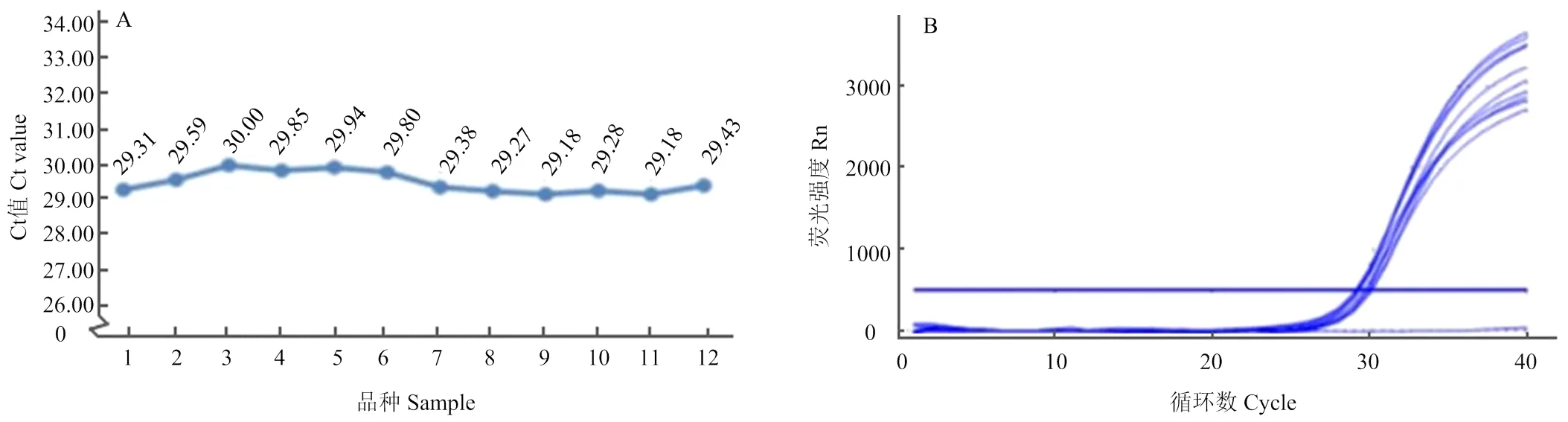
A:12个不同品种的内参基因定量PCR扩增曲线;1:黔11240-2;2:郑麦583;3:中麦14;4:德宏福麦2号;5:瑞泉麦168;6:中合-75;7:益科麦5号;8:内麦836;9:阳光838;10:安农1020;11:新麦31;12:黔090304-4。B:12个不同普通小麦品种的Ct值
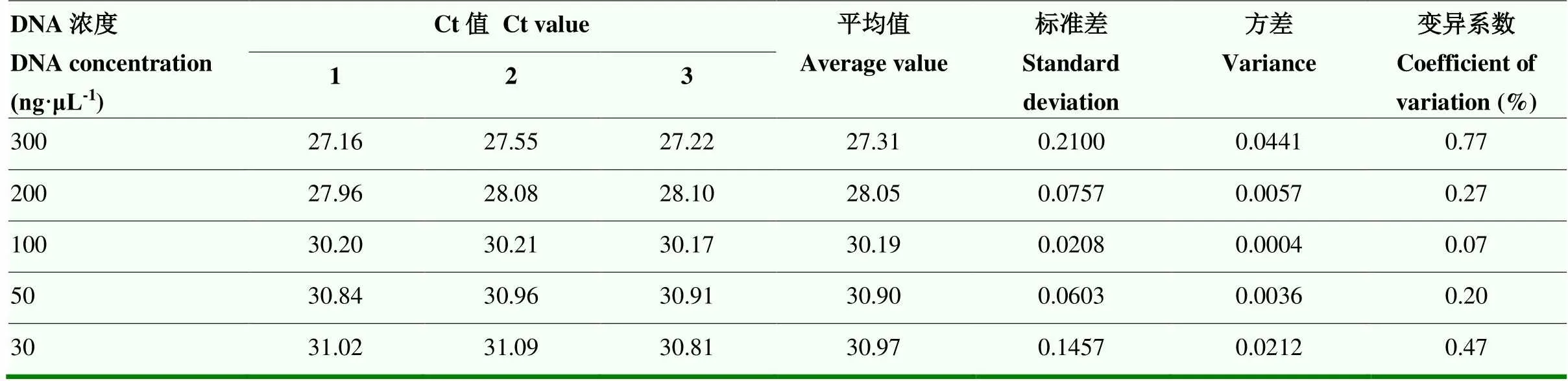
表2 内参基因重复性检测Ct值变化情况
从普通小麦近缘物种(乌拉尔图小麦AA、一粒小麦AA、拟斯卑尔脱山羊草BB、节节麦DD和圆锥小麦AABB)及普通小麦AABBDD中国春、安农0711中提取基因组DNA,分别用引物Glu-A3F/R、Glu-B3F/R、Glu-D3F/R进行PCR扩增,结果显示,Glu-A3F/R只能从含A基因组的材料中扩增出目的基因,Glu-B3F/R只能从含有B基因组的材料中扩增出目的基因,Glu-D3F/R只能从含D基因组的材料中扩增出目的基因(图3)。
2.5 2种拷贝数变异分析方法的比较
qRT-PCR与ddPCR均能对目的基因拷贝数进行测定,选用8个普通小麦品种,分别采用qRT-PCR和ddPCR 2种方法来测定普通小麦位点上的LMW-GS基因拷贝数。
以烟农19全基因组DNA(1 066.71 ng·μL-1)为模板,稀释成6个梯度,进行qRT-PCR反应;以水作为空白对照,得出Ct值与DNA初始浓度的对数值之间的标准曲线(图4),并得到内参基因的标准曲线方程(=-2.780+33.098,²=0.998,E=106.1%)和目的基因的标准曲线方程(=-2.823+33.993,²=0.998,E=108.9%),这两个基因扩增曲线的相关系数(²)均接近1,且扩增效率(E)也均接近于1,表明所构建的标准曲线可以用于拷贝数的测定。
选取8个小麦品种的基因组DNA(浓度均为200 ng·μL-1),进行qRT-PCR反应,每个样品3次重复,并且与绘制标准曲线的样品同时扩增,扩增结束后得到8个样品的Ct值(表3),结果显示,8个小麦品种中拷贝数最高为5,最低为3,有6个品种的拷贝数为3,并且利用qRT-PCR测定时重复间的变异系数为3.14%—12.72%。
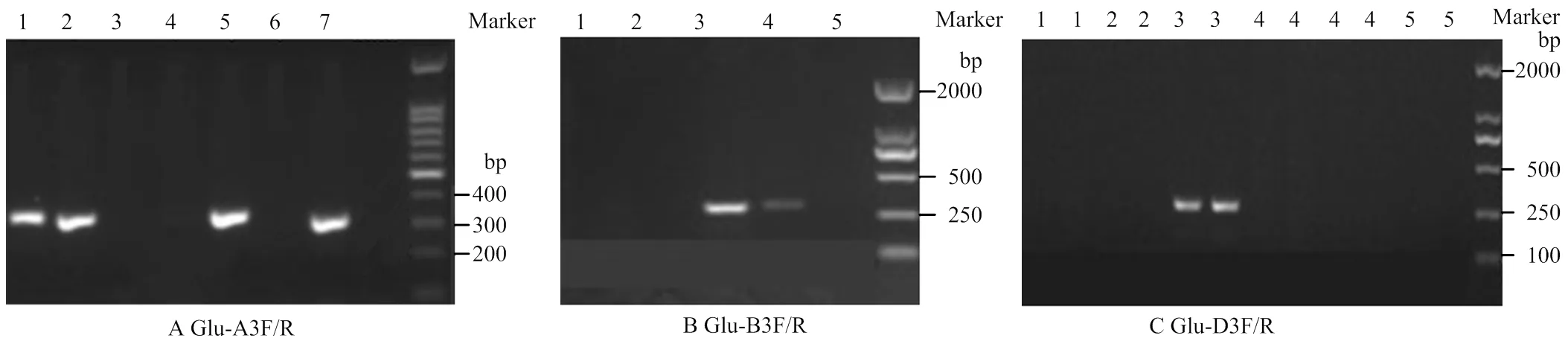
A:Glu-A3F/R引物检测。1:中国春;2:一粒小麦;3:拟斯卑尔托山羊草;4:节节麦;5:乌拉尔图小麦;6:水;7:安农0711。B:Glu-B3F/R引物检测。1:一粒小麦;2:节节麦;3:圆锥小麦;4:中国春;5:水。C:Glu-D3F/R引物检测。1:乌拉尔图小麦;2:拟斯卑尔托山羊草;3:节节麦;4:圆锥小麦;5:水

表3 8个不同样品3次重复的Ct值及目的基因拷贝数变化情况
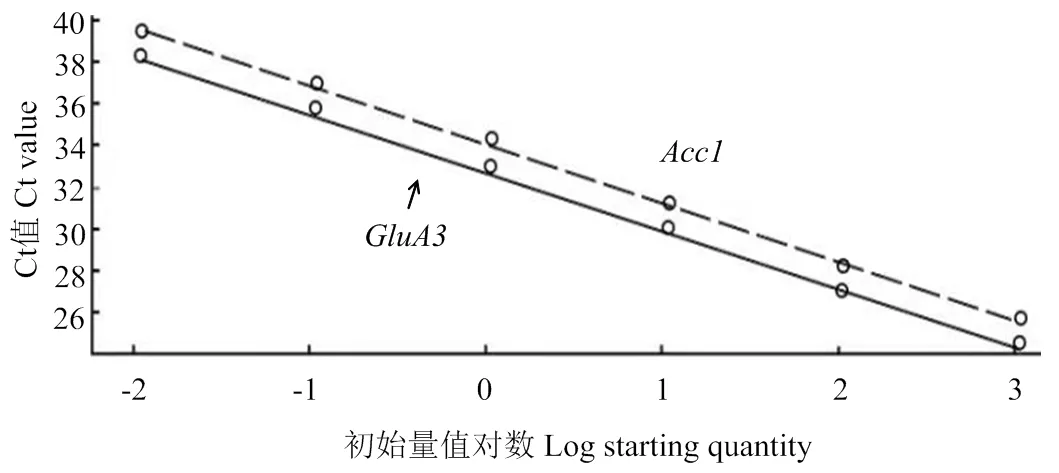
图4 目的基因和内参基因标准曲线
采用ddPCR来测定上述8个不同小麦品种的拷贝数。限制性内切酶HⅠ酶切基因组DNA(浓度为200 ng·μL-1)后,再按照ddPCR操作步骤进行试验,最后得出了8个品种的拷贝数测定结果(图5),其中,安农92484W拷贝数最高为5,小偃6号拷贝数为4,其余6个品种的拷贝数为3,且重复间的变异系数仅在0.30%—1.67%。
综上所述,利用qRT-PCR和ddPCR 2种方法对8个样品目的基因拷贝数的检测结果是一致的,但是ddPCR重复间的变异系数要远低于qPCR,表明用ddPCR方法来测定基因拷贝数更稳定、准确。
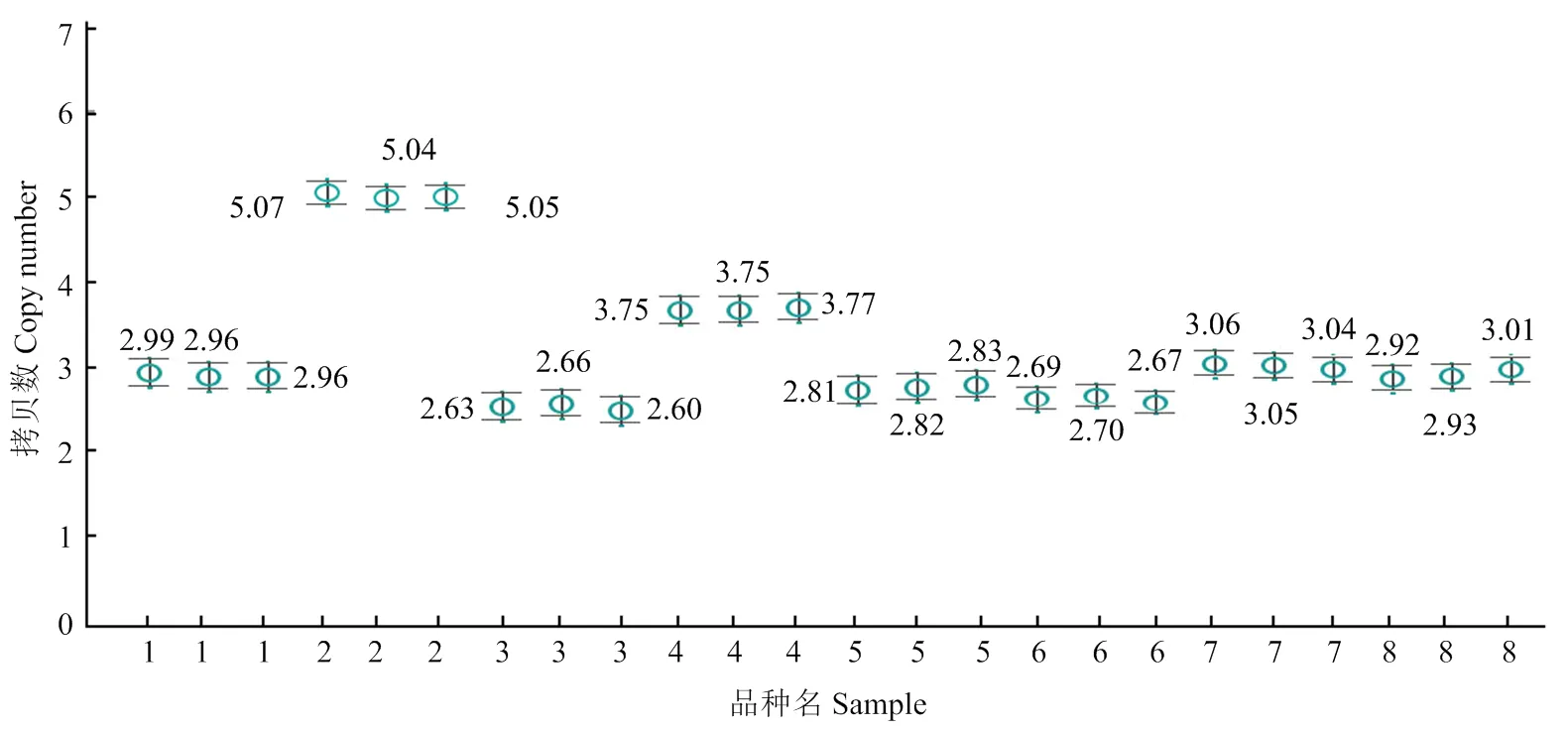
1:烟农15;2:安农92848W;3:百农64;4:小偃6号;5:周麦16;6:02P67;7:邯郸6172;8:周麦18
2.6 微滴式数字PCR测定普通小麦Glu-3位点LMW-GS拷贝数
利用微滴式数字PCR对231份不同品种的普通小麦的、和位点LMW-GS基因分别进行拷贝数检测(电子附表1),对检测结果进行了统计分析(图6),结果显示,在231份材料中,大多数小麦品种的位点LMW-GS基因拷贝数为4,所占频率51.95%,最高拷贝数为8,最低为3,不同品种间拷贝数变异系数为28.40%;大多数小麦品种的位点LMW-GS基因拷贝数为4,所占频率为32.03%,拷贝数最高为9个拷贝,最低为3个拷贝,不同品种间拷贝数变异系数为29.30%;大多数小麦品种的位点LMW-GS基因拷贝数为4,所占频率为28.57%,拷贝数最高为9个拷贝,最低为3个拷贝,不同品种间拷贝数变异系数为30.07%。
综合、和3个位点的LMW-GS基因拷贝数测定结果,可以计算出小麦位点基因的总拷贝数(图7)。结果显示,在231份材料中,大多数小麦品种的位点LMW-GS基因拷贝数为12—18,所占频率为85.90%,拷贝数最高为21,最低为10,不同品种间拷贝数变异系数为16.10%。
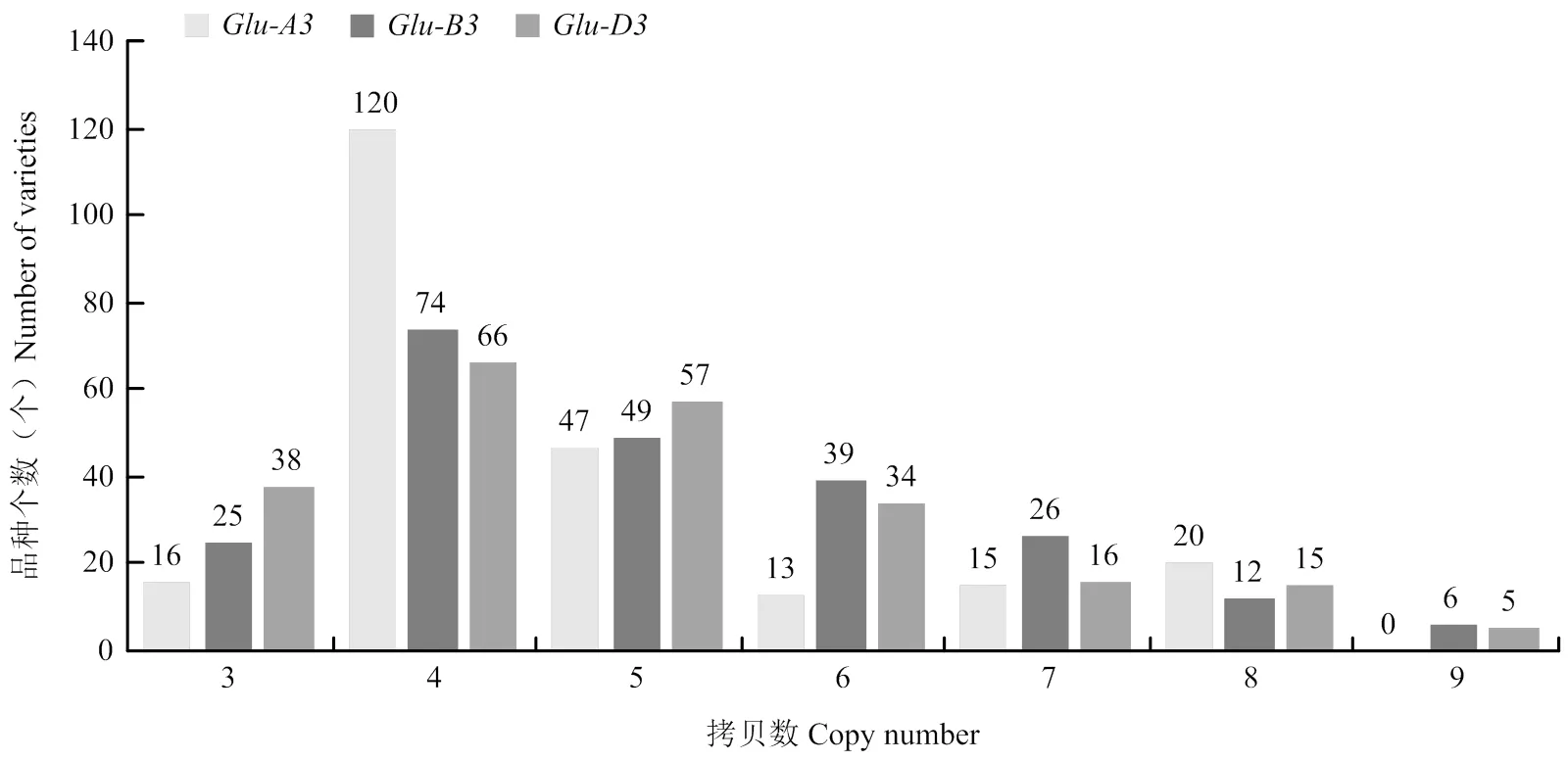
图6 231份小麦品种Glu-A3、Glu-B3和Glu-D3位点基因拷贝数频数分布条形图

图7 231份小麦品种Glu-3位点基因拷贝数频数分布条形图
3 讨论
微滴式数字PCR(ddPCR)技术是近几年迅速发展起来的第三代PCR技术,已被越来越多地应用于生命科学研究中,比如致病微生物检测[26]、稀有突变检测[27]、基因表达分析[28]、基因拷贝数变异分析[29]等。在利用ddPCR来测定目的基因拷贝数时需要一个已知拷贝数的单拷贝或低拷贝基因作为内参基因。一些研究以单拷贝的、和作为内参基因来检测转基因小麦外源基因拷贝数[30]。而本研究所选择的内参基因为质体乙酰辅酶A羧化酶(plastid ACCase)基因()。Gornicki等[31]利用中国春缺四体材料将定位在了第二同源群2A、2B和2D染色体靠近端粒的短臂上,并利用Southen印迹杂交分析得出六倍体小麦中在每个染色体组都为单拷贝,因此,在六倍体小麦中有3个拷贝。作为内参基因,还要求其在不同的品种中拷贝数是稳定的,并且内参基因的扩增还要具有重复性。本研究借鉴Mayu等[32]的研究方法,用不同品种相同浓度的DNA验证了内参基因拷贝数的稳定性,并用相同品种不同浓度的DNA验证了内参基因扩增的可重复性。因此,本研究设计的内参系统(Acc1F/R和T-Acc1)可作为检测六倍体小麦中目的基因拷贝数的内参引物和探针。
目前,研究植物基因拷贝数的方法有很多,但利用ddPCR技术来检测小麦目的基因拷贝数的报道很少。经典的植物基因拷贝数检测方法主要有Southern印迹杂交和qRT-PCR 2种[33-34]。其中传统的Sounthern印迹杂交技术对DNA样品的质量和纯度有一定的要求,其试验周期长,杂交步骤繁琐,并且对试验技术条件要求较高;可能因存在等位基因,而导致检测结果不能真实反映该基因的拷贝数;另外,试验中所用的探针标记若采用同位素标记,对人体健康和环境安全存在严重威胁[35]。qRT-PCR检测方法的步骤较为繁琐,其需要准备标准DNA样品,再利用标准DNA样品绘制标准曲线,标准曲线的建立又需对反应条件和反应体系进行不断摸索与优化,所需试验周期较长[36];借助标准曲线进行定量本身就是一种相对定量的方法,检测结果可能不准确[37]。而ddPCR作为一种新兴的、准确的绝对定量技术,与qRT-PCR相比,ddPCR的优势非常明显,主要体现在以下几个方面[38-39]:(1)灵敏度可达单个核酸分子,检测限低至0.001%;(2)无需绘制标准曲线即可对DNA样品进行绝对定量;(3)终点PCR检测,不依赖Ct值,不依赖扩增曲线,特别适合基质复杂样品的检测;(4)具有更好的准确度、精密度和重复性,可用于精确测定靶基因的拷贝数。本文以为内参基因,通过qRT-PCR和ddPCR 2种技术对8个样品的位点LMW-GS基因进行拷贝数测定,结果基本一致,但ddPCR重复间的变异系数更小,且不依赖于标准曲线。在本研究中以为内参基因,使用特异性引物和探针系统,采用ddPCR技术分别对231份普通小麦、和位点LMW-GS基因拷贝数进行测定,3个位点的基因拷贝数分别在3—9、4—9、4—9范围内,位点总拷贝数为10—21,这与前人通过Southen印迹杂交技术研究发现普通小麦的LMW-GS基因在总基因组中的拷贝数可能在10—15个和35—40个范围内基本相符[40-41]。
当拷贝数可变区域的剂量敏感基因或涉及调控作用的基因发生拷贝数变异时,也会对表型产生显著的影响[42-43]。关于基因拷贝数变异对表型的影响在一些植物性状上已有不少报道。小麦矮秆基因()由于拷贝数的增加而导致植株高度降低,其致矮能力是单拷贝的3倍多[44]。在大麦中,硼转运基因拷贝数的增加会使大麦Sahara具备硼毒性耐受性[45]。(C-重复结合因子)位于小麦和大麦的抗霜性位点2(),其拷贝数变异与低温耐受性相关,冬小麦的拷贝数要高于春小麦,四倍体硬粒小麦和六倍体面包小麦的位点上大型基因的缺失(包括、和)会导致小麦耐寒性降低[46-49]。在小麦中随着拷贝数的增加,会延长小麦春化时间导致开花推迟,而大麦中高拷贝数能够加速开花时间[50-51]。以上研究表明,某些基因的拷贝数变异对相应的表型有着重要的影响。
小麦低分子量麦谷蛋白(LMW-GS)赋予面筋强度和延展性,在小麦的营养品质和面粉加工品质形成中具有重要影响[2-3],已有大量研究表明LMW-GS基因的组成和表达量与小麦加工品质密切相关,不同种质的小麦含有的LMW-GS等位基因不同,其编码的LMW-GS亚基组合不同,因此对小麦面粉品质产生的影响也不同[7-9]。而关于位点LMW-GS基因拷贝数变异对小麦面粉品质的影响还未有报道。本研究将测定的231份普通小麦的、和位点LMW-GS基因拷贝数与小麦4个品质性状进行相关性分析,除位点基因拷贝数与2012年小麦面团稳定时间和形成时间达到显著负相关性外,其余均未达到显著相关性,说明位点的LMW-GS拷贝数多样性不是影响上述小麦品质性状的主要原因。
4 结论
以为内参基因,构建了内参系统(引物和探针),它在品种间和同一品种不同浓度间均具有稳定性和重复性,可用于小麦和其他目的基因拷贝数研究。qRT-PCR和ddPCR均可用于小麦中特定基因或位点的拷贝数检测,但相较于前者,ddPCR方法变异系数更低,且具有操作简单、结果更准确、检测通量高等优点。
[1] 赵献林, 夏先春, 刘丽, 何中虎, 孙其信. 小麦低分子量麦谷蛋白亚基及其编码基因研究进展. 中国农业科学, 2007, 40(3): 440-446.
ZHAO X L, XIA X C, LIU L, HE Z H, SUN Q X. Review on low molecular weight glutenin subunits and their coding genes. Scientia Agricultura Sinica, 2007, 40(3): 440-446. (in Chinese)
[2] XIANG L, HUANG L, GONG F Y, LIU J, WANG Y F, JIN Y R, HE Y, HE J S, JIANG Q T, ZHENG Y L, LIU D C, WU B H.Enriching LMW-GS alleles and strengthening gluten properties of common wheat through wide hybridization with wild emmer. 3 Biotech, 2019, 9(10): 355.
[3] HAZARD B, TRAFFORD K, LOVEGROVE A, GRIFFITHS S, UAUY C, SHEWRY P. Strategies to improve wheat for human health. Nature Food, 2020, 1(8): 475-480.
[4] Gupta R B, Shepherd K W. Two-step one-dimensional SDS-PAGE analysis of LMW subunits of glutelin. Theoretical and applied genetics, 1990, 80(1): 65-74.
[5] Singh N K, Shepherd K W. Linkage mapping of genes controlling endosperm storage proteins in wheat. Theoretical and Applied Genetics,1988, 75(4): 628-641.
[6] D'Ovidio R, Masci S. The low-molecular-weight glutenin subunits of wheat gluten. Journal of Cereal Science,2004, 39(3): 321-339.
[7] CHO K, JO Y M, LIM S H, KIM J Y, HAN O, LEE J Y. Overexpressing wheat low-molecular-weight glutenin subunits in rice (L.cv. Koami) seeds.3 Biotech, 2019, 9(2): 1-8.
[8] RAI A, SINGH A M, GANJEWALA D, KUMAR R R, AHLAWAT A K, SINGH S K, SHARMA P, JAIN N. Rheological evaluations and molecular marker analysis of cultivated bread wheat varieties of India. Journal of Food Science and Technology,2019, 56(4): 1696-1707.
[9] BEOM H R, KIM J S, JANG Y R, LIM S H, KIM C K, LEE C K, LEE J Y. Proteomic analysis of low-molecular-weight glutenin subunits and relationship with their genes in a common wheat variety. 3 Biotech, 2018, 8(1): 56.
[10] Shaw-Smith C, Redon R, Rickman L. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. Journal of Medical Genetics, 2004, 41(4): 241-248.
[11] Maron L G, Guimaraes C T, Kirst M, ALBERT P S, BIRCHLER J A, BRADBYRY P J, BUCKLER E S, COLUCCIO A E, DANILOVA T V, KUDRNA D, MAGALHAES J V, PINEROS M A, SCHATZ M C, WING R A, KOCHIAN L. Aluminum tolerance in maize is associated with highergene copy number. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(13): 5241-5246.
[12] Vogelstein B, Kinzler K W. Digital PCR. Proceedings of the National Academy of sciences of the United States of America, 1999, 96(16): 9236-9241.
[13] Sykes P J, Neoh S H, Brisco M J. Quantitation of targets for PCR by use of limiting dilution.Biotechniques, 1992, 13(3): 444-449.
[14] YANG Q, Xi J, CHEN X X, HU S H, CHEN N, QIAO S L, WAN S G, BAO D K. The development of a sensitive droplet digital PCR for quantitative detection of porcine reproductive and respiratory syndrome virus.International Journal of Biological Macromolecules, 2017, 104(Part A): 1223-1228.
[15] LIN Q, FU X Z, LIU L H, LIANG H R, NIU Y J, WEN Y Y, HUANG Z B, LI N Q. Development and application of a sensitive droplet digital PCR (ddPCR) for the detection of infectious spleen and kidney necrosis virus. Aquaculture, 2020, 529: 735697.
[16] ANTKOWIAK M, NOWACKA-WOSZUK J, SZCZERBAL I, SWITONSKI M, SZYDLOWSKI M. AMY2B gene copy-number variation studied by droplet digital PCR (ddPCR) in three canids: Red fox, arctic fox, and chinese raccoon dog. Folia Biologica, 2020, 68(2): 51-55.
[17] KONISHI T, SHINOHARA K, YAMADA K, SASAKI Y. Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant and Cell Physiology, 1996, 37(2): 117-122.
[18] CHEN Q, SONG J, DU W P, XU L Y, JIANG Y, ZHANG J, ZHANG M, YU G R. Phylogenetic analyses of four Chinese endemic wheat landraces based on two single copy genes. Cereal Research Communications, 2018, 46(2): 191-200.
[19] 雷映霞. 鹅观草属及其近缘属物种的分子系统与进化研究[D]. 雅安: 四川农业大学, 2018.
LEI Y X. Phylogenetic and evolution analysis of Roegneria and its related genera () [D]. Yaan: Sichuan Agricultural University, 2018. (in Chinese)
[20] GORNICKI P, PODKOWINSKI J, SCAPPINO L A, DIMAIO J, WARD E, HASELKORN R. Wheat acetyl-CoA carboxylase: cDNA and protein structure. Proceedings of the National Academy of Sciences of the United States of America,1994, 91(15): 6860-6864.
[21] Podkowinski J, Sroga G E, Haselkorn R, GORNICKI P. Structure of a gene encoding a cytosolic acetyl-CoA carboxylase of hexaploid wheat.Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(5): 1870-1874.
[22] Gornicki P, Faris J, King I, PODKOWINSKI J, GILL B, HASELKORN R. Plastid-localized acetyl-CoA carboxylase of bread wheat is encoded by a single gene on each of the three ancestral chromosome sets. Proceedings of the National Academy of Sciences of the United States of America, 1997, 94(25): 14179-14184.
[23] 缪青梅, 汪小福, 陈笑芸, 彭城, 徐晓丽, 魏巍, 徐俊锋. 基于双重微滴数字PCR精准定量转基因水稻G6H1的方法研究. 农业生物技术学报, 2019, 27(1): 159-169.
MIAO Q M, WANG X F, CHEN X Y, PENG C, XU X L, WEI W, XU J F. Studies on accurate quantification of genetically modified rice () G6H1 based on duplex droplet digital PCR. Journal of Agricultural Biotechnology, 2019, 27(1): 159-169. (in Chinese)
[24] 蔡教英, 姚丽锋, 王小玉, 游淑珠, 丁琦. 基于双重微滴式数字PCR对转基因油菜RF1品系的定量方法. 现代食品科技, 2018, 34(6): 282-287.
CAI J Y, YAO L F, WANG X Y, YOU S Z, DING Q.Quantitative analysis of genetically modified rapeseed of RF1 by duplex droplet digital polymerase chain reaction (duplex-ddPCR).Modern Food Science and Technology, 2018, 34(6): 282-287. (in Chinese)
[25] Weng H b, Pan A h, Yang L t, ZHANG C M, LIU Z L, ZHANG D B. Estimating number of transgene copies in transgenic rapeseed by real-time PCR assay with HMG I/Y as an endogenous reference gene.Plant Molecular Biology Reporter, 2004, 22(3): 289-300.
[26] Caviglia G P, Abate M L, Tandoi F, CIANCIO A, AMOROSO A, SALIZZONI M, SARACCO G M, rizzetto m, ROMAGNOLI R, smedile a. Quantitation of HBV cccDNA in anti-HBc-positive liver donors by droplet digital PCR: a new tool to detect occult infection. Journal of Hepatology, 2018, 69(2): 301-307.
[27] Dyavar S R, Ye Z, BYRAREDDY S N, SCARSI K K, WINCHESTER L C, WEINHOLD J A, FLETCHER C V, PODANY A T. Normalization of cell associated antiretroviral drug concentrations with a novel RPP30 droplet digital PCR assay. Scientific Reports, 2018, 8(1): 3626.
[28] Elmahalawy S T, Halvarsson P, Skarin M, HÖGLUND J. Genetic variants in dyf-7 validated by droplet digital PCR are not drivers for ivermectin resistance incontortus. International Journal for Parasitology: Drugs and Drug Resistance, 2018, 8(2): 278-286.
[29] YU R L, XUAN W J, ZHOU L, LUO Y, LIU X Y, XIONG P W, REN X Y. Detection of HER2 amplification in formalin-fixed paraffin- embedded breast carcinoma tissue with digital PCR using two TFF3 sequences as internal reference. Experimental and Molecular Pathology, 2018, 104(3): 235-238.
[30] 琚鹏举, 孙黛珍, 宁蕾, 葛林豪, 许成杰, 史华伟, 梁凯歌, 马亮, 刘陶然, 陈明. 采用优化的数字PCR方法分析转基因小麦外源基因拷贝数. 中国农业科学, 2020, 53(10): 1931-1939.
JU P J, SUN D Z, NING L, GE L H, XU C J, SHI H W, LIANG K G, MA L, LIU T R, CHEN M. Analysis of foreign gene copy number in transgenic wheat by optimized digital PCR. Scientia Agricultura Sinica,2020, 53(10): 1931-1939. (in Chinese)
[31] KAUTBALLY S, LEPROPRE S, LERIGOLEUR A, GINION A, BEAULOYE C. Platelet acetyl-coa carboxylase phosphorylation: a risk stratification marker that reveals platelet-lipid interplay in coronary artery disease patients. Archives of Cardiovascular Diseases Supplements, 2019, 11(2): 185-186.
[32] LIda M, Yamashiro S, Yamakawa H, HAYAKAWA K, KURIBARA H, KODAMA T, FURUI S, AKIYAMA H, MAITANI T, HINO A. Development of taxon-specific sequences of common wheat for the detection of genetically modified wheat. Journal of Agricultural and Food Chemistry, 2005, 53(16): 6294-6300.
[33] SINGH R, DUBEY A K, SANYAL I. Optimisation of adventitious shoot regeneration and agrobacterium-mediated transformation in Canna × generalis (). Horticultural Plant Journal, 2019, 5(1): 39-46.
[34] YIN Y C, HOU J M, TIAN S K, YANG L, ZHANG Z X, LI W D, LIU Y. Overexpressing chalcone synthase () gene enhanced flavonoids accumulation inuralensis hairy roots. Botany Letters, 2020, 167(2): 219-231.
[35] Yang L T, Ding J Y, Zhang C M, JIA j W, weng H B, LiN W X, ZHANG D B. Estimating the copy number of transgenes in transformed rice by real-time quantitative PCR.Plant Cell Reports, 2005, 23(10/11): 759-763.
[36] WHALE A S, HUGGETT J F, COWEN S, SPEIRS V, SHAW J, ELLISON S, FOY C A, SCOTT D J.Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Research, 2012, 40(11): e82.
[37] 姜羽, 胡佳莹, 杨立桃. 利用微滴数字PCR分析转基因生物外源基因拷贝数. 农业生物技术学报, 2014, 22(10): 1298-1305.
JIANG Y, HU J Y, YANG L T. Estimating the exogenous genes copy number of genetically modified organisms by droplet digital PCR.Journal of Agricultural Biotechnology,2014, 22(10): 1298-1305. (in Chinese)
[38] Gao F g, PFEIFER E, FARAH H, KARAMPINI E, DUA D, KAMAI N, CANE P, ToBAL K, SETHI T, SPICER J, MCCAUGHAN F.Microdroplet digital PCR: detection and quantitation of biomarkers in archived tissue and serial plasma samples in patients with lung cancer. Journal of Thoracic Oncology, 2015, 10(1): 212-217.
[39] Hindson C M, Chevillet J R, Briggs H A, GALLICHOTTE E N, RUF I K, HINDSON B J, VESSELLA R L, TEWARI M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nature Methods, 2013, 10(10): 1003-1005.
[40] Harberd N P, Bartels D, Thompson R D. Analysis of the gliadin multigene loci in bread wheat using nullisomic-tetrasomic lines. Molecular and General Genetics, 1985, 198(2): 234-242.
[41] Cassidy B G, Dvorak J, Anderson O D. The wheat low molecular weight glutenin genes: characterization of six new genes and progress in understanding gene family structure. Theoretical and Applied Genetics, 1998, 96(6/7): 743-750.
[42] KORBEL J O, KIM P M, CHEN X Y, URBAN A E, WEISSMAN S, SNYDER M, GERSTEIN M B. The current excitement about copy-number variation: how it relates to gene duplications and protein families. Current Opinion in Structural Biology, 2008, 18(3): 366-374.
[43] CHEN C, WANG W, YUAN J X, CHEN J, MOU L M. Analysis of HMW-GS and LMW-GS in spring wheat varieties and key parental materials cultivated in Gansu dryland. Acta Agriculturae Boreali- occidentalis Sinica, 2018, 27(11): 1598-1605.
[44] Li Y Y, Xiao J H, Wu J J, DUAN J L, LIU Y, YE X G, ZHANG X, GUO X P, GU Y Q, ZHANG L C, JIA J Z, KONG X Y. A tandem segmental duplication (TSD) in green revolution generegion underlies plant height variation. The New Phytologist, 2012, 196(1): 282-291.
[45] Sutton T, Baumann U, Hayes J E, COLLINS N C, SHI B J, SCHNURBUSCH T, HAY A, MAYO G M, PALLOTTA M A, TESTER M A. Adelaide research and scholarship: boron toxicity tolerance in barley arising from efflux transporter amplification. American Association for the Advancement of Science, 2007, 318(5855): 1446-1449.
[46] Francia E, Morcia C, Pasquariello M, MAZZAMURRO V, MILC J A, RIZZA F, TERZI V, PECCHIONI N. Copy number variation at thegenomic segment is a major component of frost resistance in barley. Plant Molecular Biology, 2016, 92(1/2): 161-175.
[47] Knox A K, Dhillon T, Cheng H M, TONDELLI A, PECCHIONI N, STOCKINGER E J. CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theoretical and Applied Genetics, 2010, 121(1): 21-35.
[48] Pearce S, Zhu J, BoldizsÁr Á, VÁGÚJFALVI A, BURKE A, KIMBERLEY G C, GÁBOR G, DUBCOVSKY J. Large deletions in the CBF gene cluster at thelocus are associated with reduced frost tolerance in wheat. Theoretical and Applied Genetics, 2013, 126(11): 2683-2697.
[49] AURORA D, MELULEKI Z, ADRIAN S T, PETER L, DAVID A L. Copy number variation affecting theandgenes is associated with altered flowering time in wheat (). PloS ONE,2012, 7(3): e33234.
[50] Armour J A, Sismani C, Patsalis P C, CROSS G. Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic acids research, 2000, 28(2): 605-609.
[51] NITCHER R, DISTELFELD A, Tan C T, YAN L L, DUBCOVSKY J.Increased copy number at thelocus is associated with accelerated flowering time in barley. Molecular Genetics and Genomics, 2013, 288(5): 261-275.
Analysis of copy number variation oflocus in Common wheat
CHEN Can, HAN Nannan, LIU Yang, SHI XiaoWei, SI HongQi, MA ChuanXi
College of Agronomy, Anhui Agriculture University/Key Laboratory of Wheat Biology and Genetic Improvement on Southern Yellow & Huai River Valley, Ministry of Agriculture and Rural Affairs, Hefei 230036
【】The variations in numbers of copies of a gene is a common and important gene structure variation, which often effects individual phenotype. Low-molecular-weight glutenin subunit (LMW-GS) located at theloci, is an essential part of storage protein in wheat. As a hetero-hexaploid, the huge and complex wheat genome makes it difficult to detect numbers of gene copies by classical methods. A limited information is available about CNV analysis of LWM-GS. To screen reliable and stable internal reference genes and systems for complex genome and explore the CNV determination technology suitable in different wheat varieties, the techniques used for detection of gene copy numbers, based on droplet digital PCR (ddPCR) which improve the detection flux of target gene and determine the particular numbers of gene copies ofloci.【】In this paper,was used as the internal reference gene. The internal reference primers and probes were designed by using the corresponding gene sequence. In order to analyze stability of numbers of copies ofqRT-PCR was used to analyze the repeatability and accuracy ofinternal reference system (primers and probes).The corresponding specific primers and probes were designed according to the LMW-GS gene sequence ofloci. Two different methods, qRT-PCR and ddPCR, were used to detect the numbers of copies ofloci in eight wheat varieties, in order to determine which method is more suitable for high-throughput detection ofloci gene. The specific primers and probes were also designed according to the sequences of LMW-GS gene atandloci. The numbers of copies of LMW-GS gene at,andloci of 231 varieties were determined and analyzed by ddPCR.【】The results showed that the gene copy numbers of Acc1 was consistent among varieties and different DNA concentrations of the same variety. The coefficient of variation (CV) between repeats was 0.07%-0.77%. It is also indicated from results that theinternal reference system constructed in this paper has good stability and repeatability. The results of qRT-PCR and ddPCR were consistent in detecting the numbers of copies of LMW-GS gene atloci in 8 wheat varieties, which were 3, 5, 3, 4, 3, 3, 3 and 3, respectively. However, the CV among repeats detected by ddPCR was 0.30%-1.67%, which much lower than that by qRT-PCR. It showed that using ddPCR method to detect the gene copy numbers ofloci gene is more stable and reliable. The numbers of copies of LMW-GS gene were 4 at,andhaving a frequency of 95%, 32.03% 28.57% respectively in 231 wheat varieties. The total variation range in numbers of copies ofwas 10-21, and the CV was 16.12%.【】With good stability and repeatability,could be used as internal reference gene for numbers of gene copies detection in wheat. Both qRT-PCR and ddPCR could be used in gene copy numbers detection with wheat genes, but the ddPCR is more simple, stable, reliable and has high detection flux.
wheat; low-molecular-weight glutenin subunit (LMW-GS); droplet digital PCR (ddPCR); copy number variation (CNV)

10.3864/j.issn.0578-1752.2021.06.002
2020-07-28;
2020-09-27
国家重点研发计划(2017YFD0100804,2016YFD0101802)、国家小麦产业技术体系专项(CARS-03)、安徽高校协同创新项目(GXXT-2019-033)、江苏现代作物协同创新中心项目(JCIC-MCP)
陈璨,E-mail:chencan-L@163.com。韩南南,E-mail:1565635050@qq.com。陈璨和韩南南为同等贡献作者。通信作者司红起,E-mail:sihq2002@163.com。通信作者马传喜,E-mail:machuanxi@ahau.edu.cn
(责任编辑 李莉)
