Susceptibility and tissue specificity of Spodoptera frugiperda to Junonia coenia densovirus
2021-02-25CHENZuwenYANGYanchaoZHANGJianfengJlNMinghuiXlAOYutaoXlAZhichaoLlUYuanyuanYUSaizhenYANGYongboWANGYuanLlYiLlUKaiyu
CHEN Zu-wen ,YANG Yan-chao ,ZHANG Jian-feng ,JlN Ming-hui ,XlAO Yu-tao ,XlA Zhi-chao ,LlU Yuan-yuan,YU Sai-zhen,YANG Yong-bo,WANG Yuan,Ll Yi,LlU Kai-yu
1 School of Life Sciences,Central China Normal University,Wuhan 430079,P.R.China
2 Agricultural Genomics Institute,Chinese Academy of Agricultural Sciences,Shenzhen 518120,P.R.China
3 Medical College,Hubei University of Arts and Science,Xiangyang 441053,P.R.China
4 Hubei Engineering Research Center of Viral Vector,Wuhan University of Bioengineering,Wuhan 430415,P.R.China
Abstract The fall armyworm,Spodoptera frugiperda,which destroys many economic crops such as rice and maize,has recently invaded China.Insect viruses as biological control agents play important roles in killing pests.One potential viral insecticide is the environmentally highly infective and virulent densovirus.We successfully rescued Junonia coenia densovirus (JcDV)using its infectious clone in different insect cell lines and larvae of three insect species.Results showed that the lysate of cultured insect cells transfected by the JcDV infectious clone killed the 2nd instar S.frugiperda.The LD50 of homogenate from JcDV-infected Spodoptera litura to the 2nd instar S.frugiperda (1.76×108 viral genome copies per larva during 10 d post infection) was higher than that of the 2nd instar S.litura (7.39×107 JcDV genome copies) or Helicoverpa armigera larvae(9.71×107 JcDV genome copies).The LT50 of the S.litura homogenate (2.60×109 viral genome copies each larva) to the 2nd instar S.frugiperda was 6.96 d,longer than that of the S.litura (6.18 d) or the 2nd instar H.armigera (5.94 d).JcDV could infect the fat body of H.armigera,but not S.frugiperda or S.litura.Although JcDV can infect all three lepidopteran species,their susceptibility to the virus differs.JcDV has great potential as a biological control agent against pests such as S.frugiperda.
Keywords:Spodoptera frugiperda,Junonia coenia densovirus,rescue of virus,susceptibility,biological control
1.lntroduction
The fall armyworm,Spodopterafrugiperda(J.E.Smith) is a polyphagous noctuid pest that destroys maize and rice crops in Americas,causing substantial economic lost (Nagoshiet al.2017).The pest recently migrated into Africa and Asia,reaching India (Sharanabasappaet al.2018) before entering the Yunnan Province in China (Sunet al.2019; Zhanget al.2019).Now the pest has been observed in most provinces(autonomous regions,municipalities) in China.
Insect pathogens are important in controlling pest populations.Bacteria,viruses,fungi and some insect predators have been widely used as bio-insecticides (Jinet al.2021).In particular,the insect viruses baculovirus and densovirus have been successfully applied to controlling many pests (Fédièreet al.1986; Jianget al.2007; Sun 2015;Kumaret al.2019).It was reported that theS.frugiperdanuclear polyhedrosis virus (Shapiroet al.1991; Escribanoet al.1999; Villamizaret al.2010) and theS.frugiperdagranulovirus (Cuartaset al.2015; Ferrelliet al.2018) could efficiently killS.frugiperda.More than 30 densoviruses,such asCasphaliaextraneadensovirus andPeriplaneta fuliginosadensovirus,have been identified and applied to pest control (Fédièreet al.1986; Bellonciket al.1990; Jianget al.2007),but none has been isolated fromS.frugiperdathus far.Junoniacoeniadensovirus (JcDV) -first isolated from the natural hostJ.coenia(Rivers and Longworth 1972) with a relatively high LD50(1011viral particles) to the 4th instarJ.coenia(Smilanichet al.2018) -could infectS.frugiperdawith high larvae mortality (Croizieret al.2000;Bruemmeret al.2005; Mutuelet al.2010; Liuet al.2011;Phamet al.2013).However,studies on bioassay of JcDV and bio-control applications are very limited (Gasmiet al.2018,2019).JcDV belongs to theambidensovirusgenus in the subfamily Densovirinae within the Parvoviridae family(Cotmoreet al.2014).It is an autonomous,non-enveloped DNA virus,20–25 nm in diameter with icosahedral symmetry that contains a single-stranded linear DNA genome.The full length of the JcDV genome is 5 908 nucleotides; and thens1gene is 1 638 nucleotides,encoding 545 amino acid residues.JcDV,Mythimnaloreyi(maize worm)densovirus andGalleriamellonelladensovirus might replicate in theLymantria disparLd652 cell line with low or medium infectivity (El-Mergawyet al.2003; Tijssenet al.2003; Fédièreet al.2004; Wanget al.2013),but most densoviruses cannot replicate in insect cell lines.At most,the densoviruses can only persistently infect insect cell lines at very low levels (Barreauet al.1996; Bergoin and Tijssen 2000).
In the present study,we rescued JcDV using its infectious clone in widely-used insect cell lines and economical insect species,assayed the susceptibility ofS.frugiperdato JcDV and compared its pathogenicity toSpodopteralituraandHelicoverpaarmigera.Results showed that the JcDV has great potential as a biological control agent againstS.frugiperda.
2.Materials and methods
2.1.lnsect cell lines and insects
TheTrichoplusianiBTI-Tn-5B1-4 cell line (Hi5) andS.frugiperdaSf9 cell line were established from the ovaries(Daviset al.1993); theH.armigeraHa-EM-5 cell line (Ha)was from the embryos (Zhenget al.2010); and theS.lituraSl-HP cell line was from the Sl-ZSU-1 cell line (Zhanget al.2008).All the cell lines,except Ha,were cultured in Grace’s insect cell culture medium (Life Technologies,Inc.,Grand Island,NY,USA) supplemented with 10% fetal bovine serum(FBS) and antibiotics (100 U mL–1penicillin and 100 µg mL–1streptomycin) at 28°C under normal atmospheric condition.The Ha cell line was cultured in Sf900 II insect cell culture medium (Life Technologies,Inc.,Grand Island,NY,USA) supplemented with 1% FBS and antibiotics (100 U mL–1penicillin and 100 µg mL–1streptomycin).The larvae ofH.armigeraandS.liturawere purchased from Baiyun Industry Inc.(Jiyuan,China).TheS.frugiperdastrain (DH19)-collected in Dehong Autonomous Prefecture,Yunnan Province of China,in January 2019 -was reproduced in the laboratory beyond eight generations.All three species of insects were fed an artificial diet,the main component of which was wheat germ or corn meal at (27±2)°C and(75±10)% relative humidity (RH) with a 14 h light:10 h dark photoperiod.Adults were provided with a 10% sucrose solution supplemented with 0.5% honey for nutrition.
2.2.Plasmids and reagents
The JcDV infectious clone pBRJH construct carrying the full-length genome of JcDV (GenBank accession number:NC_004284.1) (Jourdanet al.1990; Dumaset al.1992) and rabbit antibody against the non-structural JcDV protein,NS1 protein (Dinget al.2002; Abd-Allaet al.2004),were kindly gifted by Dr.Bergoin from University of Montpellier II,France.The plasmid pIZ-EGFP-SfGATAe-V5-His expressing theS.frugiperdaGATAe transcription factor (GenBank accession number:GCTM01006721.1) was constructed in the Institute of Entomology,School of Life Sciences,Central China Normal University.The transfection reagent FuGENE HD and the DyLight 549 and DyLight 488 goat anti-rabbit fluorescence antibodies were purchased from Promega corporation and Abbkine Inc.,respectively.The plasmid extraction kit was from Omega Bio-tek Inc.(Georgia,USA),and the real-time quantitative PCR (qPCR) mixture from US Everbright Inc.(Suzhou,China).
2.3.Cell transfection and larval injection
The plasmid pBRJH was extracted from theEscherichia coliSure 2 strain cultured at 28°C.The plasmid was mixed with the FuGENE HD transfection reagent (3 µg plasmid:7.5 µL transfection reagent) to transfect Hi5,Ha,Sl-HP and Sf9 cells cultured individually in a six-well plate(Corning Inc.,China) at 5×105cells/well,according to the method previously described in Gaiet al.(2013).For larvae transfection,the mixture (1 µg plasmid:2 µL transfection reagent) was injected into the 3rd instarH.argimera,S.lituraandS.frugiperda(3 µL mixture per larva) using a microinjector (Nikon Inc.,Tokyo,Japan),according to the method published in Wanget al.(2013).
2.4.lmmunofluorescence and histochemistry assay
The immunofluorescence of insect cells was performed according to the method described in Vendevilleet al.(2009).Briefly,insect cells were seeded onto coverslips in a 24-well plate (Corning Inc.,China) at 1×105cells/well,cultured overnight and then used to transfect with the plasmid pBRJH and FuGENE HD transfection reagent as described in Section 2.3.At 72 h post transfection,the cells were fixed using 4% paraformaldehyde (Sigma-Al drich Inc.,St.Louis,MO,USA) for 15 min,penetrated by 1% Triton X-100 for 1 h and blocked with 2% BSA diluted in PBS containing 0.1% Tween 20 for 2 h.The cells were then incubated with rabbit anti-NS1 antibody (1:500) in PBS containing 1% BSA and 0.1% Tween 20 for 2 h.The cells were washed three times with PBS containing 1% BSA and 0.1% tween 20 before further incubation for 1 h with goat anti-rabbit fluorescence antibody labeled with DyLight 549 (1:1 000).Finally,the cells were stained with 1 µg mL–1Hoechst 33342 (Sigma) for 10 min and imaged with a fluorescence microscope (Nikon).SfGATAe was expressed as fusions to the C-terminus of EGFP.
The 2nd instar larvae were fed an artificial diet (3 mm×3 mm×3 mm) containing the homogenate (about 150 µg wet weight per larva) ofS.lituralarvae dying from JcDV infection.At 72 h post feeding,the larvae were cut into three parts from head to tail,and the middle parts containing the midgut were fixed in picric acid fixation solution for more than 24 h.The samples were dehydrated using ethanol,penetrated using dimethylbenzene,embedded with paraffin and sectioned into 4-µm thick slices.The slices were fixed on glass slides and rinsed with dimethylbenzene,ethanol and water.Immunofluorescence was performed as described in the previous paragraph;DyLight 488 goat anti-rabbit fluorescence antibody was used as the secondary antibody,and green fluorescence was analyzed using fluorescence microscopy.Detailed protocols can be found in our previous article (Wanget al.2019).For histochemistry,sample slices were stained with H&E staining after being fixed on glass slides and rinsed with dimethylbenzene,ethanol and water.
Histochemistry and immunofluorescene were also performed on fat bodies dissected from larvae infected without or with JcDV to assess whether JcDV can infect the fat body.
2.5.qPCR
DNA was extracted from insect cells and larvae transfected with or without the pBRJH or fed a diet containing a homogenate (150 µg wet weight per larva) of JcDV-infecting 2nd instarS.lituraat different time points post treatments(24 and 72 h) according to Mutuelet al.(2010).Briefly,the cell pellet was washed with PBS and then treated with DNA extraction buffer containing 20 mg mL–1of proteinase K for 5 h at 55°C before phenol extraction and precipitation.The healthy or infected larvae were washed with PBS,homogenized and prepared using the method as described just above.Each sample pool contained four larvae.qPCR for JcDV genomic DNA was performed using the primers of the JcDVns3gene listed in Table 1,according to the protocol described in previous reports (Fédièreet al.2004;Mutuelet al.2010; Salascet al.2016; Gasmiet al.2019).The number ofribosomalproteins3A(rps3A) copies of a few insects (GenBank accession numbers are listed in Table 1)for the genomic DNA per cell was used as an internal control and set as 4.0 U.The number of JcDV genomic copies was normalized against that of therps3Agene in order to obtain the number of viral genome copies per cell based on the method reported in Salascet al.(2016).The 2–∆∆CT method was used to quantify the number of viral genomic copies per cell in the cell lines or larvae (Schmittgen and Livak 2008).Data were collected from three biological replicates and three technical replicates.

Table 1 Primer sequences for real-time quantitative PCR for different rps3A genes
To quantify the copy number of JcDV genome per microgram (wet weight) of the homogenate or per microliter of the lysate of cells,six different concentrations (10-fold dilution) of the infectious clone plasmid pBRJH were used to establish a standard curve using the primers of JcDVns3with qPCR.The number of JcDV genomic copies was set as 2 per plasmid.The number of JcDV genome copies per microgram or microliter sample was calculated by comparing Ct with the standard curve (Mutuelet al.2010;Salascet al.2016).
2.6.Bioassay of lepidopteran insect susceptibility to JcDV
Insect cells (Hi5,Ha,Sl-HP and Sf9) cultured in six-well plates (approximately 1×106cells/well) were frozen and thawed three times in 1 mL PBS at 96 h post transfecting with pBRJH.The lysate was added directly onto the surface of 10 µL artificial diet (3 mm×3 mm×3 mm) containing different amounts of JcDV genomic copies per larva at the stage of the 2nd instar.The mortalities of the larvae fed on the diet mixed with transfected cell lysates were recorded across 12 days after feeding.In the control group,the larvae were fed a diet containing the same volume (10 µL) of lysate of cells without transfection.
The dead larvae ofS.liturainfected by JcDV through injection of pBRJH were weighed and homogenized in 10 mL PBS,filtered using a 0.45-µm syringe filter after centrifuging at 10 000×g for 10 min and stored at–80°C.The supernatant was serially diluted in PBS at 1:2 for the bioassy.A total of 10 µL of supernatant was added onto the surface of the artificial diet (3 mm×3 mm×3 mm) in a six-well plate.Each 2nd instar larva (S.frugiperda,S.lituraandH.armigera)was cultured separately in the individual wells of a six-well plate.There were 39 larvae per treatment for one virus titer.The diet was replaced with a fresh one without the supernatant every day.The number of dead larvae was recorded twice per day across 10 days post infection,and the 50% lethal dose (LD50) and 50% lethal time (LT50) were calculated according to the methods described in Escribanoet al.(1999).
2.7.Statistics analysis
The LD50and LT50values and the corresponding 95%confidence limits were calculated through Probit analysis of the mortality data with SPSS 16.0 Software.All experiments were independently performed in triplicates.P<0.05 was considered as statistically significant in the Student’st-test or one-way ANOVA test used on the SPPS 16.0 Software.
3.Results
3.1.JcDV was recovered in four insect cell lines transfected with the pBRJH
Immunofluorescence assay revealed that the JcDVns1gene was expressed at different levels in the four cell lines(Hi5,Ha,Sl-HP and Sf9) when the JcDV infectious clone plasmid pBRJH was transfected into these cell lines.The expression levels based on the intensity of red fluorescence were high in the Hi5 and Sf9 cell lines,but low in the Ha and Sl-HP cells (Figs.1 and 2).TheS.frugiperdaGATAe transcription factor expressed using plasmid pIZ-EGFPSfGATAe-V5-His significantly enhanced the expression of thens1gene in the Sl-HP cell line (Fig.2).The lysate from the four insect cell lines transfected with pBRJH was used to feed the 2nd instarS.frugiperdalarvae; mortality reached up to 50–100% at day 12 post feeding (Table 2),suggesting that the four insect cell lines could produce infectious virions of JcDV recovered by transfection.However,the immunofluorescence assay demonstrated that the lysate from the four insect cell lines transfected with pBRJH could not reinfect insect cell lines,suggesting that JcDV virions might not enter these insect cell lines.
3.2.JcDV was recovered in three lepidopteran species
The larvae of three insect species,H.armigera,S.lituraandS.frugiperda,began to die from day 6 after injecting the plasmid pBRJH into the 3rd instar larvae.The larvae injected with PBS or empty vector survived until adult stage.As shown in Table 3,the mortalities of the three lepidopteran species were more than 80% at day 12 post injection with the plasmid pBRJH.In the three lepidopteran species,the qPCR demonstrated that JcDV genomic DNA replicated and increased about 67–90 folds per cell at 72 hvs.24 h post feeding with homogenate ofS.lituradying from JcDV infection (Table 4).
3.3.Susceptibility of S.frugiperda to JcDV from homogenate of JcDV-infected S.litura larvae
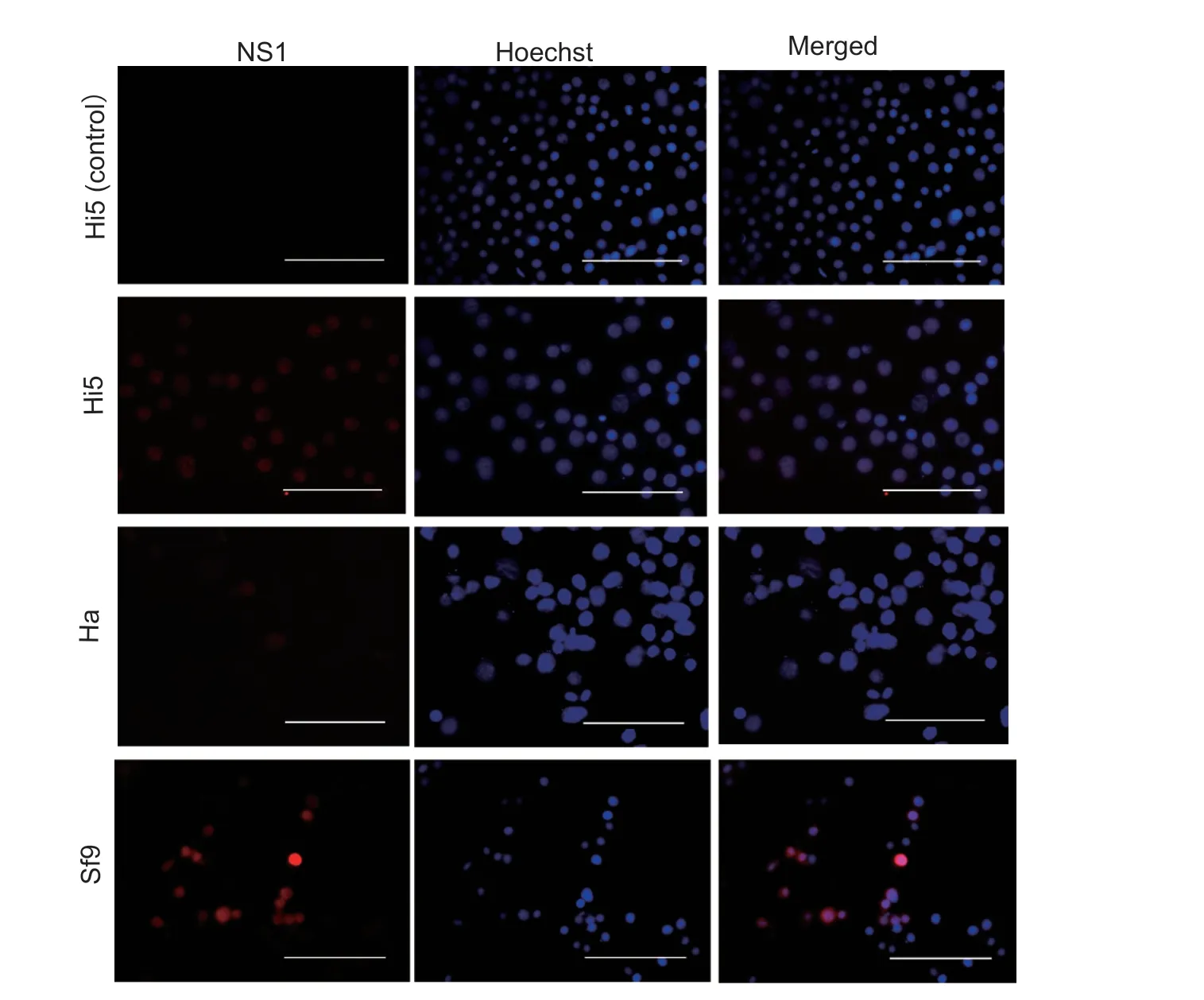
Fig.1 Indirect immunofluorescence assay of Junonia coenia densovirus (JcDV) ns1 gene expression.JcDV NS1 protein was detected in Trichoplusia ni Hi5,Helicoverpa armigera Ha and Spodoptera frugiperda Sf9 cells at 72 h post transfection using rabbit anti-NS1 polyclonal antibody.Hi5 cells in the control group were transfected using empty vectors.Bar=100 µm.
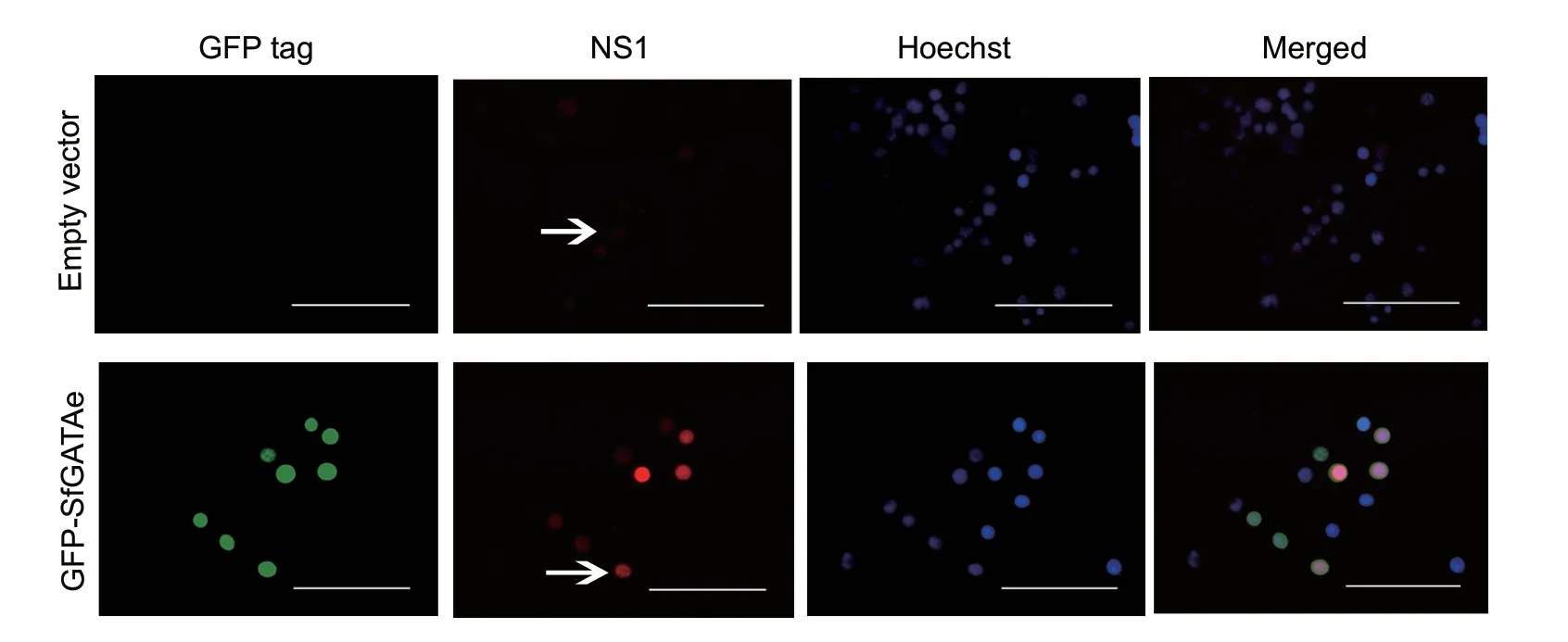
Fig.2 Spodoptera frugiperda GATAe (sfGATAe) enhanced expression of Junonia coenia densovirus (JcDV) ns1 in Spodoptera litura Sl-HP cells.The pBRJH and the plasmid expressing GFP-SfGATAe (1:1) were co-transfected into Sl-HP cells.The expression of JcDV NS1 (red fluorescence) was detected using immunofluorescence at 72 h post transfection.The upper and lower arrows indicate weak and strong signal of NS1,respectively.Bar=100 µm.
The supernatant (10 µL) of the homogenate prepared from JcDV-infectedS.liturawas serially diluted at 1:2 in PBS and added onto the surface of the diet fed to the 2nd instarS.frugiperdalarvae.They began to die at day 4 post feeding.The mortalities of the larvae fed with the artificial diet containing different dosages of the homogenate were calculated each day across 10 days post feeding.The LD50of the homogenate toS.frugiperdalarvae was 1.76× 108JcDV genome copies per larva,higher than that of theS.lituralarvae (7.39× 107JcDV genome copies) andH.armigeralarvae (9.71× 107JcDV genome copies) (Table 5).The LT50to the 2nd instarS.frugiperdalarvae was 6.96 d when feeding on the homogenate (2.60× 109viral genomic copies per larva),significantly longer than that ofS.litura(6.18 d) andH.armigera(5.94 d) (Table 5).The results indicated that all the three lepidopteran species were susceptible to JcDV produced byS.lituralarvae.
3.4.Tissue susceptibility of three lepidopteran species to JcDV
Immunofluorescence assay of the NS1 showed that JcDV was able to infect multiple tissues in the larvae of all the three insects (Fig.3).However,JcDV could infect the fat body ofH.armigeraonly (Fig.3).Histochemistry also demonstrated that only the nuclei of the fat body of JcDV-infectedH.armigeralarvae were much larger than those of the control larvae (Fig.4).Although the fat body isolated from JcDV-infectedS.frugiperdaorS.lituralarvae showed only a background green fluorescence signal (data not shown),the fat body isolated fromH.armigeralarvae infected with JcDV showed a strong JcDV NS1 signal (Fig.5).All the results revealed that JcDV could not infect the fat body ofS.frugiperdaandS.litura,but onlyH.armigera.
4.Discussion
The present study demonstrated that JcDV could be successfully rescued in multiple insect cell lines,includingS.frugiperda,S.lituraandH.armigeracell lines.Among the three lepidopteran insect species,H.armigerais the most susceptible to JcDV and the only species whose fat body could be infected by JcDV.1)The number ofJunoniacoeniadensovirus (JcDV) genome copies was measured by real time quantitative PCR using the total DNA extracted from 10 µL of cell line lysates transfected with pBRJH.
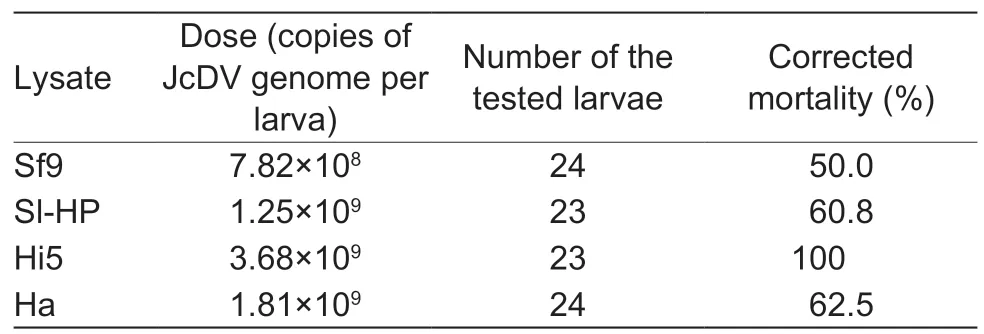
Table 2 Mortality of 2nd instar Spodoptera frugiperda fed with lysates of cells transfected with pBRJH at day 12 post feeding1)
It was reported that JcDV infected the Ld652 cell line by clathrin-mediated endocytosis (Liet al.1996; Abd-Allaet al.2004; Vendevilleet al.2009; Salascet al.2016).The present study found that the lysate from insect cells transfected with pBRJH could infect larvaeviafeeding,but the supernatant collected from the transfected cells failed to reinfect insect cell lines (data not shown).
We further observed that JcDV NS1 did not express in the midgut and fat body ofS.frugiperdaandS.litura,which was consistent with previous reports (Wanget al.2013; Salascet al.2016; Kemmerer and Bonning 2018).However,the NS1 protein was expressed in the fat body ofH.armigera(Figs.3–5),suggesting that JcDV could infect the fat body ofH.armigera(Fig.5).
Aside from insect cell lines,JcDV was also rescued in multiple lepidopteran species.The insect pests were dying from day 4 to 12 post infection; and the 2nd instar larvae ofS.frugiperda,S.lituraandH.armigera-important lepidopteran pests in China -were susceptible to the JcDV virus.Few reports with bioassays of densovirus exist.Approximately 80% of the 2nd instarS.exigualarvae were killed by 5× 1010JcDV virions per larva on day 9 post oral infection (Gasmiet al.2018).The LC50ofPeriplaneta fuliginosadensovirus toP.fuliginosawas approximately 6×104virions g–1on day 28 post infection (Jianget al.2008).The LD50of JcDV to the three lepidopteran insects was7.39×107to 17.59×107viral genome copies on day 10 post oral infection.Our results suggest that there are multiple insect hosts susceptible to JcDV and that JcDV has high potential as a bio-insecticide in the future.
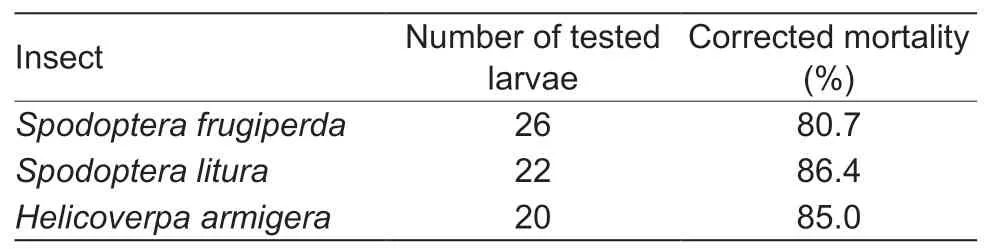
Table 3 Mortality of 3rd instar larvae injected with the infectious clone pBRJH at day 12 post injection

Table 4 Real-time quantitative PCR assay for the number of Junonia coenia densovirus (JcDV) genomic DNA copies per cell in the infected larvae at different time points
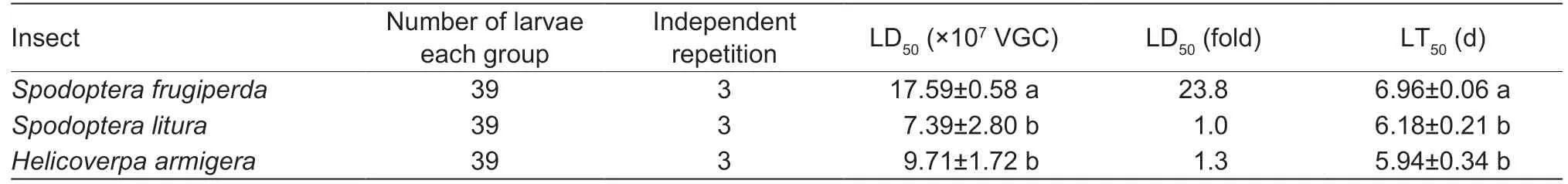
Table 5 LD50 and LT50 of Junonia coenia densovirus (JcDV) 10 days post oral infection1)
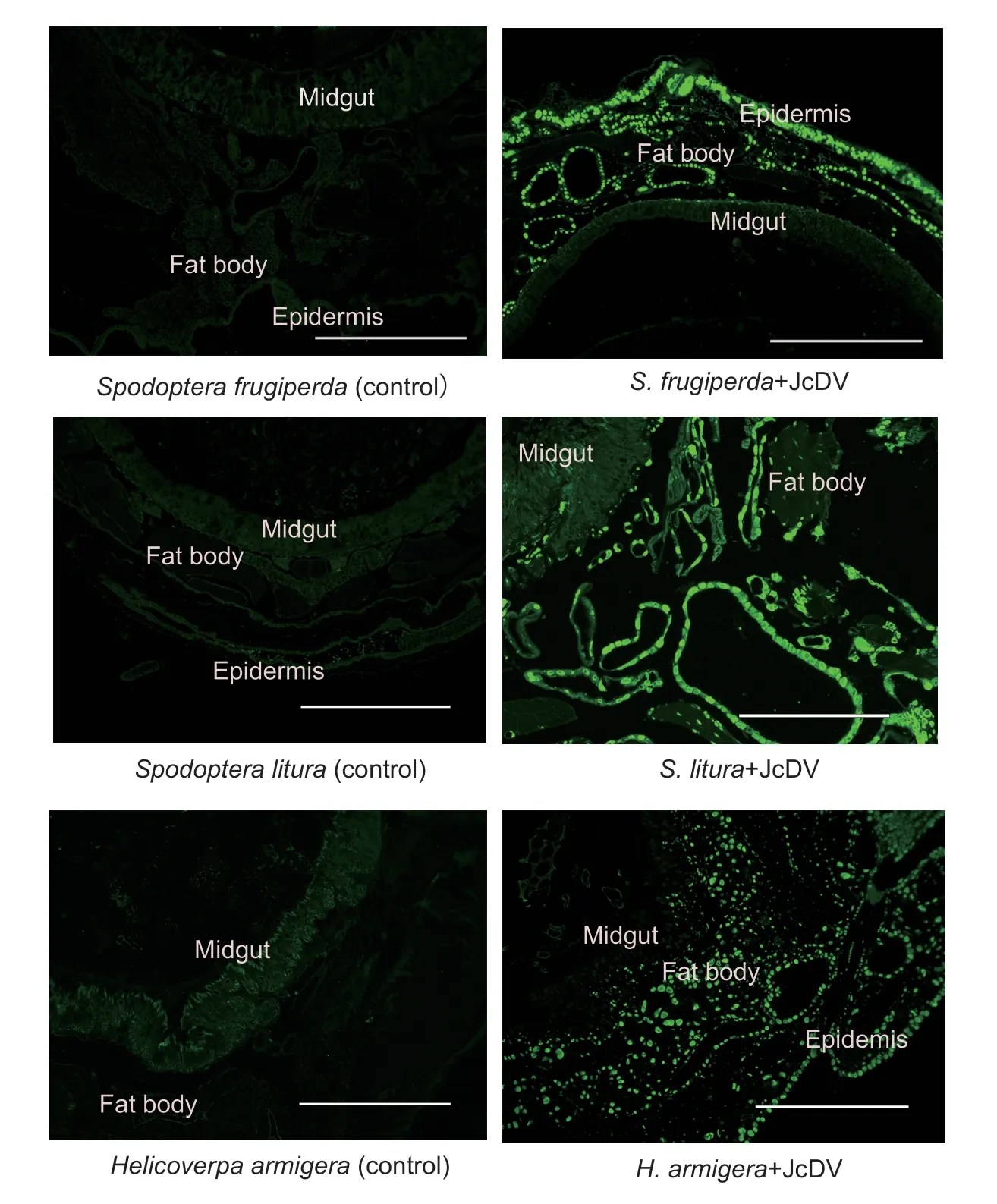
Fig.3 Indirect immunofluorescence assay of tissues susceptible to Junonia coenia densovirus (JcDV).JcDV NS1 expression(green fluorescence) was assayed using rabbit anti-NS1polyclonal antibody at 72 h post feeding.The controls (left column) were sample slices of larvae 72 h post feeding with a homogenate of healthy Spodoptera litura larvae.The treated samples (right column) were sample slices of larvae 72 h post feeding with a homogenate of S.litura larvae dying from JcDV infection (150 µg wet weight per larva; corresponding to 7.22×109 JcDV genome copies).Bar=400 µm.
GATAe protein is a master transcription factor regulating the differentiation of insect midguts.It can trigger expression of a set of midgut-tissue specific genes in other tissues,such as the fat body or S2 cell line (Okumuraet al.2005,2016;Weiet al.2019).In this study,we demonstrated that GATAe enhanced JcDV NS1 expression in some insect cell lines,which is essential to virus replication.JcDV could not infect the midgut of insect species,indicating that the virions of JcDV might not release viral genomic DNA in midgut cells when they pass through the midgut tissue by transcytosis(Wanget al.2013).
Large scale production of virus is a significant bottle neck to using insect virus for bio-control.One possible choice is to useS.liturafor virus replication because the larvae can be cultured together,whileH.armigerawill eat each other.Spodoptera lituralarva is also much bigger thanS.frugiperdaandH.armigeralarva,suggesting that eachS.lituralarva could produce more virions than any other insect species.Lastly,we found that the homogenate of deadS.lituralarvae infected with JcDV can kill multiple lepidopteran species,suggesting that JcDV can be used to control multiple insect pests.All these data indicate thatS.lituracan efficiently produce JcDV virions for pest control.
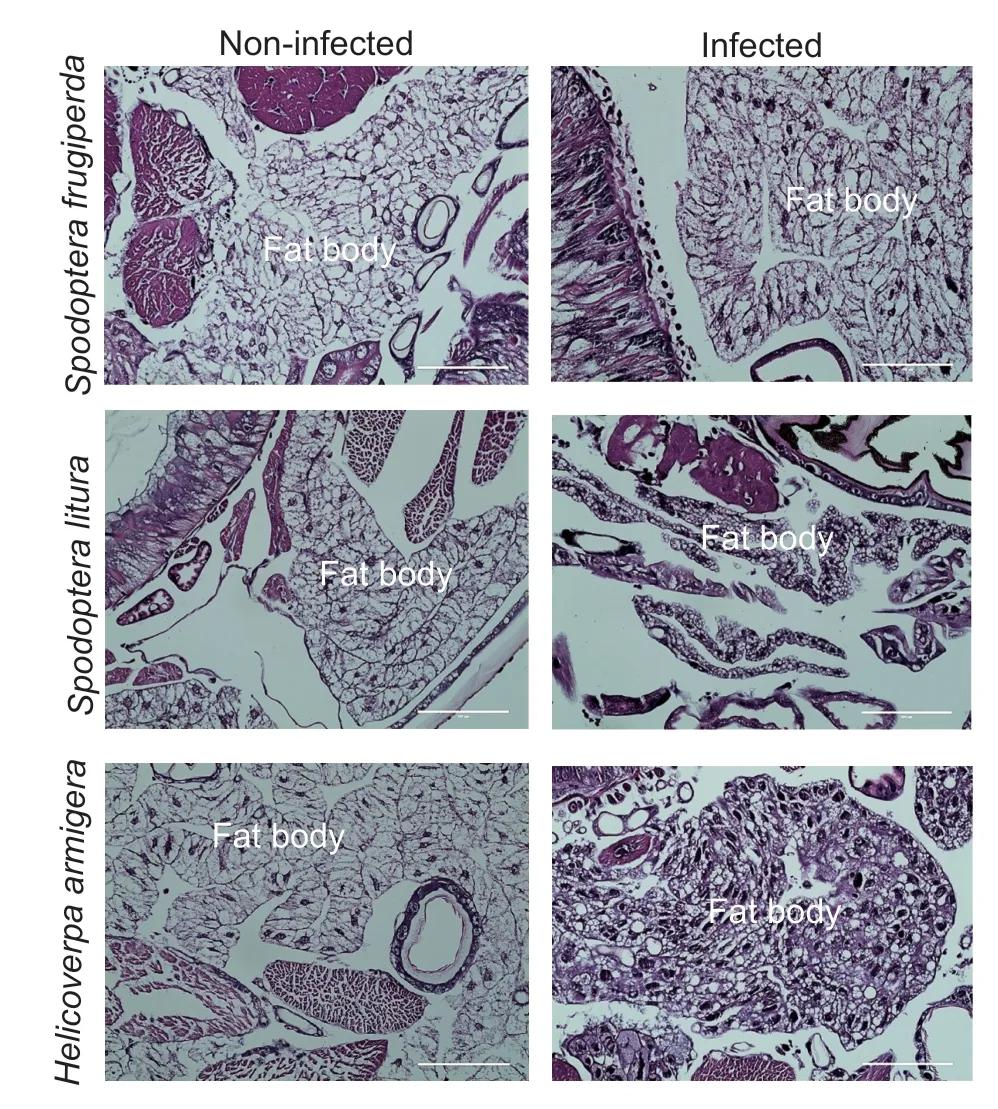
Fig.4 Histochemistry of larvae slices with or without Junonia coenia densovirus (JcDV) infection.Larvae fat body is shown with H&E staining.The nuclei of JcDV-infected cells were much larger than those of non-infected cells.Bar=100 µm.
While the current study focused on the bioassay of JcDV in the laboratory,subsequent bioassays of JcDV in insect field populations using homogenates of dead larvae or purified virions of JcDV are required.Further investigations on the impact of JcDV on the ecosystem is urgently needed before the virus can be applied to field populations of insects.Additionally,the combined application of JcDV with other insect pathogens -such as nuclear polyhedrosis virus,granulovirus and other pathogens -might enhance the efficiency of JcDV killingS.frugiperda(Gasmiet al.2019).
5.Conclusion
AllS.frugiperda,S.lituraandH.armigeraare susceptible to JcDV.JcDV can infect the fat body ofH.armigerabut notS.frugiperdaorS.litura.Several insect cell lines and lepidopteran larvae can successfully rescue JcDV virions with the infectious clone plasmid.Spodoptera lituralarva is particularly efficient at producing JcDV virions.
Acknowledgements
We thank Dr.Xu Pengjun (Tobacco Research Institute,Chinese Academy of Agricultural Sciences) and Dr.Li Changyou(Qingdao Agricultural University,China) for their experimental suggestions.We also thank Miss Feng Zhu (School of Life Sciences,Central China Normal University) for rearing the insects.This work was supported by the National Key R&D Program of China (2017YFD0200400) and the Natural Science Foundation of Hubei Province,China (2017CFB241).
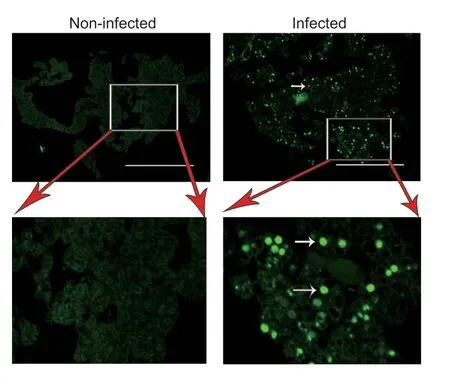
Fig.5 Immunofluorescence of fat body isolated from Helicoverpa armigera larvae.NS1 fluorescence (white arrows)was present in the isolated fat body cells of larvae infected with Junonia coenia densovirus (JcDV),but not in the non-infected larvae.Bar=400 µm.
Declaration of competing interest
The authors declare that they have no conflict of interest.
杂志排行
Journal of Integrative Agriculture的其它文章
- Research on the invasive pest of fall armyworm (Spodoptera frugiperda)in China
- Migration of invasive Spodoptera frugiperda (Lepidoptera:Noctuidae)across the Bohai Sea in northern China
- Fitness of fall armyworm,Spodoptera frugiperda to three solanaceous vegetables
- Two-way predation between immature stages of the hoverfly Eupeodes corollae and the invasive fall armyworm (Spodoptera frugiperda J.E.Smith)
- Analysis of phototactic responses in Spodoptera frugiperda using Helicoverpa armigera as control
- Genome editing of the SfABCC2 gene confers resistance to Cry1F toxin from Bacillus thuringiensis in Spodoptera frugiperda
