Molecular mechanism of Turmeric in the treatment of liver cancer based on network pharmacology
2021-01-04YuanYuanZhangWenJingZouXuLiPengZhangZiJunLiao
Yuan-Yuan Zhang, Wen-Jing Zou, Xu Li, Peng Zhang, Zi-Jun Liao
1.Xi' an Medical University, Xi' an 710068, Shaanxi, China;
2.Xi'an Fifth Hospitaly, Xi' an 710082, Shaanxi, China;
3.Department 1 of Shanxi Cancer Hospital Affiliated to Medical College of Xi'an Jiaotong University Department of First Internal Medicine, Xi' an 710061, Shaanxi, China
4.Department of traditional Chinese medicine, fengxiang hospital, Baoji 721400, Shaanxi, China
5.Department of Shanxi Cancer Hospital Affiliated to Medical College of Xi'an Jiaotong University Department of First Internal Medicine, Xi' an 710061, Shaanxi, China
Keywords:Network pharmacology Turmeric Liver cancer Mechanism
ABSTRACT Objective: The aim of this study was to use network pharmacology to predict the targets and related signaling pathways of turmeric in the treatment of liver cancer. Methods: The active ingredients of turmeric turmeric and their corresponding targets were screened and collected through the traditional Chinese medicine components and systematic Chinese medicine pharmacology database and analysis platform (TCMSP) database. Through the online human Mendelian network (OMIM), the human genome annotation database (Genecards) and the GAD database, the targets related to liver cancer were collected and compared with the targets corresponding to the drug components, the common parts were screened out to obtain the potential target genes that overlap between turmeric and liver cancer. Cytoscape was used to construct the "compound - target" action network, and the protein interaction (PPI) network was constructed through STRING software to screen the key components and key targets of turmeric for the treatment of liver cancer, and the GO enrichment and KEGG enrichment analysis were conducted on the key targets to analyze their potential mechanism of action. Results: There were 15 active components and 45 target genes in the treatment of liver cancer with turmeric turmeric, and the drug-component-target-disease network showed that the key genes mainly included: MAPK1, MAPK3, AKT1, JUN, RELA, BCL2, CASP8, ESR1, ADRB2, etc. GO functional enrichment showed that biological processes and functions were concentrated in cofactor binding, phosphatase binding, amide binding, g-proteincoupled amide receptor activity, antioxidant activity, steroid activity, nuclear receptor activity, transcription factor activity, direct ligand regulation of sequence-specific DNA binding, and steroid hormone receptor activity. KEGG functional enrichment showed that the enriched pathways mainly included hepatitis b, human immunodeficiency virus 1 infection, apoptosis, hepatitis c and some cancer signaling pathways. Conclusion: The role of turmeric in the treatment of liver cancer may be realized through the above molecular mechanism, providing theoretical evidence for subsequent studies and clinical applications.
1. Introduction
The incidence of liver cancer in China accounts for more than 50% of the world [1]. At present, the treatment of liver cancer is mainly based on comprehensive treatment, but many studies have shown that traditional Chinese medicine plays an equally important role in reducing the recurrence and metastasis of liver cancer, reducing the side effects of radiotherapy and chemotherapy, and improving the quality of life of patients with advanced liver cancer [2]. Yujin is currently clinically used as an auxiliary anti-tumor drug for gastric cancer, intestinal cancer, melanoma, liver cancer and so on. Chinese medicine believes that turmeric tastes spicy, cold, heart, liver, gall bladder, has the effect of cooling blood to stop bleeding, eliminating gall bladder and yellowing, and is mainly used clinically in the treatment of jaundice and amenorrhea [3]. This study uses network pharmacology to predict the multi-target and multi-pathway synergy of turmeric treatment for liver cancer, which further provides a reference for the clinical and basic research of liver cancer. The research results are reported below.
2. Materials and methods
2.1 Collection of turmeric active ingredients and target prediction
In Traditional Chinese Medicine Systems Pharmacology (TCMSP), enter turmeric to query the composition and the corresponding target and filter the composition information. Select the threshold oral availability (Oral bioavailability, OB) ≥ 30% and drug-like (druglikeness, DL) ≥ 0.18 ingredients and do further research.
2.2 Collection of targets of active compounds
Use TCMSP to input Yujin for searching and sorting to obtain the target of active compound. Cytoscape 3.6.1 software (http://www.cytoscape.org/) was used to construct the compound-target network by collecting the collected traditional Chinese medicine compounds and targets. In this network, nodes with a degree of centrality (DC) of 0.9 and corresponding closeness centrality (CC) are screened for topological analysis to identify key compounds or targets.
2.3 Obtaining the intersection target
Genecard (https://genecard.org/) database, OMIM database (http://omim.org/), GAD database (https://geneticassociationdb.nih.gov/) three keywords can be input by keyword Obtain and analyze related human genes. In this study, "liver cancer" and "Hepatocellular Carcinoma" were used as keywords to search and screen for known disease targets, delete duplicate values, and finally obtain known targets. Using R language drawing to match the targets related to drug active ingredients and disease targets, and draw a Venn diagram (venn) to obtain the potential anti-liver cancer targets in turmeric active ingredients.
2.4 Construction of Turmeric Target Protein Interaction (PPI) Network
The target gene corresponding to the active ingredient is matched with the target gene related to liver cancer to obtain the common gene of the two is the key target of turmeric anti-liver cancer. Cytoscap software can build an active ingredient-target network according to the network relationship of nodes, and can edit and analyze the network. In this study, Cytoscape 3.6.1 software was used to build a "drug-co-active ingredient-key target gene" relationship network. The peripheral nodes of the network represent the key target genes of diseases and active ingredients of drugs; the active ingredients of drugs are shown in the network. The entire network demonstrates the connection between drug-co-active ingredients and disease targets, and the mechanism of turmeric treatment of liver cancer is explored by constructing this network.
2.5 Construction of key target protein-related networks
Use the STRING database platform (https://string-db.org/Version 10.5) to predict protein interactions. Import the key target of turmeric action into the String database, select the research species as human (Human sapiens), obtain the protein interaction relationship, and export the results in TSV format. The light blue line represents the interprotein interaction from the database, and the purple line represents the experimentally verified interprotein interaction.
2.6 Gene ontology (GO) enrichment analysis
The R gene was used to perform GO enrichment analysis on the obtained genes. First, install the Bioconductor software package "org.Hs.eg.db" in the installation package in R and run it to convert the key target genes of turmeric treatment for liver cancer into entrezID for the enrichment analysis of key target genes GO function (P <0.05 , FDR <0.05), and output the result as a bubble chart.
2.7 KEGG pathway enrichment analysis
The R gene was used to analyze the KEGG pathway. First, install the Bioconductor software package "org.Hs.eg.db" in the installation package in R and run it to convert the key target genes of Sophora flavescens to treat liver cancer into entrezID for functional enrichment analysis of key target genes KEGG (P <0.05 , FDR <0.05), and output the results as a bar graph.
3. Results
3.1 Turmeric active ingredients and their target genes
A total of 46 chemical components of turmeric were retrieved through the database of TCMSP platform, and 15 potential active ingredients of Sophora flavescens were further screened through oral availability (OB)> 30% and analogy (DL)> 0.18, see Table 1.

Table 1 Active ingredients of Turmeric
3.2 Liver cancer related genes
Database and GAD database, a total of 8992 liver cancer-related genes were screened out, and then each liver cancer-related gene was mapped to the target gene corresponding to turmeric respectively, resulting in 46 The target genes of turmeric active ingredients for the treatment of liver cancer are shown in Figure 1.
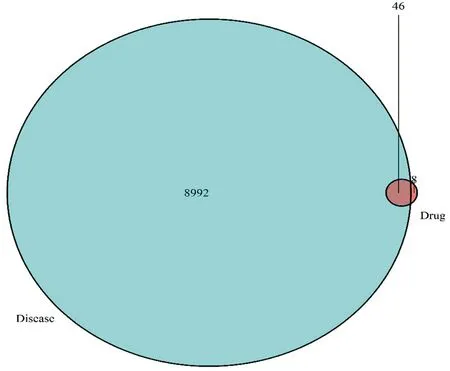
Figure1: Turmeric activity-related genes and liver cancer-related genes
3.3 "Drug-component-target-disease" network construction and analysis
The key target genes related to turmeric and liver cancer were imported into Cytoscape software to construct a "drug-co-active ingredient-key target gene" relationship network (Figure 2). The results show that the potential active molecules with top 3 connectivity in turmeric are sitosterol, B-sitosterol and naringenin. The above three components have important effects on the treatment of liver cancer.

Figure2: "Drug-component-target-disease" network diagram
3.4 Construction of co-expression network
Import the potential key target genes for the treatment of primary liver cancer into the String database platform, select the species as human, obtain the protein interaction relationship, and use the platform to draw the protein relationship network (Figure 3). Excluding free (no interacting proteins), the size and color of the circle, ie the node, represent the size of the Degree value. The larger the node, the greater the Degree value, and the greater the Degree value, the color changes from red to blue. The thickness of the side represents the size of the Combine score. The thicker the side, the greater the value of Combine score. The first 7 key protein nodes are made according to the nodes with Degree ≥ 10 (as shown in Figure 4). The key nodes are MAPK1, MAPK3, AKT1, JUN, RELA, BCL2, and CASP8. These target genes have important significance in the treatment of liver cancer.

Figure3: Co-expression network of related genes and target proteins

Figure4: Key gene nodes in the liver cancer-related gene protein network
3.5 GO function enrichment analysis
By key target genes, and then sorted according to the corrected P value, select the top 20 P value for bubble chart display, see Figure 5. The signal pathways of turmeric treatment for liver cancer include cofactor binding, phosphatase binding, amide binding, G protein coupled amide receptor activity, antioxidant activity, steroid activity, nuclear receptor activity, transcription factor activity, direct ligand regulation Signal pathways such as sequence-specific DNA binding and steroid hormone receptor activity.

Figure5: GO function enrichment of key target genes
3.6 KEGG pathway enrichment analysis
KEGG pathway enrichment analysis, select the signal pathway whose P value is in the top 20, and perform KEGG pathway enrichment analysis on the genes that have been obtained, see Figure 6. It is suggested that turmeric's anti-hepatoma effect may be related to hepatitis B, human immunodeficiency virus 1 infection, apoptosis, hepatitis C and some cancer signaling pathways.

Figure6: KEGG function enrichment of key target genes
4.Disscusion
In this study, the active ingredients of turmeric were obtained through the TCMSP platform database, and the liver cancer-related genes were obtained through the GeneCards database, OMIM database, and GAD database. The "drug-component-target-disease" network was constructed through Cytoscape software to analyze the turmeric The potential active molecules in the top 3 of the active ingredients are then constructed through the String database for coexpression network to obtain key target genes. Finally, GO function enrichment and KEGG function analysis are performed to obtain their related signal pathways. Zhang et al [4] showed that sitosterol inhibited the growth and proliferation of tumor cells by inducing apoptosis, but had no significant inhibitory effect on normal liver cells.
This study found that the seven key target nodes for turmeric treatment of liver cancer are MAPK1, MAPK3, AKT1, JUN, RELA, BCL2, and CASP8. Zhao et al [5] showed that the activation of MAPK pathway can mediate the malignant biological behavior of liver cancer. Our research shows that the key targets of turmeric to treat liver cancer are MAPK1 and MAPK3, but the current research on turmeric inhibiting the growth of liver cancer cells by inhibiting MAPK1 and MAPK3 has not been reported. Studies have shown that AKT can affect the biological behavior of tumors through the following two aspects: One is that AKT can phosphorylate the proapoptotic protein Bad, activate NF-κB, and then inhibit the activity of transcription factors, thereby inhibiting the release of apoptosis factors [6-8]: Second, the activation of AKT can affect the tumor cell cycle and promote tumor cell proliferation [9]. Previous studies have confirmed that turmeric has an anti-tumor effect by inhibiting the activation of NF-κB to block the signaling pathway [10], and our research also shows that turmeric is a key target for liver cancer including AKT, which further validates our research. Correctness.The c-jun gene, a member of the JUN family, is a component of transcription factor 1 and is involved in biological activities such as cell proliferation, apoptosis, migration, and invasion [11, 12], and may be involved in the regulation of VEGF, thereby promoting tumors. Angiogenesis [13-15]. At present, there is no report about the effect of turmeric on the anti-liver cancer effect by affecting the expression of the JUN gene, and our research found that the JUN gene is a key target for turmeric to function, so it is worthy of our further research. Abnormal expression of BCL2 gene can promote cell proliferation, participate in tumor formation and other processes, and participate in biological behaviors such as tumor cell migration and invasion [16, 17]. There are also reports that BCL2 can inhibit tumor cell apoptosis and promote the occurrence and development of liver cancer cells [18]. However, there is no report on the research of turmeric inhibiting the occurrence of liver cancer by inhibiting BCL2.
This study found that the enrichment of GO functions of core genes mainly focuses on cofactor binding, phosphatase binding, amide binding, G protein coupled amide receptor activity, antioxidant activity, steroid activity, nuclear receptor activity, transcription factor activity, and direct The ligand regulates signaling pathways such as sequence-specific DNA binding and steroid hormone receptor activity. Previous studies have reported that CINP as a cofactor is involved in cell proliferation and tumor growth [19], but current research on turmeric in this area has not been reported. Phosphatase, as a key enzyme in cell signal transduction, participates in various processes such as cell metabolism, DNA replication, translation and transcription, cell division and apoptosis [20]. However, the current research on Yujin in this area has not been reported yet, so further research is possible. It has been reported that the combination of alcohol extracts of melanogaster and VCR can reverse the resistance of human gastric cancer heterotopic xenografts to VCR and inhibit the growth of xenografts, and the mechanism may be the effect on GCS to increase the sensitivity of gastric cancer cells to VCR [21] .At present, there is no report about the effect of turmeric on GCS against liver cancer. The body continuously produces and scavenges active oxygen radicals during the metabolism, thereby maintaining a dynamic balance between oxidation and reduction. If this equilibrium state is broken, it will destroy the functions of DNA and proteins, thereby inducing tumors [22]. At present, studies have reported that endophytic fungi extracted from different parts of turmeric have anti-inflammatory and antioxidant effects [23], but no literature has reported that turmeric can inhibit the growth of liver cancer cells through antioxidant effects. Gu et al. [24] have shown that nuclear receptor (Nur77) may promote the migration and invasion of liver cancer cells, but the specific mechanism is not yet clear, and our research shows that turmeric can exert anti-cancer effects by affecting the activity of nuclear receptors The role of liver cancer, it is worth further digging its mechanism of action.
This study shows that turmeric's anti-hepatoma effects may involve hepatitis B, human immunodeficiency virus 1 infection, apoptosis, hepatitis C, and some cancer signaling pathways. Bao et al [25] studies have shown that the occurrence of liver cancer in China is mainly related to viral infections (mainly B virus) infections. Zhang et al [26] also proved that HBV positive is an independent risk factor for hepatitis B cirrhosis complicated with liver cancer, and our research also shows that hepatitis B is the most enriched pathway. The process of liver fibrosis is usually accompanied by shunting of intrahepatic portal veins and regeneration of hepatocytes, which leads to nodules and eventually leads to cirrhosis [27], which can further induce liver cancer. Li et al. [28] demonstrated through relevant experiments that Guiyujin may play an anti-hepatic fibrosis role by protecting free radicals, resisting lipid peroxidation, and resisting liver damage.Previous studies have shown that [29] coinfection is common in both HBV and HIV, so co-infection is more common in the world. HBV can promote HIV replication and the course of AIDS, and our research is also enriched To the human immunodeficiency virus 1 infection is related to the occurrence and development of liver cancer, and turmeric may play an anti-liver cancer role through the influence of this pathway, but there is no research report at present, it is worth further research.
In summary, through the research method of network pharmacology, a "drug-component-target-disease" network was constructed to analyze the multi-component, multi-target, and multipath of turmeric, which initially verified the basics of turmeric treatment of liver cancer The pharmacological effect and mechanism of action provide ideas for further research on turmeric treatment of liver cancer.
杂志排行
Journal of Hainan Medical College的其它文章
- Mechanism of therapeutic effects of Huangbo on ureteral stones based on network pharmacology
- Meta-analysis of randomized controlled trials of traditional Chinese medicine in the treatment of obstructive sleep apnea syndrome
- Therapeutic efficacy of traditional Chinese medicine compound in the treatment of membranous nephropathy: A systematic review and metaanalysis
- Correlation between CT dynamic enhanced scanning parameters and serum tumor markers before and after radiofrequency ablation in patients with lung cancer
- Effects of fumigation and washing with Shujin Huoluo decoction combined with Duhuo Parasitic decoction on TIMP-1 and PON1 levels in KOA patients
- The effect of Xuebijing on burn patients and its effect on pain and wound healing
