Hypothalamic-Pituitary-Gonadal (HPG) Axis and Transcriptional Regulatory Elements Regulate piwil2 Gene Expression During Gametogenesis and Gonadal Development in Japanese Flounder (Paralichthys olivaceus)
2020-11-30NIFeifeiYUHaiyangQUJiangboMENGLihuiLIUXiumeiYANWeijieCHANGJingZHANGQuanqiWANGXuboandYUHaiyang
NI Feifei, YU Haiyang, QU Jiangbo, MENG Lihui, LIU Xiumei, YAN Weijie,CHANG Jing, ZHANG Quanqi, WANG Xubo, and YU Haiyang, *
Hypothalamic-Pituitary-Gonadal (HPG) Axis and Transcriptional Regulatory Elements RegulateGene Expression During Gametogenesis and Gonadal Development in Japanese Flounder ()
NI Feifei1), YU Haiyang1), QU Jiangbo1), MENG Lihui1), LIU Xiumei2), YAN Weijie1),CHANG Jing1), ZHANG Quanqi1), WANG Xubo1), and YU Haiyang1), *
1),,266003,2),,264005,
The P-element induced wimpy testis (Piwi) proteins, which are associated with PIWI-interacting RNAs (piRNAs), play important roles in meiosis, germ cell division, and germline maintenance. In this study, we identified and characterized thegene, a constituent factor of the piRNA pathways involved in the biogenesis of reproductive development. The biological analysis indicated that, which contains PAZ and PIWI domains, was highly conserved between teleosts and tetrapods. Thedistribution profile in different tissues confirmed a sexually dimorphic expression pattern, with a higher expres- sion level in testis.hybridization demonstrated thatwas expressed in the oogonia and oocytes of the ovaries as well as in the Sertoli cells and spermatocytes of the testes. Geneshowed a maternally inherited expression pattern during embryonic development, and was highly expressed during the early embryonic development. Different luciferase reporters were constructed to determine the transcriptional regulatory mechanisms of. Thecore promoter region was located at −360bp to −60bp. Furthermore, some representative sex hormones, including human chorionic gonadotropin, 17α-methyltestosterone, and estradiol-17β had distinct regulatory effects on. In a summery, these results indicate that, regulated by sex hormones and transcrip- tional elements, has vital functions in the reproductive cycle and gonadal development.
; gametogenesis and gonadal development; HPG axis; transcriptional regulatory elements;
1 Introduction
Gametogenesis, including spermatogenesis and oogene- sis, is a complex process that is regulated by internal and external factors (Zhang., 2014). Internal factors, in- cluding the regulation of spatial and temporal genes and the formation of an RNA-induced silencing complex, play major roles in vertebrate gametogenesis (Schultz., 2003). The Argonaute family of proteins forms a typical element of the RNA-induced silencing complex (RISC) and plays a crucial role regulating small RNA in gene silencing. The Argonaute family proteins are divided into the Ago- and Piwi subfamilies (Carmell., 2002). The Ago-subfamily proteins exist in all types of cells. They can combine with small interfering RNAs and micro- RNAs (He and Hannon, 2004), and have crucial role in RNA interference. The Piwi-subfamily proteins are usu- ally abundant in germ cells. They tend to bind with PIWI-interacting RNAs (piRNAs), and inhibit the expression of transposable elements (TEs) further ensuring the sta- bility of genome (Aravin., 2008).
genes, which belong to the Piwi-subfamily, are crucial components of the RISC involved in RNA inter- ference and play an essential regulatory role in gameto- genesis and gonadal development (Hammond., 2001).genes are conserved and characterized by sequences encoding two domains: a PAZ (Piwi-Argonaute-Zwille)- encoding sequence in the center of the nucleotide se- quence and a C-terminus PIWI domain-encoding sequence(Filipowicz, 2005;Pan., 2012). Both of these do- mains are responsible for the interaction between Piwi proteins and piRNA.
genes are enriched in germ cells, and mutations ingenes can cause failure of germline cell division. After combining with specific piRNAs, Piwi proteins first- ly affect the expression at the post-transcriptional level and then regulate the transcriptional silencing (Pandey., 2018). The functions ofgenes in gametogenesis have been well investigated, particularly in some model organ- isms. For example, three homologs (,, and) have been identified in mice, all of which are enriched in germ cells (Taborska., 2019). Additionally,,, andmutations lead to obvious defects in spermato- genesis that cause infertility (Manakov., 2015). In some lower vertebrates and fishes, such as zebrafish,() and() are highly expressed in the germ- line (Houwing., 2007). Amutation causes fail- ure of germ cell maintenance, while a lack ofaffects differentiation of germ cells (Houwing., 2008). Stud- ies involvinghomologous genes in teleosts, such as,, and, have reported thatgenes are involved in the reproductive cycle and influence sex differentiation (Zhou., 2012; Xiao., 2013; Wang.,2017).
The hypothalamic-pituitary-gonadal (HPG) axis regu-lates the Piwi-piRNA pathway and affects reproductive activities (Wang., 2018). Multiple hormones in the HPG axis interact to regulate individual reproductive ac- tivities. Gonadotropin-releasing hormone, which is syn- thesized and secreted by the hypothalamus, stimulates pi- tuitary gonadotropic cells to release gonadotropins, such as follicle-stimulating hormone and luteinizing hormone (LH), which regulate the secretion of steroids (García-Lopez., 2010). Exogenously administered human cho- rionic gonadotropin (hCG), which is similar to LH, promotes maturation of the gonads and the ovulation during the spawning period in teleosts (Zhou., 2015; Zhao., 2018). As steroids, 17α-methyltestosterone (17α-MT) regulates sex determination and differentiation in males (Lee., 2017), and estradiol-17β (E2) is a critical hor- mone stimulated by gonadotropins that controls growth and maturation of oocytes (Gauthier-Clerc., 2006). Previous studies have shown that steroids negatively af- fectedin mice (Pan., 2012), suggesting thatgenes might be important in the pathways that the hor- mones are included in regulating the reproductive deve- lopment. In teleosts, the HPG axis hormones suppressexpression in carp (Zhou., 2014), but the de- tailed regulatory functions ofin other teleostshave rarely been studied.
is an important cultured fish species in northern China, Korea, and Japan (Sun., 2013). The sex ratio ofis close to 1:1 in the natural environment. Considering that females sexually mature earlier and are substantially larger than males, the culture of primarily females would help optimize produc- tion. Increasing the female sex ratio has become a hot topic in the field of breeding (Yamamoto 1999; Tian., 2009). Genetic manipulation and hormonal treatment are major methods for regulating sex differentiation in fish (Zhang., 2010). Asis an important gene regu- lating gonadal development and gametogenesis, clarify- ing the molecular mechanisms ofwill help to under- stand its important role in the regulation of reproduction and gender differentiation. Furthermore, exploring the me- chanism of hormone interaction withgenes is im- portant for culturing primarily females and thereby im- proving the economic value of the aquaculture.
In this study, we cloned thegene into elucidate its molecular and genetic features. Then, we identified the localization ofduring gametogenesis and its transcriptional activity in adult tissues and during embryonic development. The transcriptional activity of thepromoter regions was analyzed to investigate the transcriptional regulatory elements. Furthermore, we ana- lyzed the effects of hormones including hCG, 17α-MT, and E2 on, to understand the regulatory mecha- nisms ofduring gonadal development. In general, we analyzed the gene structure and evolutionary status of thegene, and further explored its expression pat- tern and regulatory factors. The results will provide a foundation for further study of gonadal development in.
2 Materials and Methods
2.1 Fish and Embryo Collection
Experimental fish were provided by a commercial hat- chery in Haiyang, Shandong Province, China. Animal ex- periments were performed in accordance with the Regu- lations for the Administration of Affairs Concerning Ex- perimental Animals (State Science and Technology Com- mission of China for No. 2, October 31, 1988. http://www. gov.cn/gongbao/content/2011/content_1860757.htm). Six 1.5-year-old adult fish (three females and three males) were obtained for tissue distribution studies, which in- cluded specimens from the heart, liver, spleen, kidney, brain, gills, muscle, intestines, testes, and ovaries. Body lengths were 310–420mm and body weights were 520– 720g. Gonads forhybridization (ISH) were col- lected and fully measured. In addition, ten different larval stages, including unfertilized eggs, 1-cell, 8-cell, 64-cell, blastula, gastrula, neurula, heart-beating, hatching, and 1- day post-hatch (dph) embryos and larvae were gathered for experiments. All tissues and larvae were frozen in liquid nitrogen and stored at −80℃ for further experiments.
2.2 RNA Extraction and cDNA Synthesis
Total RNA from differenttissues or em- bryoswas extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s proto- col. Then, RNase-free DNase I (TaKaRa, Dalian, China) was used to dislodge any remaining DNA. The concentra- tion and quality of the DNA were determined by 1.5% aga- rose gel electrophoresis and spectrophotometry, respective- ly. cDNA was synthesized with total RNA and random hexamer primers using the Reverse Transcriptase M-MLV Kit (TaKaRa).was selected as reference gene to verify the success of reverse transcription and the quality of the cDNA.
2.3 Gene piwil2 Molecular Cloning
Primers (-core-Fw/Rv, Table 1) designed based on conserved sequences of other teleosts were used to obtain the core cDNA fragment of.Next, agarose gel electrophoresis and a Zymoclean Gel DNA Recovery Kit (Zymo Research, Orange, CA, USA) were used to separate and purify the amplified polymerase chain reac- tion (PCR) products. Then, thePCR products were li- gated into the pMD-19T vector for sequencing (TaKaRa). Thetranscriptome which was previously se- quenced by our laboratory (Wang., 2014) was used to ensure sequence accuracy.
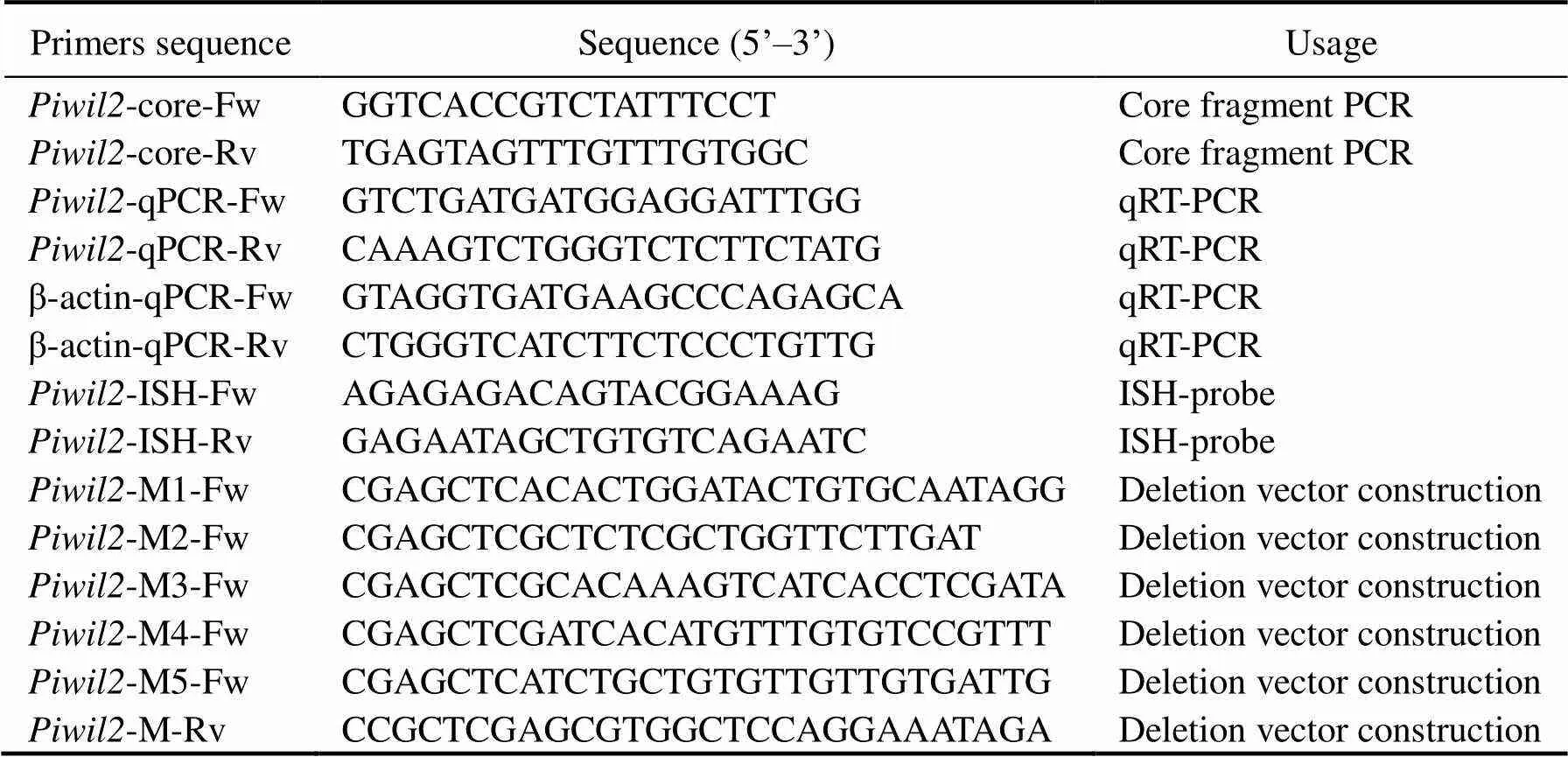
Table 1 Primers used to analyze the Paralichthys olivaceus piwil2 gene
2.4 Sequence Analysis
The BLAST server (http://www.ncbi.nlm.nih.gov/ BLAST/)was used to carry out the homology search for thenucleotide sequence. The highly conservedprotein domains, including the PAZ and PIWI ami- no acid (aa) sequences, were deduced through the SMART program (http://smart.embl-heidelberg.de/). The neighbor- joining (NJ) method was applied to establish the phylo- genetic tree according to the alignment ofamino acid sequences using MEGA 6.0 software (Tamura., 2013). The scale bar is a numerical scale representing the differences between the amino acid sequences of different species. The branch support statistic was 500.
2.5 Quantitative Real-Time PCR (qRT-PCR)
qRT-PCR was used to detect the expression profiles ofstages, and hormone treatments.was selected as a refe- rence gene to measuremRNA expression levels. All primers, including-qPCR-Fw/Rv and- Fw/Rv (Table 1), were designed by the Integrated DNA Technologies website (http://sg.idtdna.com/Primerquest/ Home/Index) in non-conserved domains. The pre-experi- mental amplification was performed to verify primer spe- cificity. qRT-PCR was performed with 2× SYBR Green qPCR MasterMix (Novoprotein, Shanghai, China) using a LightCycler 480 (Roche, Forrentrasse, Switzerland). The reaction volume was 20μL, including 10μL of 2× SYBR qPCR SuperMix, 0.4μL of each primer (10μmolL−1), 1μL of cDNA (20ngμL−1), and 8.2μL of nuclease-free water. The PCR reaction conditions were 95℃ (5min) for pre-incubation followed by 40 cycles at 95℃ (15s), 60℃ (15s) and 72℃ (45s). At least three templates were ana- lyzed to guarantee the accuracy of the experimental re- sults. All data were subjected to the 2−ΔΔCtmethod.
2.6 In situ Hybridization (ISH)
ISH was performed to localize themRNA in gonadal tissues as described previously. The specific pri- mers (-ISH-Fw and-ISH-Rv, Table 1) were designed according towhole-genome sequencingThe sense and anti-sense probes were synthesized using a DIG RNA Labeling Kit (SP6/T7) (Roche, Mannheim, Ger- many), following the manufacturer’s instructions. The paraffin sections were stained and combined with theantisense probe according to a previous study (Gao., 2013).
2.7 Construction of Deletion Vectors and the Luciferase Assay
To reveal thepromoter’s transcriptional regula- tory mechanisms, deleted vectors were constructed by li- gating thepromoter fragments with different sizes, including pGL3-M1 (2083bp), pGL3-M2 (1541bp), pGL3- M3 (1117bp), pGL3-M4 (798bp), and pGL3-M5 (300bp), into the luciferase reporter vector pGL3-Basic. The pri- mer sequences are shown in Table 1. The human 293T cellline was used in the luciferase assay considering the trans- fection efficiency of cells. The human 293T cell line was donated by the Key Laboratory of Marine Genetics and Breeding, Ministry of Education, College of Marine Life Sciences, Ocean University of China. LipoGene™ 2000 PLus Transfection Reagent (US Everbright Inc., Suzhou, China) was used for the transient transfection experiments. The Dual-Luciferase Reporter Assay System (Promega, Mannheim, Germany) was used to perform the dual‐luci- ferase reporter assays after 24h of transfection according to the manufacturer’s instructions. All samples in the dual-luciferase reporter assay were assessed with three replicates.
2.8 In vivo Hormone Treatment
The exogenous hormone hCG is highly similar to HPG axis hormone LH, and has been widely used to manipu- late for spawn in production because of the abundant raw materials. In the same way, 17α-MT and E2 are also read- ily available and have similar function to the steroids in HPG axis. Thus, hCG, 17α-MT, and E2 (Sigma-Aldrich, St. Louis, MO, USA) were selected forexperi- ments to examine the effects of hormones onex- pression. For the hCG experiment, the male and female fish were divided into three groups (=3), respectively, in- cluding a control group (injected with saline), a low con- centration group (injected with 600IUkg−1bodyweight), and a high concentration group (injected with 3000IUkg−1bodyweight). For the 17α-MT experiment, males were di- vided into three groups (=3), including a control group (injected with saline), a low concentration group (injected with 1.5μgkg−1bodyweight), and a high concentration group (injected with 50μgkg−1bodyweight). For the E2 experiment, males were divided into two groups (=3), including a control group (injected with saline) and an E2 group (injected with 3μgkg−1bodyweight). The hormones were injected intraperitoneally. The same hormone treat- ment was administered for the following 5 days and the treated tissue samples were obtained on day 6. Previous research on other teleosts provided the references for the treatment concentrations used in this study (Xiao., 2013;Li., 2018;Wang., 2018). Each group was maintained in a stable condition for 5 days. All gonads were collected on day 6 and were stored at −80℃for fur- ther study.
2.9 In vitro Hormone Treatment
The gonads of three male fish were dissected and wash- ed three times in phosphate-buffered saline (PBS) con- taining 1% penicillin-streptomycin solution. The tissues were minced into small pieces (mean length=1.5cm) in a Petri dish. Then the isometric segments were cultured in a 12- well culture plate for furtherhormone treatments.
Four different concentrations of 17α-MT and E2, such as 10−6, 10−8, 10−10, and 10−12molL−1, were added to the treatment groups, respectively. The tissues from each well were collected after 4h of treatment, and stored for further qRT-PCR to determineexpression.
2.10 Statistical Analysis
Data are expressed as mean±standard error (SE) of at least three independent experiments, and were analyzed by one-way analysis of variance. All qRT-PCR data were analyzed using SPSS 20.0 software (IBM, Armonk, NY, USA).-values <0.05 were considered significant.
3 Results
3.1 piwil2 Cloning and Sequence Analysis
The open reading frame (ORF) of the2 genewas obtained from thetestes by PCR. The full- length ORF was 3192bp and encoded 1063 amino acids. An analysis of the protein structure identified PAZ and PIWI as conserved domains in(Fig.1A). The PAZ domain was located at positions 477–616 at the N-ter- minal and contained 140 amino acids. The PIWI domain, which contained 292 amino acids, was located at the C- terminal at positions 758–1049. The PAZ (Fig.1B) and PIWI domains (Fig.1C) inwere highly con- served after a comparison with other teleosts, suggesting that these two domains may play an essential role main- taining functions of thegeneamong different spe- cies.
3.2 Homology and Phylogenetic Analyses
A phylogenetic tree was established using the amino acid sequences and the NJ method to demonstrate the evolutionary relationships betweenand other vertebrates (Fig.2). As expected, the twohomologs can be branched into two main clades, called theandclades in the phylogenetic tree. Only a few mammals, such as humans and mice, possess theandclades.was clearly cluster- ed with other teleosts and belonged to the teleost sub- group, while the others formed a tetrapod subgroup.
The gene structure was highly conserved betweenand other species, as the structures had the same number of exons and introns (Fig.3). The exon and intron data were uploaded to the supplementary file. As the un- translated region (UTR) sequences of various species were incomplete, only the ORF sequences were the focus of our analysis. Twenty-three exons and 23 introns were found inafter comparingcDNA and genomic DNAFor most species, there was an intron in- serted in the 5’UTR of, and the 5’UTR sequence is always part of one-to-a-few first exons. The similarity of the genomic organization revealed the conservative ofgenes among teleosts during the evolution.
3.3 piwil2 Expression Patterns in Different Tissues
ThemRNA expression levels in different tissues, including heart, brain, liver, intestines, kidney, spleen, mu- scle, gills, testes, and ovaries were analyzed by qRT-PCR (Fig.4A). The results showed thatwas particularly strongly expressed in the testes and ovaries, and showed a sexually dimorphic expression pattern. Furthermore,was also expressed in other somatic tissues at low levels.
ISH was used to further explore the tissue-specific ex- pression pattern of(Fig.5). Strong positive signals were detected throughout the cytoplasm of oogonia and oocytes in ovarian sections. The signals were concen- trated in the Sertoli cells and spermatocytes of testicular sections, with no signal in spermatids. These results indi- cate that thegene might play a crucial role in ga- metogenesis.
3.4 piwil2 Expression Pattern During Embryogenesis
To determine theexpression pattern during em- bryogenesis,mRNA levels were analyzed at dif- ferent embryonic development stages by qRT-PCR (Fig.4B).was maternally inherited as it was highly ex- pressed from the 1-cell to the blastula stage. The expres- sion levels suddenly decreased from the beginning of the gastrula stage and continued to decrease until birth. Con- sidering the difference inexpression levels at dif- ferent embryogenesis stages, we speculate thatmight perform different functions at these stages.
3.5 The piwil2 Core Promoter Region and Related Transcriptional Regulatory Elements
Five different deletion fragments of thepromo- ter region were obtained and connected to upstream of the luciferase reporter gene to detect the core promoter region (Fig.6). The transcriptional initiation site (ATG) for the pro- moter deletion assay was designated as +1 and theupstream 5’-flanking sequence from −2143bp to −60bp was analyzed after removing all introns.expres- sion increased significantly in the M5 fragment. The M4 fragment included M5, but the expression level ofexpression in the M2 region decreased rapidly and then recovered in the M1 fragment. Therefore, the dele- tion of the fragment between −360bp to −60bp seemed to be the core promoter region of.
Then, the transcription elements of these five different deletion fragments were analyzed. Some transcription ele- ments, such as SMAD, NKX6, CAAT, and NF-1F, were detected in the core promoter region (M5). M2 and M4 highly inhibitedexpression, in which GATA, CART, IRFF, and HOMF were found. BRAC, NF-1F, HNF, and AP1R were detected in the M1 and M3 regions, neutral- izing the inhibitory effect of the previously deleted frag- ments. All of these transcriptional elements may be close- ly related to regulation ofexpression and may fur- ther affect gametogenesis and gonadal development.
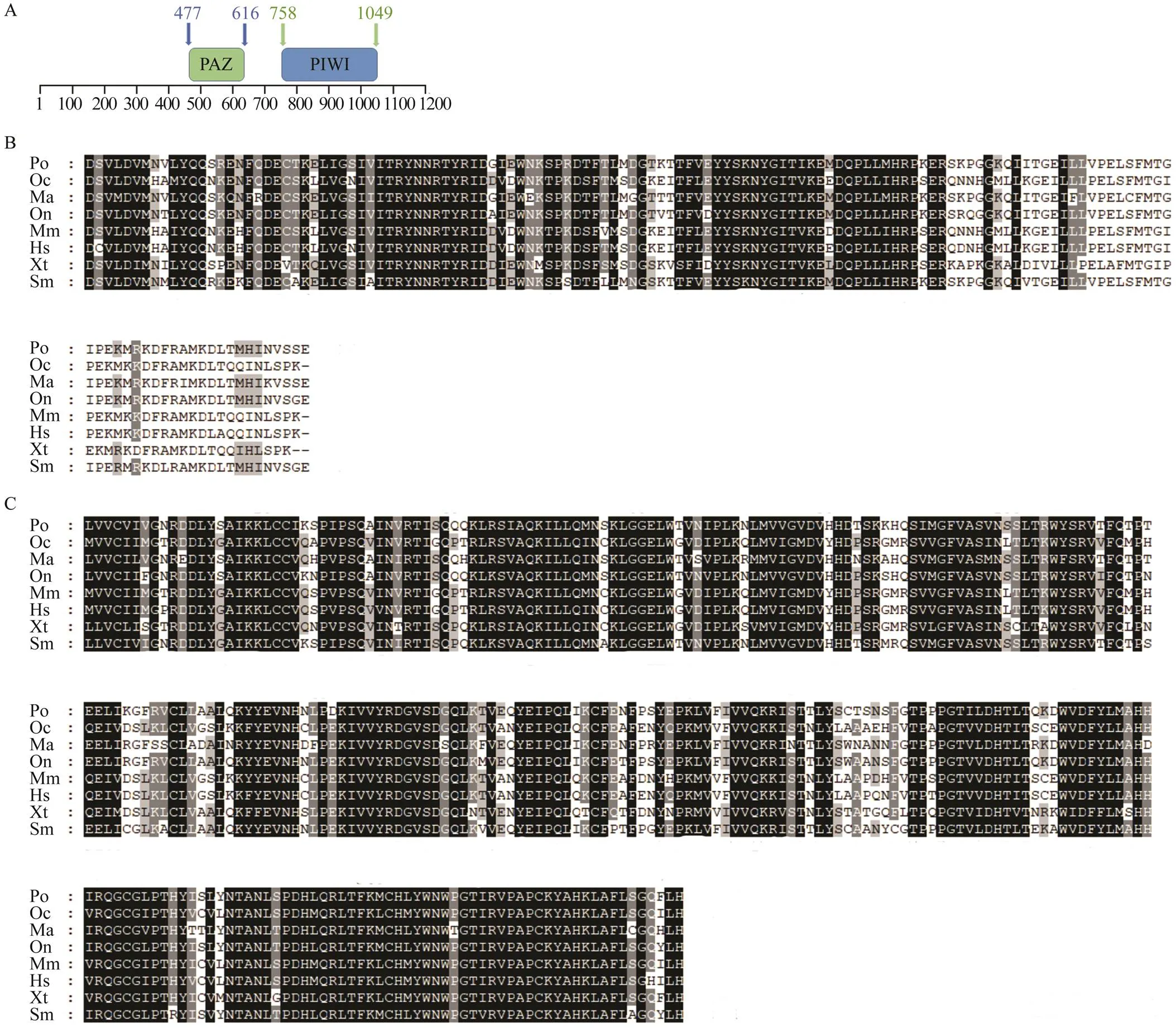
Fig.1 Amino acid sequence and structural domain analysis of the piwil2 gene. (A) The amino acid sequence analysis of the piwil2 gene. The green square represents the PAZ domain and the blue square represents the PIWI domain. The amino acids positions of the structural domain are respectively marked in the picture. (B) Alignment of the piwil2 PAZ domain among different species. Dark shadowed regions represent the same amino acid residues and the grey shadowed regions represent similar amino acids residues. (C) Alignment of the piwil2 PIWI domain among different species. The selected species include Paralichthys olivaceus (Po, XM_020094094.1); Oryctolagus cuniculus (Oc, XM_008249876.1); Mono- pterus albus (Ma, XM_020590301.1); Oreochromis niloticus (On, XM_003445662.4); Mus musculus (Mm, NM_001364321.1);Homo sapiens (Hs, NM_018068.5); Xenopus tropicalis (Xt, XM_018091922.1);and Scophthalmus maximus (Sm, KY_ 123251.1). The amino acid sequences are from the GenBank accession numbers in brackets.
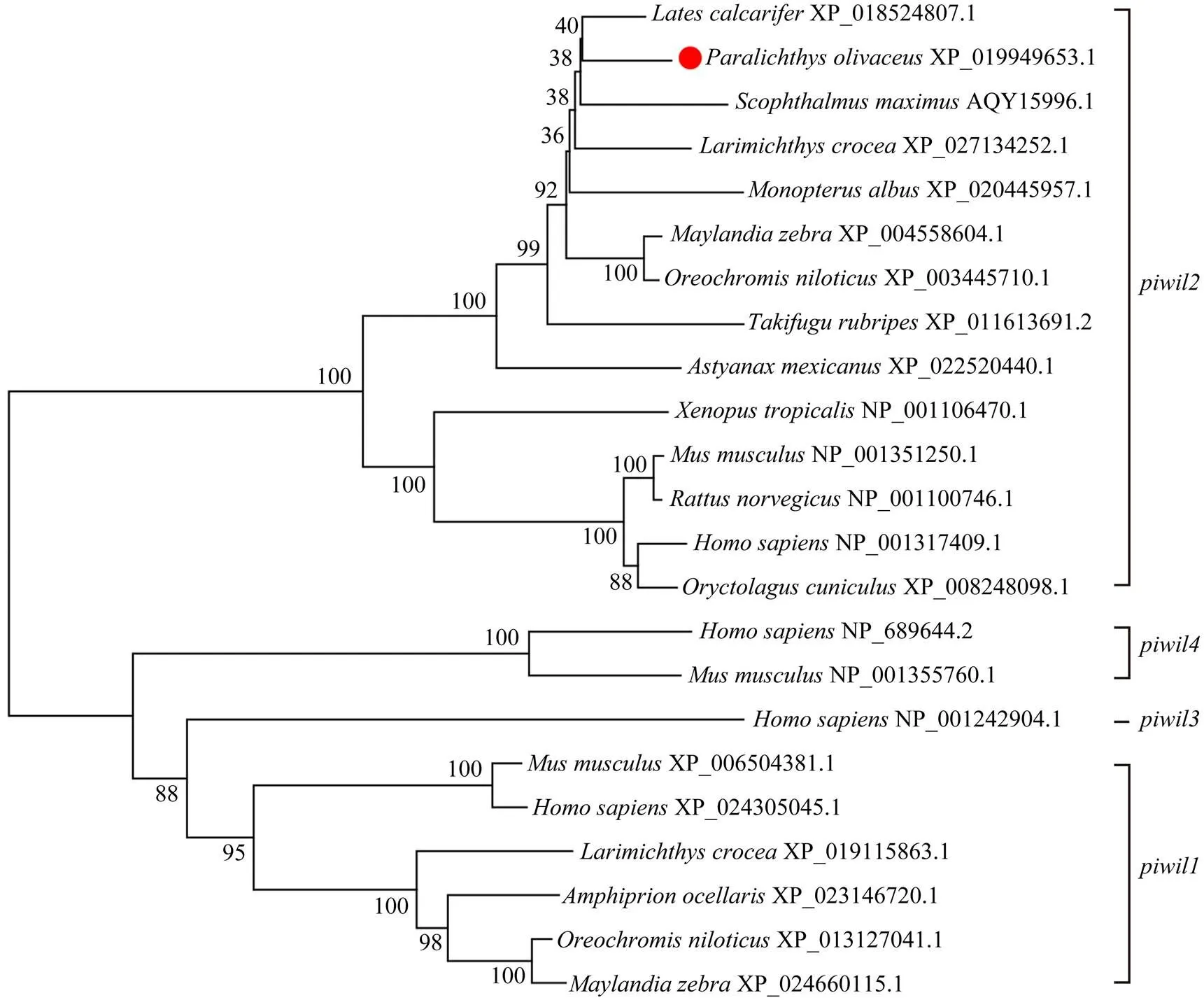
Fig.2 piwil2 phylogenetic tree in Paralichthys olivaceus with other representative vertebrates, predicted by the amino acid sequences. The species and GenBank accession numbers are shown.
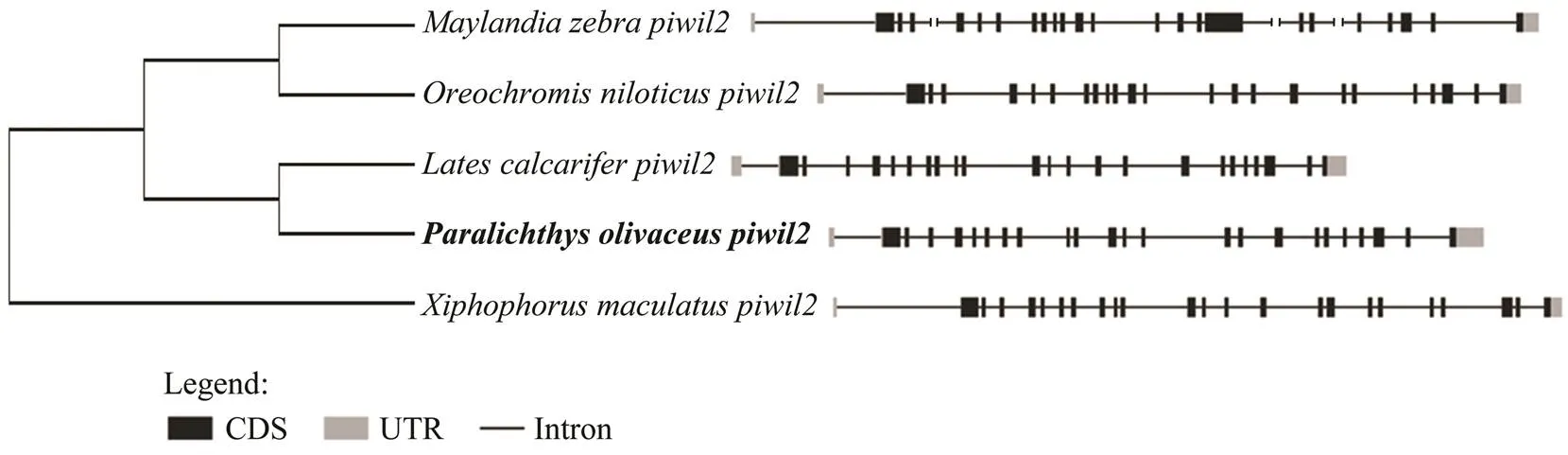
Fig.3 Comparison of piwil2 genomic structure between Paralichthys olivaceus and other teleosts. The black areas repre- sent exons, the lines represent introns, and the gray areas represent the untranslated region (UTR) area.
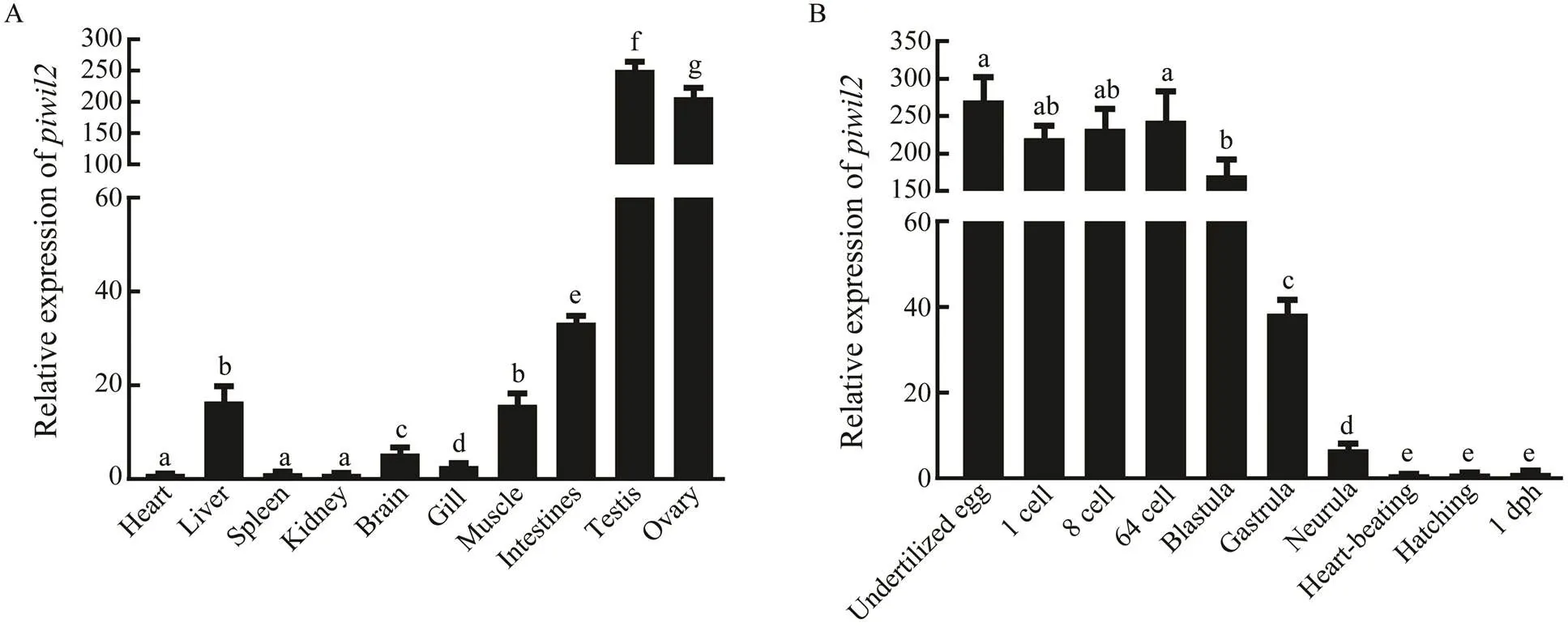
Fig.4 The relative expression levels of piwil2 in differenttissues and stages of embryonic development.(A) The piwil2 expression patterns in various tissues of males and females. (B) The piwil2 expression pattern during different stages of embryonic development. All data are expressed as mean±SEM. from three separate individuals (n=3). Different super- scripts indicate that P<0.05.
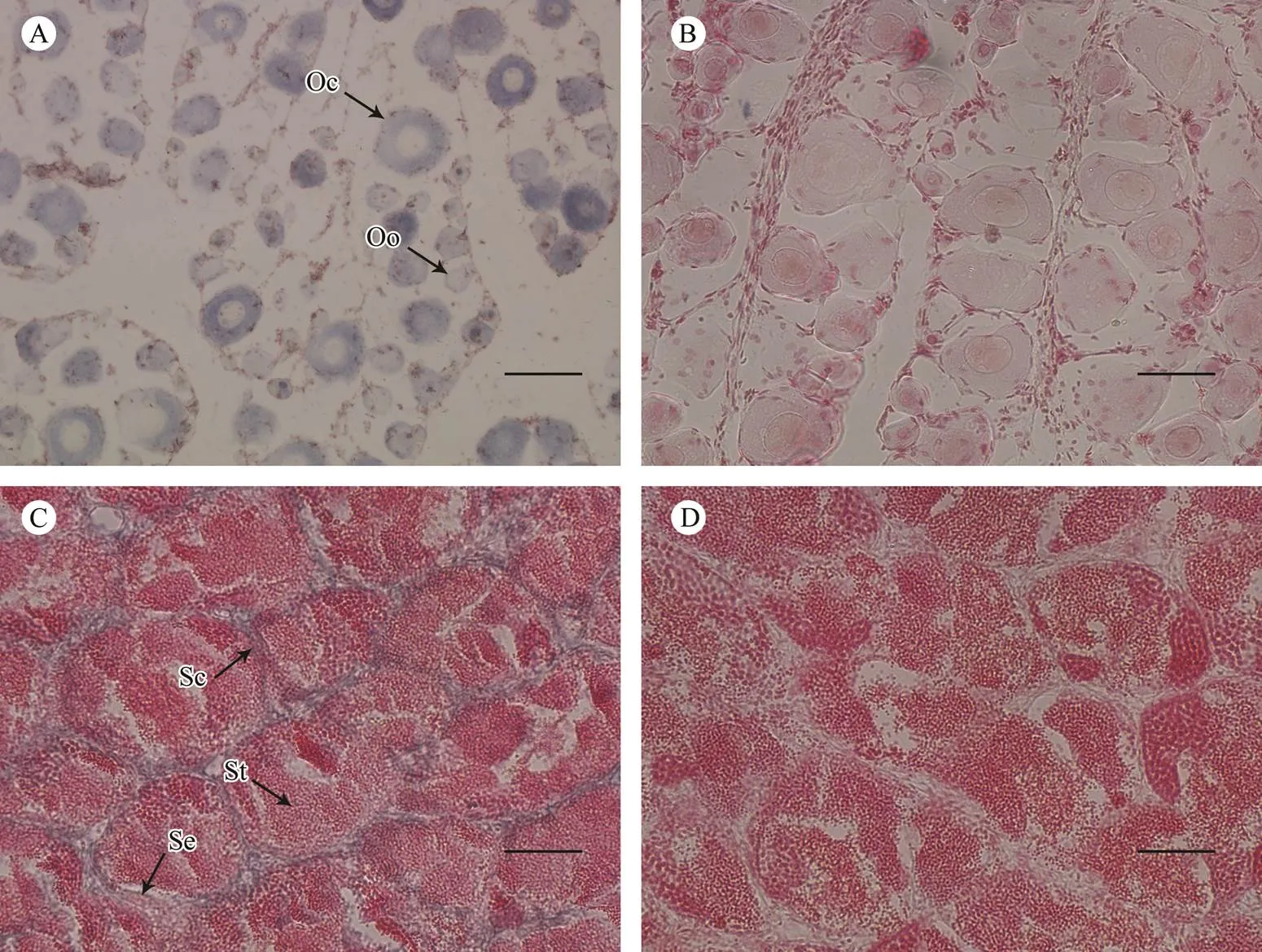
Fig.5 The mRNA distribution of piwil2 in gonadal cells. The antisense probe labeled with DIG was stained blue (A and C). The sense probe is the negative control and the hybridization was unstained (B and D). Se, Sertoli cells; Sc, spermato- cytes; St, spermatid; Oo, oogonia; Oc, oocytes. Scale bars=50µm.
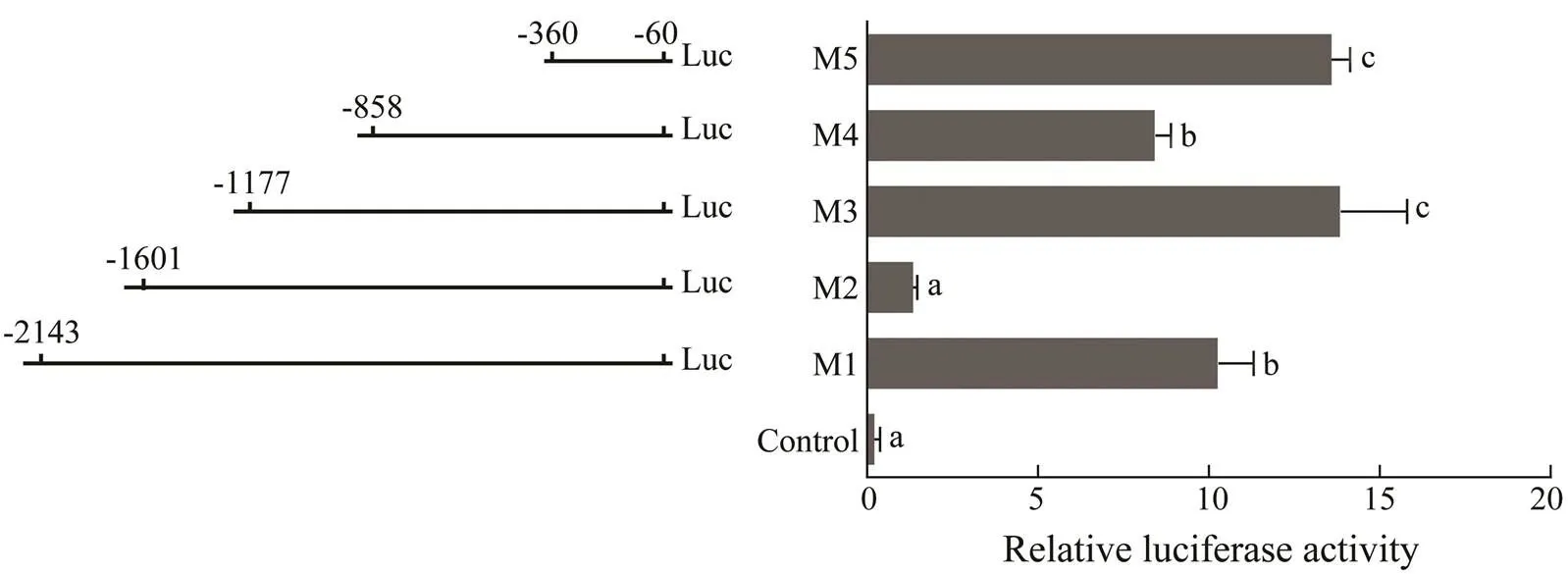
Fig.6 Transcriptional activities of deleted vectors measured by the luciferase assay. Left, A series of deletion fragments linked with the luciferase gene in the pGL3 vector. Right, Relative activities of the deleted vectors pGL3 M1-M5 deter- mined by the luciferase assay. Different superscripts indicate that P<0.05.
3.6 Effect of in vivo and in vitro Hormone Treatments on piwil2
Theexpression levels were analyzed under the hormone treatments tounderstand the effects on thegene. As the result, hCG suppressedexpres- sion in males (Fig.7A). In females, the high concentration of hCG had an inhibitory effect on, while the low concentration promotedexpression (Fig.7B). For 17α-MT, both the high and low concentrations increasedexpression in males (Fig.7C). Additionally,transcription was promoted in response to E2 (Fig.7D).
Different concentrations of sex hormones were used for thetreatment to determine themRNA le- vels. As shown in Fig.7E, all 17α-MT-treated groups show- ed a slight decrease inmRNA levels compared with the control group, particularly when the concentrationwas 10−8molL−1. However,expression declined slightly when the E2 concentration was 10−6molL−1and increased when the E2 concentration was 10−12molL−1(Fig.7F).
4 Discussion
Thegene is involved in many complex processes, such as spermatogenesis, egg activation, and fertilization (Wang., 2018). Piwi proteins play a crucial role dur- ing gonadal development and gametogenesis (Qiao., 2002), and when combined with piRNA, they participate in the Ping-Pong cycle to regulate genomic integrity and retrotransposon repression (Aravin., 2008). Thegene has been extensively researched in vertebrates and invertebrates, with some studies employing model organ- isms, including mice and zebrafish (Kuramochi-Miyaga- wa., 2001; Thomson and Lin, 2009). However, infor- mation regardingTherefore, in this study, we identified and explored the fundamental functions of
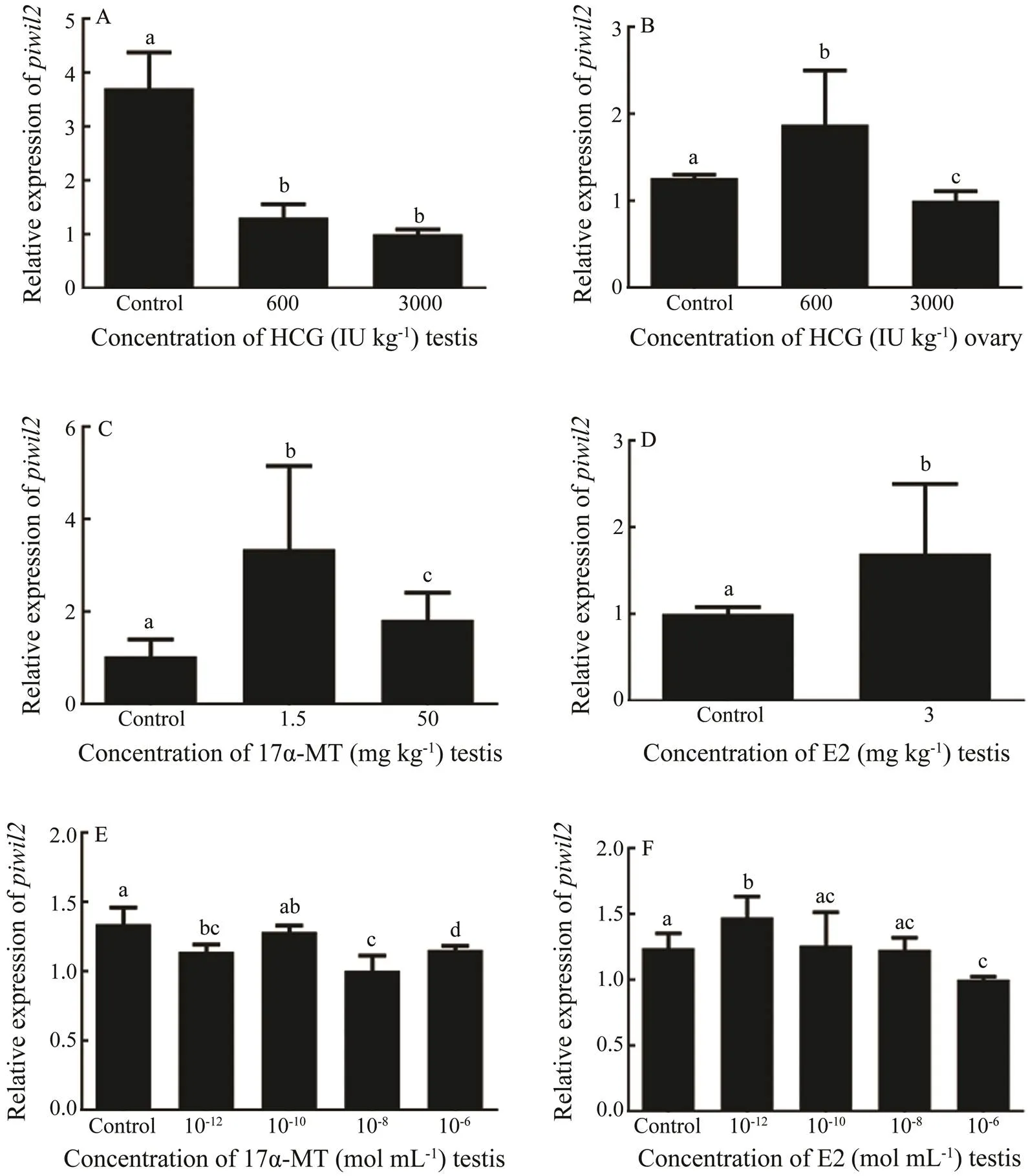
Fig.7 The relative expression levels of piwil2 after the hormone treatments.(A) In vivo hCG treatment of testis. (B)In vivo hCG treatment of ovary. (C) In vivo 17α-MT treatment of testis. (D) In vivo E2 treatment of testis. (E) In vitro17α- MT treatment of testicular cells. (F) In vitroE2 treatment of testicular cells. All data are shown as mean±SEM (n=3 in every group). Different superscripts indicate that P<0.05.
4.1 Gene piwil2 Is Highly Conserved in Vertebrates
Genefromwas cloned to investigate its potential functionsThefromcontained the same PAZ and PIWI domains as in other teleost species, indicating thatplays a conserved role in different species. Evidence gathered from a phy- logenetic tree showed thatwas more consistent with the teleost branch and clearly separate from tetrapods. Therefore, it was concluded thatis highly conserved and might participate in the same regu- latory mechanisms in different species.
4.2 Piwil2 Transcripts Are More Abundant in the Testes than in the Ovaries
isa germline-specific gene that is enriched in gonadal tissues (Wen., 2018). Previous studies have shown thatis expressed in the testes and ovaries and that its expression is higher in testes than in ovaries(Mishima., 2008). In addition,is highly ex- pressed in the cytoplasm of oocytes and oogonia of(Zhao., 2012). In this study, the expres- sion pattern ofwas similar with that described in previous studies with other species, demon- strating thathas a crucial function in germline stem cell maintenance and gametogenesis. In addition,expression was relatively lower in somatic tissues, indi- cating thathas other essential functions beyond reproduction, which need to be further explored.
4.3 Piwil2 May Be Involved in the Regulation of Embryogenesis
Some studies have reported that PIWI and piRNA re- gulate gene expression at the post-transcriptional level and participate in the regulation of embryonic development, gender determination, and other events (Schwager.,2015). It has been inferred that the piRNA pathway is highly active during early embryonic development, asis expressed at high levels at this time (Rouget., 2010). The genome recombination and chromatin remo- deling processes also occur during early embryogenesis (Sunanaga., 2010;Zhang., 2010), suggesting that the highexpression levels during this period regulate transcription and ensure stability of the genome. Theexpression pattern during embryogenesis inwas similar to those in other species, such asand(Zhang., 2010; Wang., 2017). Therefore, the present study provides strong evi- dence that the-encoded protein participates in em- bryogenesis and has conserved functions(Li., 2011).
4.4 Transcription Elements Regulate piwil2 Expression
Defining the core promoter region of thegene has importance for revealing the transcriptional regula- tory mechanisms and for further regulatingexpres- sion (Chang., 2015). In this study, the −360 to −60bp region in the5’-UTR contained some important transcriptional elements (SMAD, NKX6, CAAT, and NF- 1F) that significantly enhanced promoter activity and was, therefore, suggested to be the core promoter region of thegene. The −858 to −360bp and the −1601 to −1177bp sections in the 5’-flanking region were regions whereexpression was suppressed. As some studies have demonstrated that piwi is inhibited by transcriptional ele- ments (Sohn., 2014; Chang., 2015), the tran- scriptional elements in this fragment (GATA, CART, IRFF, and HOMF) may downregulate the activation of the pro- moter in. The transcriptional regulatory me- chanisms present in the −1177 to −858bp and −2143 to −1607bp regions played a positive role, thereby counter- acting the inhibitory effects of the previous regions. BRAC, NF-1F, HNF, and AP1R in these regions potentially pro- motedexpression. Although these results are based on a bioanalytical level of analysis and prediction, these transcriptional elements may play an important role regu- latingexpression, and their detailed mechanisms inneed to be further studied.
4.5 The HPG Axis Affects piwil2 Expression
Gametogenesis and gonadal development in teleosts are regulated by a series of complex systems. Previous stud- ies have shown that the HPG axis is an important neuro- endocrine regulatory system (Fuqua and Rogol, 2013). Therefore, in this study, hCG, 17α-MT, and E2 were se- lected to verify their effects onexpressionThey all play a crucial role in gametogenesis by regulating the piRNA pathway(Scholz and Gutzeit, 2000; Zhang., 2010; Miura., 2013; Zhou., 2015). Asis a key gene in the piRNA pathway,may be involved in the regulation of HPG axis hormones during gameto- genesis.
The expression ofdecreased in males treated withhCG. The high hCG concentration inhibitedexpres- sion in females, whereas the low concentration promoted expression. These results are similar to those previously reported in Nile tilapia (Xiao., 2013). Additionally, 17α-MT and E2 promotedexpression in males. As 17α-MT masculinizes the gonads and promotes sperma- togenesis and a lack of E2 disrupts early spermatogenesis (Lee., 1986; Murata., 2002), we hypothesized that sex hormones regulate the formation of spermatogenesis, leading to a change ofexpression (Hansson., 1976).
Thetreatment was designed to explore the ef- fect of complex regulatory system including cytokines and other factors onexpression at the cellular level after the hormone treatments. 17α-MT decreasedexpression in testes, which was opposite to theresult. We inferred that this result may be due to the complex and interrelated regulatory systems in individuals. The HPG axis, an important hormone regulatory path- way, is affected by many feedback loops (Rosvall., 2016). In thetreatment, a high 17α-MT concentra- tion not only promoted spermatogenesis, but also regu- lated the secretion of gonadotropins through negative feed- back control. All of these mechanisms masked the true level ofresponding to 17α-MT. A low E2 concentration increasedexpression, whichwas con- sistent with theexperiment. A higher E2 concen- tration had a negative effect on cell activities, leading to the inhibition ofexpression. These results suggest thatwas affected by the HPG axis and participates in gametogenesis(Bohórquez., 2017).
5 Conclusions
In this study, we investigated the basic functions and regulatory factors ofThecDNA sequence from the testes ofwas iden- tifiedA genetic structural analysis showed thatwas conserved in different species. The tissue expression pat- tern analysis demonstrated that theexpression le- vel was higher in testes than in other tissues. The tissue localization results suggested thatwas highly ex- pressed in the cytoplasm of oogonia and oocytes in the ovaries, as well as in Sertoli cells and spermatocytes in the testes. Moreover, the core promoter region was detect- ed, and some major transcriptional elements were identi- fied to explore thetranscriptional regulatory me- chanism. In addition,was maternally inherited and determined to play a vital role in the mechanism underly- ing hormonal regulation. Taken together, these results pro- vide a theoretical basis for further exploring the role of hormones in the regulation of the expression ofgene.
Acknowledgements
This study was supported by the National Natural Sci- ence Foundation of China (No. 31672646) and the Natu- ral Science Foundation of Shandong Province (No. ZR 2017MC072).
Aravin, A. A., Sachidanandam, R., Bourc’his, D., Schaefer, C., Pezic, D., Toth, K. F., Bestor, T., and Hannon, G. J.,2008. A piRNA pathway primed by individual transposons is linked toDNA methylation in mice.,31(6): 785-799, DOI:10.1016/j.molcel.2008.09.003.
Bohórquez, M. O. T., Mechaly, A. S., Elisio, M., Chalde, T.,Canosa, L. F., Miranda, L. A., and Somoza, G. M.,2017. Kiss-peptins and their receptors in the brain-pituitary-gonadal axis of: Their relationship with gameto- genesis along the reproductive cycle., 252: 209-218, DOI: 10.1016/j.ygcen.2017.06.028.
Carmell, M. A., Xuan, Z., Zhang, M. Q., and Hannon, G. J.,2002. The Argonaute family: Tentacles that reach into RNAi, deve- lopmental control, stem cell maintenance, and tumorigenesis.,16(21): 2733-2742, DOI:10.1101/gad.1026102.
Chang, G., Chen, R., Xu, L., Ma, T., Wang, H., Chen, J., Zhang, Y., Li, Z., Wan, F., Guo, X., Xu, Q., Zhao, M., and Chen, G.,2015. DNA methylation and NF-Y regulateexpression during chicken spermatogenesis.,162: 95-103, DOI: 10.1016/j.anireprosci.2015.09.016.
Filipowicz, W.,2005. RNAi: The nuts and bolts of the RISC ma-chine.,122(1): 17-20.
Fuqua, J. S., and Rogol, A. D.,2013. Neuroendocrine alterations in the exercising human: Implications for energy homeostasis.,62(7): 911-921, DOI: 10.1016/j.cell.2005.06.023.
Gao, J., Wang, J., Jiang, J., Fan, L., Wang, W., Liu, J., Zhang, Q., and Wang, X.,2013. Identification and characterization of a nanog homolog in Japanese flounder ().,531(2): 411-421, DOI: 10.1016/j.gene.2013.08.030.
García-Lopez, A., de Jonge, H., Nobrega, R. H., de Waal, P. P., van Dijk, W., Hemrika, W., Taranger, G. L., Bogerd, J., and Schulz, R. W.,2010. Studies in zebrafish reveal unusual cellu-lar expression patterns of gonadotropin receptor messenger ribonucleic acids in the testis and unexpected functional dif- ferentiation of the gonadotropins.,151(5): 2349-2360, DOI: 10.1210/en.2009-1227.
Gauthier-Clerc, S., Pellerin, J., and Amiard, J.,2006. Estradiol-17β and testosterone concentrations in male and female(Mollusca bivalvia) during the reproductive cycle.,145(2): 133-139, DOI: 10.1016/j.ygcen.2005.08.004.
Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R., andHannon, G. J.,2001. Argonaute2, a link between genetic and biochemical analyses of RNAi.,293(5532): 1146-1150, DOI: 10.1126/science.1064023.
Hansson, V., Calandra, R., Purvis, K., Ritzen, M., and French, F. S.,1976. Hormonal regulation of spermatogenesis.,34: 187-214.
He, L., and Hannon, G. J.,2004. MicroRNAs: Small RNAs with a big role in gene regulation.,5(7): 522, DOI: 10.1038/nrg1379.
Houwing, S., Berezikov, E., and Ketting, R. F.,2008.is re- quired for germ cell differentiation and meiosis in zebrafish.,27(20): 2702-2711, DOI: 10.1038/emboj.2008.204.
Houwing, S., Kamminga, L. M., Berezikov, E., Cronembold, D., Girard, A., Van, H. D. E., Filippov, D. V., Blaser, H., Raz, E., Moens, C. B., Plasterk, R. H. A., Hannon, G. J., Draper, B. W., and Ketting, R. F.,2007. A role forand piRNAs in germ cell maintenance and transposon silencing in zebrafish.,129(1): 69-82, DOI: 10.1016/j.cell.2007.03.026.
Kuramochi-Miyagawa, S., Kimura, T., Yomogida, K., Kuroiwa, A.,Tadokoro, Y., Fujita, Y., Sato, M., Matsuda, Y., and Nakano, T.,2001. Two mouse piwi-related genes:and.,108(1-2): 121-133, DOI: 10.1016/s0925-4773(01)00499-3.
Lee, C.S., Tamaru, C., Banno, J., and Kelley, C.,1986. Inf- luence of chronic administration of LHRH-analogue and/or 17α-methyltestosterone on maturation in milkfish,.,59(2): 147-159.
Lee, S. L. J., Horsfield, J. A., Black, M. A., Rutherford, K., Fisher, A., and Gemmell, N. J.,2017. Histological and transcriptomic effects of 17α-methyltestosterone on zebrafish gonad develop- ment.,18(1): 557, DOI: 10.1186/s12864-017-3915-z.
Li, D., Sun, H., Deng, W., Tao, D., Liu, Y., and Ma, Y.,2011.antagonizes Bmp signaling to regulate dorsal-ventral pattern- ing during zebrafish early embryogenesis.,28(6): 397-402, DOI: 10.2108/zsj.28.397.
Li, X., Yu, H., Wang, Y., Liu, X., Liu, Y., Qu, J., and Wang, X.,2018. Roles of twogenes during gonadal development in Japanese flounder: Sex differentiation, spermatogenesis and gonadal function maintenance.,19(2): 512, DOI: 10.3390/ijms19020512.
Manakov, S. A., Pezic, D., Marinov, G. K., Pastor, W. A., Sa- chidanandam, R., and Aravin, A. A.,2015.andhavedifferential effects on piRNA biogenesis and DNA methyla- tion.,12(8): 1234-1243, DOI: 10.1016/j.celrep.2015.07.036.
Mishima, T., Takizawa, T., Luo, S.S., Ishibashi, O., Kawa- higashi, Y., Mizuguchi, Y., Ishikawa, T., Mori, M., Kanda, T., Goto, T., and Takizawa, T.,2008. MicroRNA (miRNA) clo- ning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary.,136(6): 811-822, DOI: 10.1530/REP-08-0349.
Miura, S., Kobayashi, Y., Bhandari, R. K., and Nakamura, M.,2013. Estrogen favors the differentiation of ovarian tissues in the ambisexual gonads of anemonefish.,319(10): 560-568, DOI: 10.1002/jez.1818.
Murata, Y., Robertson, K., Jones, M., and Simpson, E.,2002. Ef- fect of estrogen deficiency in the male: The ArKO mouse mo- del.,193(1-2): 7-12, DOI: 10.1016/s0303-7207(02)00090-4.
Pan, Y., Hu, M., Liang, H., Wang, J., and Tang, L.,2012. The ex- pression of the PIWI family membersandin mice testis is negatively affected by estrogen.,350(1): 177-181, DOI: 10.1007/s00441-012-1447-z.
Pandey, R., Homolka, D., Olotu, O., Sachidanandam, R., Kotaja, N., and Pillai, R. S.,2018. Exonuclease domain-containing 1 enhancespiRNA biogenesisits interaction with.,24(13): 3423-3432, DOI: 10.1016/j.celrep.2018.08.087.
Qiao, D., Zeeman, A., Deng, W., Looijenga, L., and Lin, H.,2002. Molecular characterization of, a human member of thegene family whose overexpression is correlated to se- minomas.,21(25): 3988-3999, DOI: 10.1038/sj.onc.1205505.
Rosvall, K. A., Bergeon Burns, C. M., Jayaratna, S. P., Dossey, E. K., and Ketterson, E. D.,2016. Gonads and the evolution of hormonal phenotypes.,56(2): 225-234, DOI: 10.1093/icb/icw050.
Rouget, C., Papin, C., Boureux, A., Meunier, A., Franco, B., Ro- bine, N., Lai, E. C., Pelisson, A., and Simonelig, M.,2010. Maternal mRNA deadenylation and decay by the piRNA path- way in the earlyembryo.,467(7319): 1128-1132, DOI: 10.1038/nature09465.
Scholz, S., and Gutzeit, H.,2000. 17-α-ethinylestradiol affects reproduction, sexual differentiation and aromatase gene ex- pression of the medaka ().,50(4): 363-373, DOI: 10.1016/s0166-445x(00)00090-4.
Schultz, N., Hamra, F. K., and Garbers, D. L.,2003. A multitude of genes expressed solely in meiotic or postmeiotic sperma- togenic cells offers a myriad of contraceptive targets.,100(21): 12201-12206, DOI: 10.1073/pnas.1635054100.
Schwager, E. E., Meng, Y., and Extavour, C. G.,2015.andare required for mitotic integrity in early embryogenesis in the spider.,402(2): 276-290, DOI: 10.1016/j.ydbio.2014.08.032.
Sun, P., You, F., Ma, D., Li, J., and Zhang, P.,2013. Sex steroid changes during temperature‐induced gonadal differentiation in(Temminck & Schegel, 1846).,29(4): 886-890.
Sunanaga, T., Inubushi, H., and Kawamura, K.,2010.‐ex- pressing hemoblasts serve as germline stem cells during post- embryonic germ cell specification in colonial ascidian,.,52(7): 603-614, DOI: 10.1111/j.1440-169X.2010.01196.x.
Taborska, E., Pasulka, J., Malik, R., Horvat, F., Jenickova, I., Matosevic, Z. J., and Svoboda, B.,2019. Restricted and non-essential redundancy of RNAi and piRNA pathways in mouse oocytes.: 678177, DOI: 10.1371/journal.pgen.1008261.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S.,2013. MEGA6: Molecular evolutionary genetics analysis version 6.0.,30(12): 2725-2729, DOI: 10.1093/molbev/mst197.
Thomson, T., and Lin, H.,2009. The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect.,25: 355-376, DOI: 10.1146/annurev.cellbio.24.110707.175327.
Tian, Y., Chen, S., Xu, T., Deng, H., and Ding, H.,2009. The com- parison of growth performances of Japanese flounder () families and selection of parents with good trait.,33(6): 901-911.
Wang, H., Wang, B., Liu, J., Li, A., Zhu, H., Wang, X., and Zhang, Q.,2018.gene is regulated by hypothalamic-pituitary-gonadal axis in turbot (): A different effect in ovaries and testes.,658: 86-95, DOI: 10.1016/j.gene.2018.03.016.
Wang, H., Wang, B., Liu, X., Liu, Y., Du, X., Zhang, Q., and Wang, X.,2017. Identification and expression ofturbot, with implications of the in-volvement in embryonic and gonadal development.,208: 84-93, DOI: 10.1016/j.cbpb.2017.04.007.
Wang, W., Wang, J., You, F., Ma, L., Yang, X., Gao, J., He, Y., Qi, J., Yu, H., Wang, Z., Wang, X., Wu, Z., and Zhang, Q.,2014. Detection of alternative splice and gene duplication by RNA sequencing in Japanese flounder,.,4(12): 2419-2424, DOI: 10. 1534/g3.114.012138.
Wen, X., Wang, D., Li, X., Zhao, C., Wang, T., Qian, X., and Yin, S.,2018. Differential expression of twoorthologs during embryonic and gonadal development in pufferfish,.,219: 44-51, DOI: 10.1016/j.cbpb.2018.03.005.
Xiao, J., Zhou, Y., Luo, Y., Zhong, H., Huang, Y., Zhang, Y., Luo, Z., Ling, Z., Zhang, M., and Gan, X.,2013. Suppression effect of LHRH-A and hCG onexpression in testis of Nile tilapia.,189: 43-50, DOI: 10.1016/j.ygcen.2013.04.021.
Yamamoto, E.,1999. Studies on sex-manipulation and produc- tion of cloned populations in hirame,(Temminck et Schlegel).,173(1-4): 235-246.
Zhang, D., Duarte-Guterman, P., Langlois, V. S., and Trudeau, V. L.,2010. Temporal expression and steroidal regulation of piRNA pathway genes (,,) during Silurana () tropicalis embryogenesis and early larval development.,152(2): 202-206, DOI: 10.1016/j.cbpc.2010.04.005.
Zhang, L., Liu, W., Shao, C., Ning, Z., Li, H., Liu, K., Dong, Z., Qi, Q., Zhao, W., and Chen, S.,2014. Cloning, expression and methylation analysis of).,18: 45-54, DOI: 10.1016/j.margen.2014.04.004.
Zhao, C., Zhu, W., Yin, S., Cao, Q., Zhang, H., Wen, X., Zhang, G., Xie, W., and Chen, S.,2018. Molecular characterization and expression ofandduring gonadal develop- ment and treatment with HCG and LHRH-A2 in.,647: 181-191, DOI: 10.1016/j.gene.2018.01.038.
Zhao, H., Duan, J., Cheng, N., and Nagahama, Y.,2012. Speci- fic expression ofand) germ cells.,418(4): 592-597, DOI: 10.1016/j.bbrc.2011.12.062.
Zhou, Y., Hao, G., Zhong, H., Wu, Q., Lu, S., Zhao, Q., and Liu, Z.,2015. Human chorionic gonadotropin promotes expression of protein absorption factors in the intestine of goldfish ().,14(3): 8306-8313, DOI: 10.4238/2015.July.27.19.
Zhou, Y., Wang, F., Liu, S., Zhong, H., Liu, Z., Tao, M., Zhang, C., and Liu, Y.,2012. Human chorionic gonadotropin sup- presses expression ofin common carp () ovaries.,176(2): 126-131, DOI: 10.1016/j.ygcen.2011.11.044.
Zhou, Y., Zhong, H., Liu, S., Yu, F., Hu, J., Zhang, C., Tao, M., and Liu, Y.,2014. Elevated expression ofand piRNAs in ovaries of triploid crucian carp.,383(1-2): 1-9, DOI: 10.1016/j.mce.2013.11.019.
. Tel: 0086-532-82031986
E-mail: yuhaiyang@ouc.edu.cn
January 7, 2020;
March 3, 2020;
June 19, 2020
(Edited by Qiu Yantao)
杂志排行
Journal of Ocean University of China的其它文章
- Numerical Simulation and Risk Analysis of Coastal Inundation in Land Reclamation Areas: A Case Study of the Pearl River Estuary
- Variation of Yellow River Runoff and Its Influence on Salinity in Laizhou Bay
- Cold Water in the Lee of the Batanes Islands in the Luzon Strait
- Preliminary Design of a Submerged Support Structure for Floating Wind Turbines
- Inversion of Oceanic Parameters Represented by CTD Utilizing Seismic Multi-Attributes Based on Convolutional Neural Network
- Trace-Norm Regularized Multi-Task Learning for Sea State Bias Estimation
