Identification of protein targets for the antidepressant effects of Kai-Xin-San in Chinese medicine using isobaric tags for relative and absolute quantitation
2020-09-18XianZheDongDongXiaoWangTianYiZhangXuLiuPingLiuYuanHu
Xian-Zhe Dong , Dong-Xiao Wang Tian-Yi Zhang Xu Liu Ping Liu Yuan Hu
1 Department of Clinical Pharmacology and Pharmacy, Center of Pharmacy, Chinese PLA General Hospital, Beijing, China 2 Department of Pharmacy, Xuanwu Hospital of Capital Medical University, Beijing, China
Abstract
Key Words: brain-derived neurotrophic factor signal pathway; depression; isobaric tags for relative and absolute quantitation; Kai-Xin-San;neurogenesis; protein network; proteomics analysis; synaptic plasticity; traditional Chinese medicine
Introduction
Major depressive disorder, a mental disorder, affects more than 300 million people worldwide and is a leading cause of disability (Ignácio et al., 2019). Although the pathogenesis of depression has been extensively studied, the underlying mechanisms of antidepressant drugs are not yet clear (Seki et al., 2018; Soza et al., 2018). The monoaminergic system is targeted in most antidepressant drugs to regulate treatment responses. However, such drugs have been found to have a delayed onset of action, measured in weeks or even months(Insel and Wang, 2009), and a relatively low percentage of remission (< 50%) (Mann, 2005). Another important problem is the existence of treatment-resistant depression (Collo G, Merlo Pich, 2018; Zhou et al., 2018). There is therefore an urgent clinical need for antidepressant therapies with multiple targets, such as neurogenic and neurotrophic factors, intracellular signaling cascades, synaptic transmission, glutamate signaling, and modulation of glucocorticoids and cytokines,to overcome the delayed clinical effects (Hodes et al., 2010; Su et al., 2012; Ignácio et al., 2014).
Multi-target therapy with herbal drugs has been suggested as an alternative and complementary treatment for depression. In a recent review, Butler and Pilkington (2013) summarized the application and efficacy of Chinese herbs in depression, anxiety, and insomnia. Kai-Xin-San (KXS), a traditional Chinese medicine, has been widely used to supplement Qi, nourish the heart, and calm the mind in patients since the time of the Tang dynasties (i.e., 652 A.D.). KXS consists of Ginseng Radix (rhizome and root of Panax ginseng C. A. Mey.), Acori Tatarinowii Rhizoma (rhizome of Acorus tatarinowii Schott), Polygalae Radix (root of Polygala tenuifolia Wild.), and Poria (sclerotium of Poria cocos (Schw.) Wolf). KXS has effects on depressive-like symptoms including grief, sorrow, moodiness, and forgetfulness (Dong et al., 2014). We have previously demonstrated that KXS relieves depression-like symptoms by increasing the levels of neurotransmitters and neurotrophic factors in the brain of a mouse model of behavioral despair and a rat model of chronic mild stress (CMS) (Zhou et al., 2012; Hu et al., 2014b). In addition, various factors, including monoamine neurotransmitters,brain-derived neurotrophic factor (BDNF), the hypothalamic-pituitary-adrenal axis, glutamate, and the cAMP response element-binding protein (CREB) pathway, may all participate in the mechanism of action underlying the antidepressive effect of KXS (Hu et al., 2009, 2010a, 2010b, 2011, 2012, 2014a).However, as yet no studies have been performed to identify the key proteins implicated in KXS treatment.
Proteomics is a powerful platform for the comprehensive profiling of drug-regulated proteins, and has been widely applied to investigate the mechanisms of different drugs,including in traditional Chinese medicine (Liu et al., 2018).Isobaric tags for relative and absolute quantitation (iTRAQ)-based quantitative proteomics has been widely used to explore protein alterations in patients with depressive disorders (Henningsen et al., 2012; Zhan et al., 2014; Han et al., 2015; Shao et al., 2015). In the present study, we aimed to examine the differential expression of hippocampal proteins in CMS rats treated with KXS, using an iTRAQ-based method of proteomics.
Subjects and Methods
KXS preparation
Ginseng Radix, Polygalae Radix, Acori Tatarinowii Rhizoma,and Poria were mixed at a ratio of 3:3:2:2 and processed as previously reported (Hu et al., 2008). The aqueous extracts were filtered and evaporated under reduced pressure to obtain concentrates, which were freeze-dried to yield powder. A yield of 1 g powder contained 3.76 g of total original herbs. The obtained KXS powder was then dissolved in saline at 0.714 g/mL and standardized using a high-performance liquid chromatography (HPLC)-fingerprint method (Hu et al., 2008) (Figure 1). All materials necessary to formulate KXS were purchased from the LvYe Medicinal Material Company, China, and identified by Professor Ping Liu. The voucher specimens were registered in the Herbarium of Traditional Chinese Medicinal Pharmacy, Chinese PLA General Hospital, China.
Animals and CMS procedure
We purchased 45 specific-pathogen-free 6-week-old male Sprague-Dawley rats from the Laboratory Animal Breeding and Research Center in PLA General Hospital, China (license No. SCXK (Jing) 2017-0006). All animal procedures and protocols were performed in accordance with the Guide for the Care and Use of Laboratory Animals (NIH) and were reviewed and approved by the Ethics Committee for Animal Research from the PLA General Hospital (approval No. X5-2016-07)on March 5, 2016. The rats were housed individually and fed a laboratory standard diet ad libitum for 1 week to acclimate to the environment. Before the onset of CMS, rats were randomly assigned to three groups (n = 15/group), including the control group (rats without CMS), CMS group, and KXS group (CMS rats treated with KXS). The CMS rat models of depression were established using a published protocol (Hu et al., 2010b)(Figure 1). Briefly, the stressors comprised continuous illumination for 24 hours, tilted cage for 12 hours, high-speed agitation for 10 minutes, deprivation of food for 24 hours,deprivation of water for 24 hours, immobilization for 2 hours,and forced swimming in ice water for 5 minutes. CMS was conducted for 28 days (7 days/cycle for 4 cycles). In the KXS group, 600 mg/kg/d KXS was orally administered once daily for 14 days after stress stimulation (Dong et al., 2013). At the end of the experiment, all rats were sacrificed by decapitation,and hippocampal tissue was dissected quickly on ice before being transferred to liquid nitrogen.
Sucrose preference test
Following the 1-week adaptation period, all rats were deprived of drinking water and food for 24 hours. During a 1-hour window, rats were fed with a 1% sucrose solution and water.Sucrose intake was measured by comparing bottle weight before and after the 1-hour window, and expressed in relation to the total liquid consumption (sucrose intake/(sucrose intake+ water intake)) (Hu et al., 2013). The sucrose preference test was conducted again after 4 weeks of CMS and 2 weeks of KXS administration.
Protein preparation and iTRAQ labeling
Approximately 100 mg of hippocampus was homogenized in 300 μL of lysis buffer (8 mM urea, 1% DTT, and protease inhibitor cocktail (1:1000, P8340, Sigma-Aldrich, St. Louis,MO, USA) (1:5, w:v)) using a Dounce glass/Teflon homogenizer (Kimble-Chase, Rochester, NY, USA). Undissolved materials were removed by centrifugation at 4000 × g at 4°C for 30 minutes. Protein concentrations were determined using the Bradford assay (BioRad, Hercules, CA, USA). Protein extracts were denatured, alkylated, and digested with sequencing-grade modified trypsin (50:1) overnight at 37°C. Protein was labeled with iTRAQ reagent tags using 4-plex iTRAQ kits according to the manufacturer’s instructions (AB Sciex Inc.,Framingham, MA, USA). The CMS samples were labeled with iTRAQ tags 115. Protein samples from the control and KXS groups were labeled with tags 114 and 116, respectively.
Strong cation exchange chromatography
After a 2-hour incubation at room temperature, all labeled samples were mixed at equal ratios. Strong cation exchange chromatography was performed to remove excess iTRAQ reagent and interfering substances. The samples were separated using an HPLC system (Rigol, Beijing, China) equipped with a 4.6 × 250 mm C-18 HPLC column (5 μm, Agela, Tianjin,China). The fractions were collected using gradient elution with buffer A (2% acetonitrile (ACN), 98% H2O; pH 10.0)and buffer B (98% ACN, 2% H2O, pH 10.0) at a flow rate of 0.7 mL/min. Mobile phase: 30 minutes of 5-35% buffer B; 2 minutes of 35-95% buffer B; 5 minutes of 95% buffer B; 2 minutes of 95-5% buffer B; 6 minutes of 5% buffer B.
Liquid chromatography-tandem mass spectrometry analysis
The fractionated peptides were separated on a TripleTOF 5600 mass spectrometer coupled to an Eksigent Ultra HPLC(AB Sciex). The sample was dissolved in solution A (1.9%ACN, 98% H2O, 0.1% fomic acid), trapped on a precolumn(C18, 100 μm × 20 mm, 5 μm particle size), and separated in a capillary analytical column (C18, 75 μm × 120 mm, 3 μm particle size). Solutions A and B (98% ACN, 1.9% H2O, 0.1%fomic acid) were used in the gradient elution at a flow rate of 330 mL/min.
The peptides were then analyzed on a TripleTOF 5600 MS at an electrospray potential of 2.0 kV at 320°C. The Eksigent Ultra HPLC was set to perform data acquisition in the positive ion mode with a range of 300-1400 kDa in the full scan. The MS scan range was set at 300-1400 m/z and the MS/MS scan range was from 120 m/z to an auto-selected range according to the lowest m/z. Data-dependent acquisition was performed,and the top 50 precursor ions were selected to fragment using collision-induced dissociation. The collision-induced dissociation energy was automatically adjusted by the rolling collision-induced dissociation function.
Database search
The liquid chromatography-tandem mass spectrometry data were then compared against data in the NCBI database using Protein Pilot Software Beta (ABSciex, Framingham, MA,USA, version 4.5). For protein identification and quantification, peptide mass tolerance and fragment tolerance were each set at 0.3 Da. Only one missed tryptic cleavage was allowed.The false positive rates were controlled below 1%.
Protein identification and quantitation
The raw data were converted into an MGF format and compared against the Rat International Protein Index database(version 3.87, http://www.ebi.ac.uk/IPI/IPIhelp.html) using the Mascot search engine (version 2.3.01). All identified proteins were based on at least two peptides. The false positive rates obtained from a decoy database were controlled below 5%. The relative intensities of the reporter ion were used to derive quantitative information about the labeled peptides.Three criteria were used to select differentially expressed proteins: (1) more than one high-scoring peptide; (2) P value <0.05; and (3) fold-changes were ≥ 1.2 or ≤ 0.83.
Bioinformatics
DAVID Bioinformatics Resources v6.7 (http://david.abcc.ncifcrf.gov/home.jsp) (Dennis et al., 2003) were used to obtain the gene ontology (GO) terms of enrichment, and highlighted the most relevant GO and Kyoto encyclopedia of genes and genomes (KEGG) terms associated with modulated proteins obtained from the comparisons. A P value < 0.05 was considered enriched. KEGG terms with corrected P values< 0.05 were considered significant. Functional analysis was performed using a gene ontology tool (GOTERM_CC_ALL)with UniProt (Swiss-Prot/TrEMBL, http://www.uniprot. org/)accession numbers. Significant pathways were classified into hierarchical categories according to KEGG. The analysis of protein-protein interactions was performed with STRING 9.0(http://www.string-db.org/).
Statistical analysis
All data are expressed as the mean ± standard error. Differences between groups were analyzed using a one-way analysis of variance followed by Dunnett’s test. All data were analyzed statistically using Prism 5.0 (Graph Pad Software, Inc.). P-values of less than 0.05 were considered statistically significant.
Results
KXS increases sucrose consumption and body weight in CMS model rats
Figure 2 shows sucrose consumption and body weight in control, CMS, and KXS rats before CMS treatment (day 0),after CMS treatment (day 28), and after KXS treatment (day 42). Decreased sucrose consumption is a key symptom of depression-like behavior, which can reflect anhedonia (Géa et al., 2019). The CMS model that was used in the current study induced lower sucrose consumption. After 28 days of CMS exposure, sucrose intake significantly decreased (P < 0.001), and body weight also decreased in the CMS group. After chronic administration of KXS powder (600 mg/kg/d) for 14 days,sucrose consumption was higher in the KXS group than in the CMS group at day 42 (P < 0.001; Figure 2A). Furthermore,compared with the CMS group, body weight was higher in the KXS group (Figure 2B).
Effect of KXS on differential proteomics in rats with CMS
The number of differentially expressed proteins in the control,CMS, and KXS groups is shown in Figure 3. The detailed information for these proteins is listed in Table 1. We identified 88 differentially expressed proteins between the CMS and control groups (compared with the control group, 29 proteins had increased expression and 59 proteins had decreased expression in the CMS group). In addition, there were 162 differentially expressed proteins between the KXS and CMS groups (compared with the CMS group, 51 proteins had decreased expression and 111 proteins had increased expression in the KXS group).
Functional analysis of differentially expressed proteins in KXS-treated CMS rats
Biological functional analysis demonstrated that the differentially expressed proteins in the CMS group are involved in cellular and metabolic processes, the regulation of biological processes, macromolecule metabolic processes, and responses to stimuli (Figure 4A). In the KXS group, we observed a similar pattern of differentially expressed protein functions, but the number of proteins in each category was double that in the CMS group.
Molecular functional analysis identified that the differentially expressed proteins were involved in protein binding,nucleic acid binding, and structural molecule activity in both the CMS and KXS groups. Proteins for helicase activity, protein transporter activity, and ligase activity were observed in the KXS group only, and proteins for electron carrier activity were observed in the CMS group only (Figure 4B).
Potential molecular targets of KXS in CMS
We identified 33 differentially expressed proteins in the CMS group compared with both the control and KXS groups, including 7 upregulated and 26 downregulated proteins (Table 2). Functional analysis showed that these proteins participate in synaptic plasticity, neurodevelopment, and neurogenesis,and may be responsible for the major therapeutic effect of KXS. Proteins that were downregulated by CMS and upregulated by KXS are involved mainly in glutamate signaling(amino acid transporter, Arpp-21 protein, proline-rich transmembrane protein 2), synaptic plasticity (such as brain acid soluble protein 1 and secretogranin-1), mTORC1, BDNF, the cAMP pathway (eukaryotic translation initiation factor 4E binding protein 2, cAMP-regulated phosphoprotein 19), metabolic processes (thymosin beta-4/10, protein Dpm1, protein Txndc17), and cell survival processes (leucine zipper putative tumor suppressor 1, death-associated protein 1, protein LOC102548415). In contrast, proteins that were upregulated by CMS and downregulated by KXS are mainly involved in cytoskeletal/structural molecule activity (myelin-associated glycoprotein; keratin, type II cytoskeletal 72; keratin, type II cytoskeletal 1; keratin, type I cytoskeletal 10), and synaptic plasticity (deoxyguanosine kinase (predicted) and protein Sdk2) (Table 2).
Protein-protein interaction network of KXS in CMS
Our previous research and other literature were also reviewed and analyzed to explore the relevance of the proteins and genes that were affected by KXS treatment. Table 3 shows the 28 proteins that may be involved in the antidepressant effect of KXS. Figure 5 shows the network that consists of the 28 proteins. We found that the backbone network consists of BDNF, Ark, CREB, mTOR, Eef2, Eif4ebp2, and PI3K.
Discussion
In brain tissue from depressed patients, differentially expressed proteins that are involved in synaptic transmission,such as glutamate transport, have been identified in microarray studies (Choudary et al., 2005; Sequeira et al., 2009). Glutamate accumulation may not only cause cytotoxic damage to neurons and glia (Petroff, 2002), but may also perturb the ratio of excitatory neurotransmitter levels (Cryan and Kaupmann, 2005). In patients with major depressive disorder,Choudary et al. (2005) revealed a downregulation of SLC1A2,a key member of the glutamate/neutral amino acid transporter protein family. Arpp-21 protein (ARPP21) is involved in glutamate signaling through mediation of the calmodulin pathway (Lin et al., 2015). Mutant proline-rich transmembrane protein 2 (PRRT2) may affect glutamate signaling and glutamate receptor activity by the inhibition of SNAP25 (Li et al., 2015). In our study, we revealed a downregulation in proteins such as SLC1A2, APRR21, and PRRT2 in the KXS group compared with the CMS group. These results suggest that glutamate transporters and signaling, which may stop the accumulation of glutamate in the brain, may be involved in the antidepressant effect of KXS.
Because of their important roles in dendritic structure,synaptic stability, and synaptic plasticity, a disruption in specific integrin subunits, such as tubulin and actin, may cause impairments in behavior (Minamide et al., 2000; Medina et al., 2008; Davis et al., 2011). In a previous study, cytoskeletal abnormalities induced a regression of dendritic processes in hippocampal neurons, a decreased calcium concentration in dendritic spines, and caused depression-associated neurodegeneration (Lee et al., 2002). In our study, there was signifi-cantly increased expression of myelin-associated glycoprotein and three keratin (Krt) family proteins (Krt72, Krt1, Krt10) in the CMS group compared with the control and KXS groups.These four cytoskeleton-related proteins have been shown to play key roles in the axonal cytoskeleton, serving as scaffolds for various cellular processes including tissue growth and stress responses (Fruttiger et al., 1995; Marcus et al., 2002;Coulombe and Wong, 2004; Ramms et al., 2013), thus further supporting their roles in depression and in the antidepressive effects of KXS treatment.

Table 1 NCBI accession numbers of significant differential proteins between control, CMS and KXS groups

Figure 1 Chronic mild stress procedure(A) and chemical fingerprinting of KXS standardization (B).

Figure 2 Effect of KXS on behavioral changes in rats with CMS.

Figure 3 Effect of KXS treatment on differentially expressed proteins in CMS rats.
In an animal model, exposure to marked psychological stress can trigger a depressive-like disorder by downregulating the expression of neurotrophin-related genes, which has a negative impact on neurogenesis and synaptic plasticity,ultimately resulting in the loss of glial cells and glial glutamate transporters (Harvey et al., 2003; Martins-de-Souza et al., 2012). In the present study, we identified downregulated proteins that were involved in neurogenesis and synaptic plasticity, supporting previous findings in animal models.
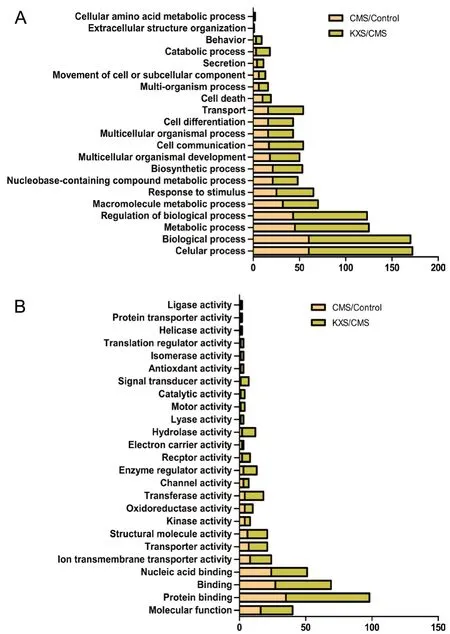
Figure 4 Gene-ontology-based functional categorization of proteins with significantly different expression levels following CMS or KXS treatments.
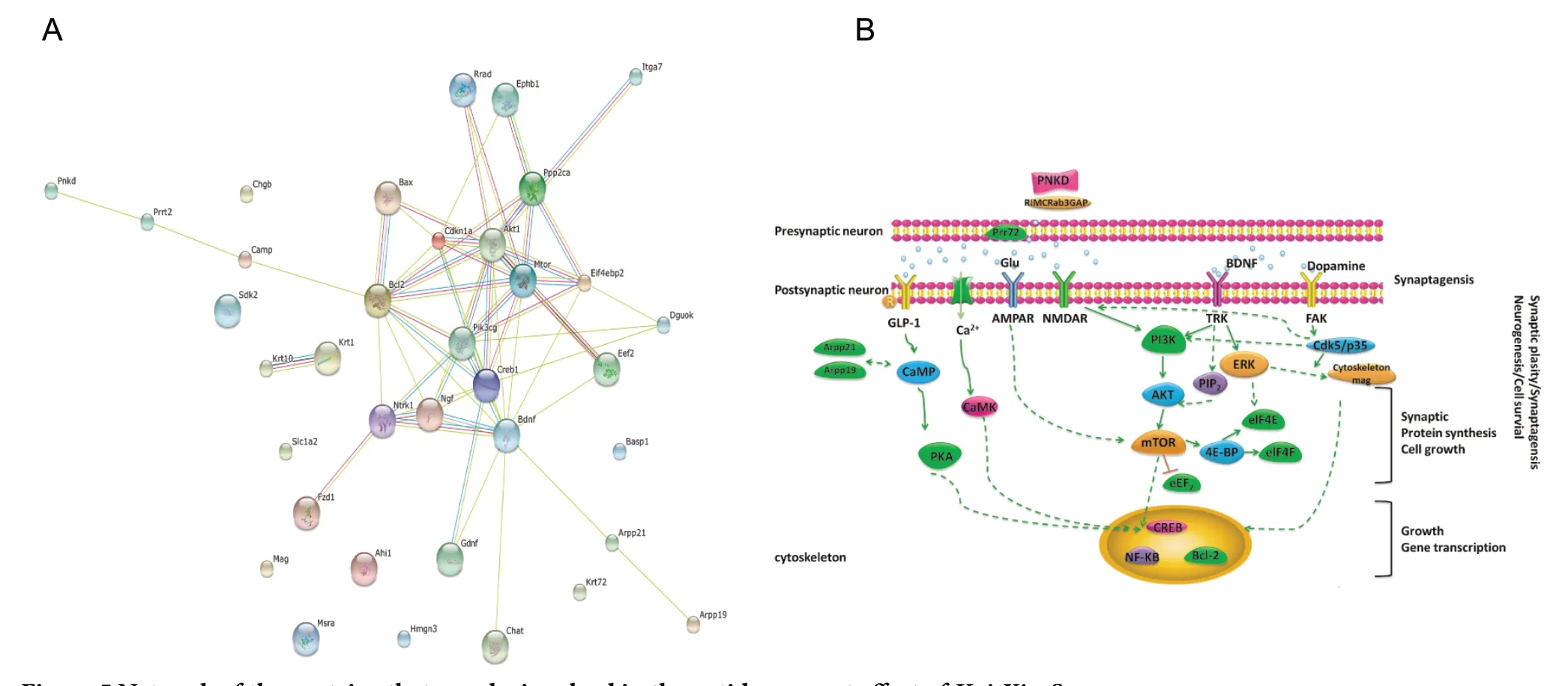
Figure 5 Network of the proteins that may be involved in the antidepressant effect of Kai-Xin-San.
The major pharmacologically active ingredients in KXS include ginsenosides (Rg1, Rb1, and Re) and oligosaccharide esters (tenuifoliside powder A and 3,6′-disinapoylsucrose) (Wang et al., 2010; Dong et al., 2014; Hu et al., 2014a).With its natural combinatorial chemistry, KXS is a potential multi-target treatment for depressive disorder. This idea is supported by the reported multiple targets involved in KXS treatment, including the excitatory neurotransmitter glutamate and its receptors, downstream signaling effectors such as calcium/calmodulin-dependent protein kinase II (CaMKII),protein kinase B (AKT), extracellular signal-regulated kinases, CREB, phosphatidyl inositol-3 kinase (PI3K), and BDNF and its receptor Trk (Hodes et al., 2010; Hu et al., 2010b, 2011,2014a; Dong et al., 2014; Liu et al., 2015).
In the current study, we identified upregulated eukaryotic initiation factor 4E binding proteins 2 (eIF4E-BP2) in the KXS group. Functional analysis identified that BDNF and its upstream PI3K-AKT-mTOR pathway are major mechanisms in synapses and neurogenesis. mTORC1-dependent release of IF4E-BP2 is the major form of 4E-BP in the brain, and the resulting enhancement in eIF4F formation (eIF4E-eIF4G interaction) is critical to multiple forms of translation-dependent synaptic plasticity (Richter and Klann, 2009). eIF4E-BP2 may be regulated by MAP-kinase-interacting kinase. Therefore,their interaction may contribute to BDNF-TrkB signaling, as well as the protein-synthesis-dependent structural plasticity of dendritic spines (Panja et al., 2014). Six proteins regulated by KXS were identified in our network analysis, including cAMP-regulated phosphoprotein 19 (ARPP19), three glutamate transport proteins, paroxysmal nonkinesigenic dyskinesia, and deoxyguanosine kinase, as possibly being involved in preventing excitotoxic injury in the brain (Haldipur et al., 2014) and in regulating the excitatory neurotransmitter levels as previously mentioned. Paroxysmal nonkinesigenic dyskinesia and deoxyguanosine kinase are two novel proteins that may have functions in abnormal behaviors that involve reversing the impairment of synaptic facilitation and transmission (Ronchi et al., 2012; Ansoleaga et al., 2015; Shen et al., 2015). We also found that some synaptic plasticity cluster proteins were decreased in the CMS group and activated in the KXS group, such as Sdhaf4, Ahi1, Hmgn3, Basp1, and Chgb. Protein-protein interactions provide crucial scaffolds for modeling to give insights into the complex interactivemechanisms involved in the effects of KXS on depression.

Table 2 Quantitative information of the 33 regulated proteins by both KXS and CMS treatment

Table 3 Regulation of active compounds in KXS on proteins
In conclusion, the present study identified 33 differentially expressed proteins in the CMS group compared with the control and KXS groups, including 7 upregulated and 26 downregulated proteins. Functional analysis revealed that these proteins participate in synaptic plasticity, neurodevelopment,and neurogenesis. Our results provide experimental evidence that synaptic plasticity, neurodevelopment, and neurogenesis may contribute to the antidepressant effects of KXS. This study provides further support for the antidepressant effect of KXS and its mechanisms, provides support for the application of KXS as a new antidepressant drug, and provides potential targets for antidepressant research involving traditional Chinese medicine. However, our study mainly focused on high-throughput data screening and bioinformatics prediction analysis, and further studies in different animal models and humans are warranted.
Acknowledgments:We thank Dr. Yan Li from Beijing Bio-Fly Bioscience Co., China for her contribution in mass spectrometric and bioinformatics analysis.
Author contributions:Study design and writing: XZD, YH; manuscript review and revision for intellectual content: PL, YH; study assessments and evaluations: XZD, DXW, TYZ, XL; manuscript review for intellectual content: YH. All authors have read and approved the final manuscript.
Conflicts of interest:The authors declare no competing interests.
Financial support:This work was supported by the National Natural Science Foundation of China, No. 81573876 (to YH). The funder had no roles in the study design, conduction of experiment, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional review board statement:The study was approved by the Ethics Committee for Animal Research from the Chinese PLA General Hospital (approval No. X5-2016-07) on March 5, 2016.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-Non-Commercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Open peer reviewer:Peng Luo, Air Force Medical University, China.
Additional file:Open peer review report 1.
杂志排行
中国神经再生研究(英文版)的其它文章
- Ethanol extract from Gynostemma pentaphyllum ameliorates dopaminergic neuronal cell death in transgenic mice expressing mutant A53T human alpha-synuclein
- Peripheral nerve injury induced changes in the spinal cord and strategies to counteract/enhance the changes to promote nerve regeneration
- Genetic targeting of astrocytes to combat neurodegenerative disease
- Pathological significance of tRNA-derived small RNAs in neurological disorders
- Applications of advanced signal processing and machine learning in the neonatal hypoxic-ischemic electroencephalography
- Protective effect of hydrogen sulfide on oxidative stress-induced neurodegenerative diseases
