ldentification of candidate biomarkers correlated with pathogenesis of postoperative peritoneal adhesion by using microarray analysis
2020-01-16YaoYaoBianLiLiYangYanYanMinZhaoYanQiChenYaQiZhouZiXinWangWenLinLiLiZeng
Yao-Yao Bian, Li-Li Yang, Yan Yan, Min Zhao, Yan-Qi Chen, Ya-Qi Zhou, Zi-Xin Wang, Wen-Lin Li, Li Zeng
Yao-Yao Bian, Ya-Qi Zhou, Zi-Xin Wang, School of Nursing, Nanjing University of Chinese Medicine, Nanjing 210023, Jiangsu Province, China
Li-Li Yang, Min Zhao, Yan-Qi Chen, Li Zeng, School of First Clinical Medicine, Nanjing University of Chinese Medicine, Nanjing 210023, Jiangsu Province, China
Li-Li Yang, Wen-Lin Li, Li Zeng, Jingwen Library, Nanjing University of Chinese Medicine,Nanjing 210023, Jiangsu Province, China
Yan Yan, Guang′anmen Hospital, China Academy of Chinese Medical Sciences, Beijing 100053, China
Abstract
Key words: Postoperative peritoneal adhesion; Candidate biomarkers; Molecular pathogenesis; Bioinformatics analysis
INTRODUCTION
Postoperative peritoneal adhesion (PPA) is an indefinite adhesion or fibrous cord formed among abdominal organs or tissues. It remains an urgent clinical concern due to the increasing occurrence of abdominal surgery. PPA can lead to acute or chronic complications including persistent abdominal pain and bloating, female infertility,bowel obstruction, and intestinal necrosis[1]. A previous review showed that even minimally invasive surgical techniques that are widely used to minimize surgery lesions of peritoneal trauma cannot prevent adhesion formation[2]. PPA not only increases the reoperation rate and extends hospital stay but also results in a considerable disease burden and heavy financial responsibility for individuals,families, and society[3]. PPA has an incidence rate ranging from 90% to 95%[4].Approximately 117 per 100000 people in the United States have been hospitalized for adhesion-related problems, and the direct annual hospitalization cost for adhesionrelated complications and surgery increased from US$1.3 billion in 1994 to US$50 billion in 2010 in United States[5,6].
The occurrence and development of peritoneal adhesion are complex biological processes, during which many genes and pathways are involved in the pathogenesis of adhesion formation. However, the potential biological function of adhesion formation remains limited. Therefore, there is a need to further identify the differentially expressed transcripts and related pathways associated with peritoneal adhesion formation.
In this study, we retrieved a dataset of mRNA expression microarrays from Gene Expression Omnibus (commonly known as GEO) and identified a panel of altered genes and related pathways involved in the developmental progression of peritoneal adhesion by using bioinformatics methods. To validate our findings, we also carried out preliminary verification by using molecular biology techniquesin vivo. The study aimed to identify candidate biomarkers to uncover the underlying pathogenesis of PPA at the molecular level.
MATERIALS AND METHODS
Data acquisition and identification of differentially expressed genes
The microarray expression dataset was downloaded from the GEO data base by searching the key words: “RNA,” “peritoneal adhesion,” or “abdominal adhesion”and “Mus musculus” (organism). After screening, the accession number GSE123413 was obtained for analysis. The platform was GPL 21163, Agilent-074809 SurePrint G3 Mouse GE v2 8x60K Microarray. This dataset consisted of the transcriptional profiling of cecum tissues harvested at three time points (i.e. 3 h, 12 h, and 3 d) after the models were prepared by cauterization. The raw data were preprocessed and normalized using the affy package in R (version 3.3.4). Then the differentially expressed genes(DEGs) between sham (SH) and PPA groups at three different time points were screened separately using the limma package. |log2FC| >1 andP< 0.05 were considered statistically significant for DEGs. The overlapping DEGs of the three different time points were generated using the online tool Venny (http://bioinfogp.cnb.csic.es/tools/venny). The heat map of DEGs was obtained with the online tool Morpheus (https://software.broadinstitute.org/morpheus/).
Function and pathway enrichment analysis
To understand the underlying biological phenomena, gene ontology (GO) terms were used to determine gene annotation. Kyoto Encyclopedia of Genes and Genomes(commonly known as KEGG) enrichment was performed to locate the significant enrichment pathway. Both analyses were implemented on the Database for Annotation, Visualization and Integrated Discovery (commonly known as DAVID;http://david.abcc.ncifcrf.gov/)[7].
Protein-protein interaction (PPI) network construction and modules analysis
To further predict the interaction of peritoneal adhesion-associated protein pairs, the Search Tool for the Retrieval of Interacting Genes (commonly known as STRING)[8]was performed with a confidence score > 0.7 defined as significant. Then PPI integrated networks were mapped by Cytoscape 3.4.0 software[9]. Finally, the plug-in Molecular Complex Detection (commonly known as MCODE) was used to screen the modules of hub genes from the PPI network when node degree > 30 was considered as cut-off criteria.
Animal experiments
Twenty male Sprague-Dawley rats (8 wk old, weighing 280 ± 20 g) were purchased from the Qinglongshan Experimental Animal Breeding Farm (Nanjing, China). The rats were randomly divided into two groups: SH (n= 10) and PPA (n= 10). Both groups were housed in a standard condition of 12 h light-dark cycle (light on at 07:00 a.m.) under a controlled temperature of 22 ± 2°C. All animals were provided plenty of food and water and allowed to acclimatize to this condition 3 d before use. All experiments in this study followed the Guidelines of Accommodation and Care for Animals formulated by the Chinese Convention for the protection of vertebrate animals used for experimental and other scientific purposes and were authorized by the Laboratory Animal Management Committee of the Nanjing University of Chinese Medicine.
Surgical procedures and adhesion quality
The cecum cauterization model was established by a previous study[10]. After preoperative fasting for 12 h, the rats were placed under anesthesia with 1.0%-1.5%isoflurane. A 1.5 cm midline incision was made in the abdominal wall after traditional skin preservation and sterilization under aseptic conditions. The cecum was isolated and then cauterized using bipolar forceps to inflict a coagulation function for 1 s.Finally, the cecum was restored into the abdominal cavity, and the abdominal wall was sutured. After 3 d, the rats were sacrificed. Two independent investigators who were blinded to both groups evaluated the adhesion quality on the basis of a fivestage grading score system[11,12]shown in Table 1[13].
Masson staining
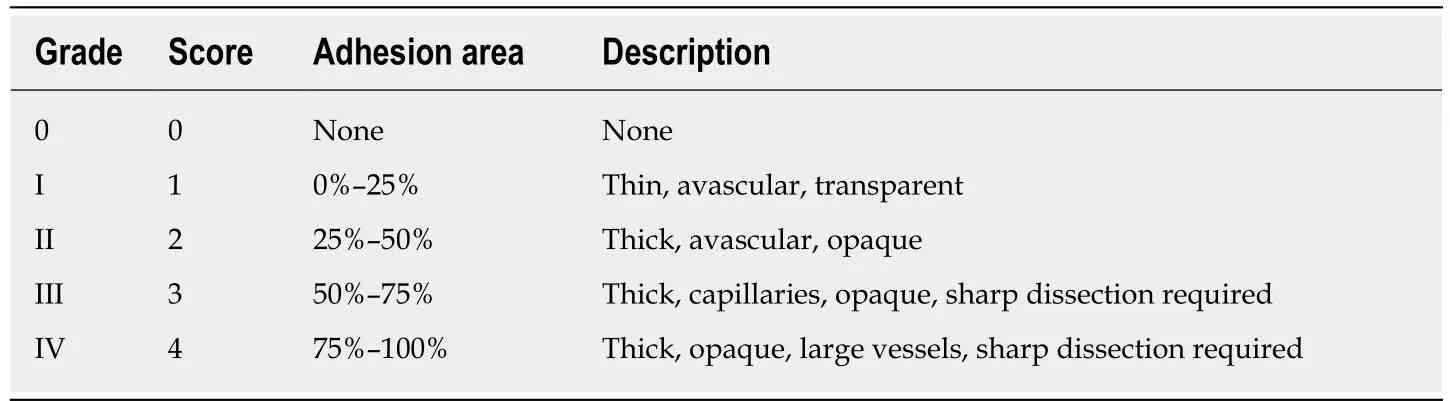
Table 1 Peritoneal adhesion scoring system
After fixation in 10% neutral formalin for 48 h, the cecum tissues were embedded in paraffin, and cut into sections. Masson staining was performed to estimate the collagen deposition. Represented views were visualized under a microscope(DM2500; Leica, Germany).
RNA extraction and quantitative PCR
Total RNA was extracted from the damaged cecum using TRIzol reagent (Invitrogen,United States). The complementary DNA (cDNA) was generatedviareverse transcription by using the First Stand cDNA Synthesis Kit (Thermo Fisher Scientific,United States). UltraSYBR One Step RT-qPCR Kit (Cwbio Technology, China) was used according to the manufacturer’s protocol. Based on GAPDH as standardization,the expression levels were conducted by using the 2-ΔΔCTanalysis method. The primer sequences are shown in Table 2.
Western blot analysis
Tissue proteins were extracted by the RIPA Protein Extraction Kit (Beyotime Biotechnology, China) according to routine protocols. The extracted proteins were added to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to PVDF membranes. Then the membranes were incubated with primary antibody at 4°C overnight against Toll-like receptor 4 (TLR4, 1:200), myeloid differentiation primary response protein 88 (MyD88, 1:200), nuclear factor kappa B (NF-κB, 1:200),and β-actin (1:2000) (all from Santa Cruz Biotechnology, United States). After incubation with secondary antibody for 1 h at 37°C, protein expression was viewed by the Chemiluminescence Imaging system (Bio-Rad, United States).
Statistical analysis
Statistical analyses were conducted using SPSS 19.0 software. All data are presented as the mean ± SD. Statistical comparisons within the two groups were made by the unpaired Student’st-test.Pvalues less than 0.05 were considered statistically significant.
RESULTS
DEG identification
The GSE123413 expression profile dataset consisted of the expression data matrix of 56743 gene probes. The raw data were processed and normalized with R software, as presented in Figure 1A. We identified the DEGs of three time points and found 457 overlapping genes. Of these genes, 446 expressed genes were altered, among which 183 were upregulated and 263 were downregulated. The expression levels of dysregulated genes are shown in Figure 1B.
Functional and pathway enrichment analyses
Analyses of the above-obtained 446 genes were performed using DAVID for biological process annotation and KEGG pathway enrichment. GO term assignment analysis suggested that the most enriched GOs were enriched in inflammatory response (GO: 0006954), neutrophil chemotaxis (GO: 0030593), cytokine activity (GO:0005125), cellular response to interleukin-1 (IL-1) (GO: 0071347), immune response(GO: 0006955), and response to lipopolysaccharide (GO: 0032496), as presented in Figure 2A. According to pathway enrichment analysis, the enriched altered genes were involved in PPA-related pathways such as the cytokine-cytokine receptor interaction and tumor necrosis factor (TNF) signaling pathway (Figure 2B).
PPI network construction and modules analysis
The 743 pairs involving 376 proteins were mapped in the PPI network through theSTRING website. The genes with relatively high degrees were considered hub genes related to peritoneal adhesion such as TNF (degree = 43), IL-6 (degree = 42), C-X-C motif chemokine ligand 2 (CXCL2; degree = 39), CXCL1 (degree = 38), and IL-1β(degree = 34). These genes may play pivotal roles in PPA progression. To further depict the complex relationship, the obtained interaction pairs were constructed using Cytoscape software (Figure 3A). After analysis using the plug-in MCODE, the top two significant modules were identified and enriched in the inflammatory response by DAVID. Module 1 involved 18 genes and 153 connections, and module 2 implicated 30 genes and 176 connections that were markedly enriched in the TLR signaling pathway, as shown in Figure 3B and Table 3.

Table 2 Primers used for qPCR
Macroscopic evaluation of adhesions
The incisions of all rats were primarily healed, without infection or other complications. The adhesion grades and scores among groups are shown in Table 4.In the SH group, the adhesion grade ranged from 0 to 1, which indicated the absence of adhesion or minor adhesion. However, the adhesion grade ranged from 2 to 4, and the score was approximately 3.4, which suggested that dense connective tissues were present in the PPA groups. The magnitude of peritoneal adhesions is presented in Figure 4.
Masson staining of adhesive tissue
Microscopically, collagen fibers appeared blue and muscle fibers were red in color.The model group displayed a deep and extensive range of blue, which indicated increased collagen fiber accumulation. However, the SH group displayed a light and narrow range of blue color, which suggested decreased collagen fiber accumulation(Figure 5).
Quantitative PCR validation of hub gene expression
To verify the bioinformatics analysis, the expression levels of the hub genes (TNF, IL-1β, IL6, CXCL1, and CXCL2) of the cecum tissues were quantified by quantitative PCR (qPCR) between the adhesion models and SH controls. The results indicated that the mRNA expression levels significantly increased (P< 0.05, Figure 6) in PPA groups compared with the SH groups.
Western blot validation of protein expression
In our study, functional enrichment of modules analysis suggested that the significant hub genes were markedly enriched in the TLR signaling pathway. Increasing evidence[14-16]suggests that TLR4, MyD88, and NF-κB play essential roles in the inflammatory pathway such as Toll-like signaling. On the basis of this evidence, we measured the expression levels of the abovementioned proteins by western blotting.The protein expression levels of TLR4, MyD88, and NF-κB in the PPA groups were significantly upregulated compared with those in the SH group (Figure 7).
DISCUSSION
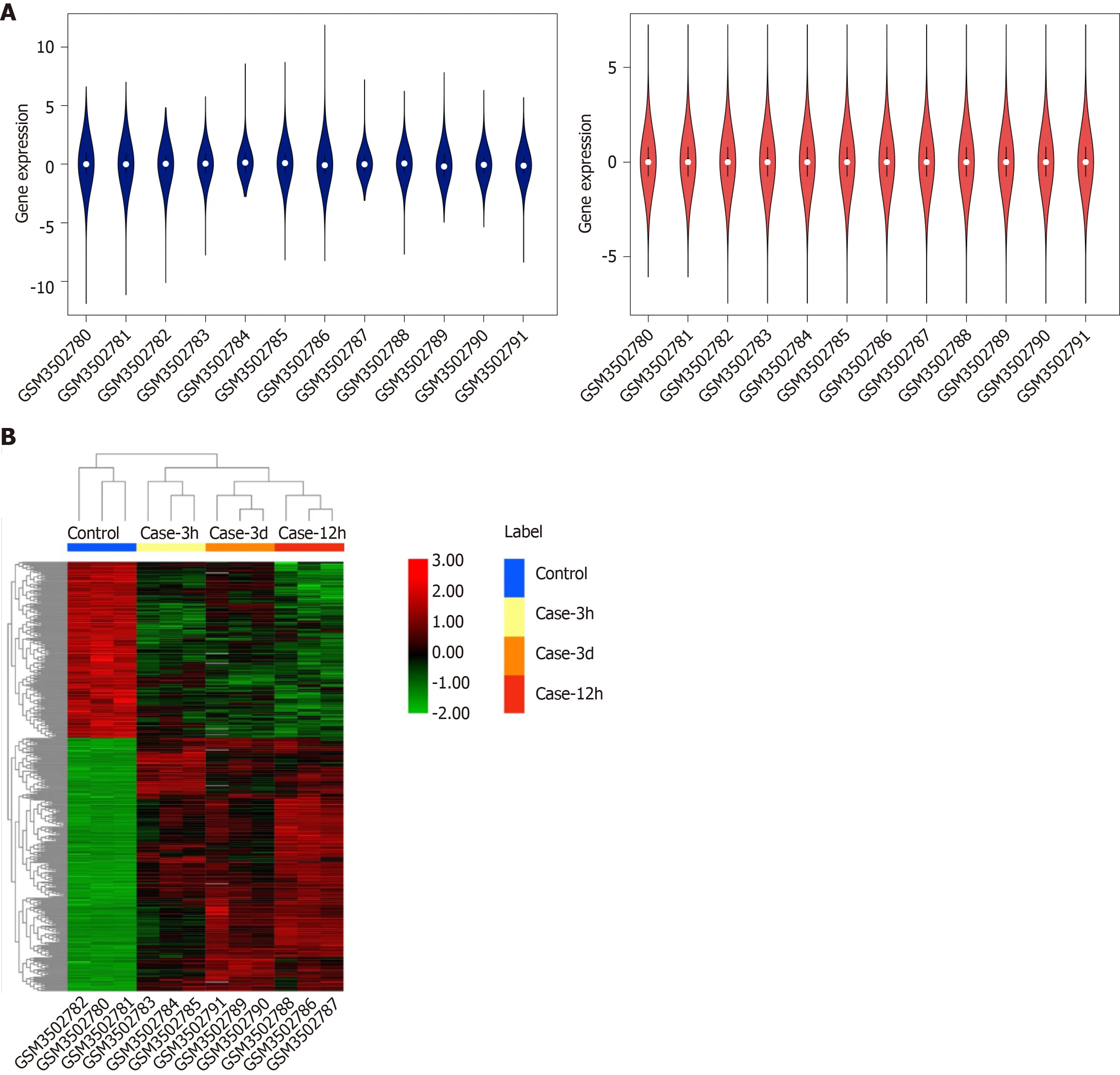
Figure 1 Box plots of data normalization and hierarchical cluster heatmap.
PPA is triggered immediately after surgery, and starts from the inflammation response, which aims to repair injured lesion. Adhesive formation is a strictly timed process. After peritoneal injury, acute inflammation occurs within minutes. Then numerous macrophages and neutrophils migrate onto the damaged lesions within hours, which can trigger a series of cascade reactions. The proliferation and migration of fibroblasts and mesothelium cell further enhance fibrinolysis activity, and the epithelial tissue is repaired within 3 d. If the repair is delayed, adhesion formation occurs from the third day. Hence, it is urgent to identify the potential biomarkers and associated pathways related to adhesion formation within 3 d at the molecular level.
To achieve this goal, we provided a bioinformatics analysis of DEGs and the associated pathways related to adhesion formation on the basis of the available GEO dataset. Consequently, 446 DEGs at three time points were identified. Based on the top 20 GO terms, the altered genes displayed a variety of functionalities in inflammatory and immune response. The results were supported by a previous study[17], thereby indicating that the importance of the inflammatory immune response in the abdominal microenvironment is involved in adhesion formation.Given the implication of gene function enrichment, the DEGs were enriched in the cytokine-cytokine receptor interaction and TNF signaling pathway. Thus, the importance of the biological process and associated inflammatory pathways may be related to adhesion formation.
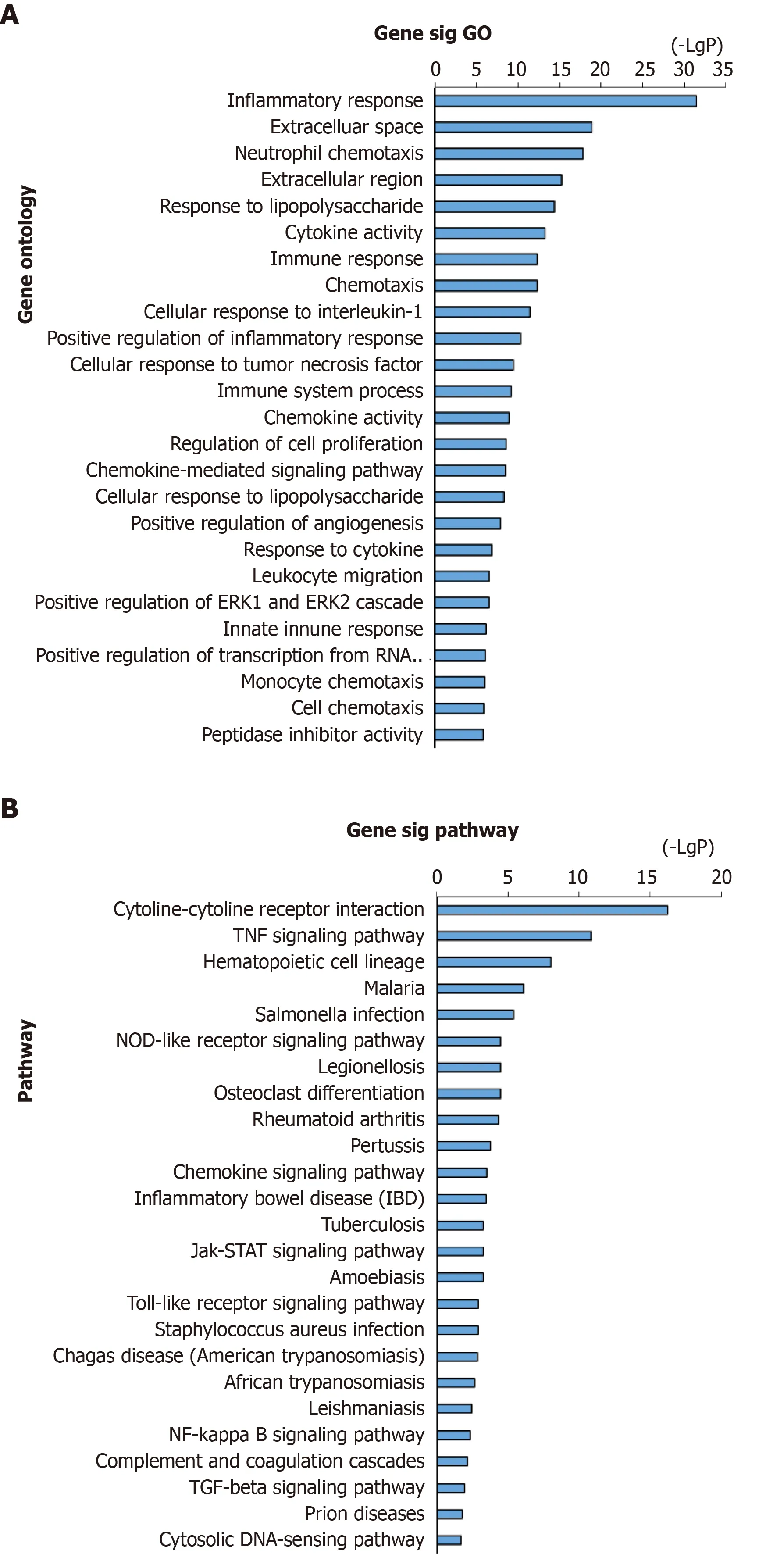
Figure 2 GO annotation and pathway enrichment of DEGs in adhesion formation.
The pathophysiological process of adhesion formation is rapid, cascaded, and complex, during which many genetic and epigenetic modifications of driving genes occur. Among the hub genes, TNF, IL-1β, IL-6, CXCL1, and CXCL2, which may act as important regulators of wound inflammatory, were marked with a high degree. Proinflammatory and anti-inflammatory cytokines expressed by these genes had double effects on the balance of the microenvironment. Once the imbalance is broken,adhesion occurs. We primarily validated the crucial roles of these mRNAs in the progression of adhesion formation by using qPCR analysis in the animal experiments.The results were consistent with those of microarray analysis. These signatures thatare associated with peritoneal adhesion have also been extensively reported[18-20]. For example, IL-1β is used as an important short-term medium and can be regarded as a reliable biomarker in peritoneal adhesion formation. IL-6 contributes to epithelial cell proliferation and the accumulation of inflammatory cells and fibers into injury sites,following the promotion of the pathological process of adhesion, thereby inducing adhesion formation. The preoperative application of anti-IL-1β and anti-IL-6 antibodies can effectively reduce the occurrence of postoperative abdominal adhesion[19,21]. TNF-α, as an immune regulator factor, combined with TNFR2 can activate the NF-κB signaling pathway in either a classical or non-classical manner,which further mediates the imbalance of inflammatory adhesion and eventually leads to adhesion formation. TNF-α is a pro-fibrogenic cytokine that can promote fibroblast proliferation and stimulate the peritoneal mesothelium cells to increase PAI-1 synthesis, which can further inhibit plasminogen activation and promote fiber accumulation, thereby resulting in adhesion formation. Adhesion-associated CXCL1 and CXCL2 also recruits circulating leukocytes to the injury sites that are involved in the inflammatory response[22]. Thus, these genes might act as key biomarkers in adhesion formation.
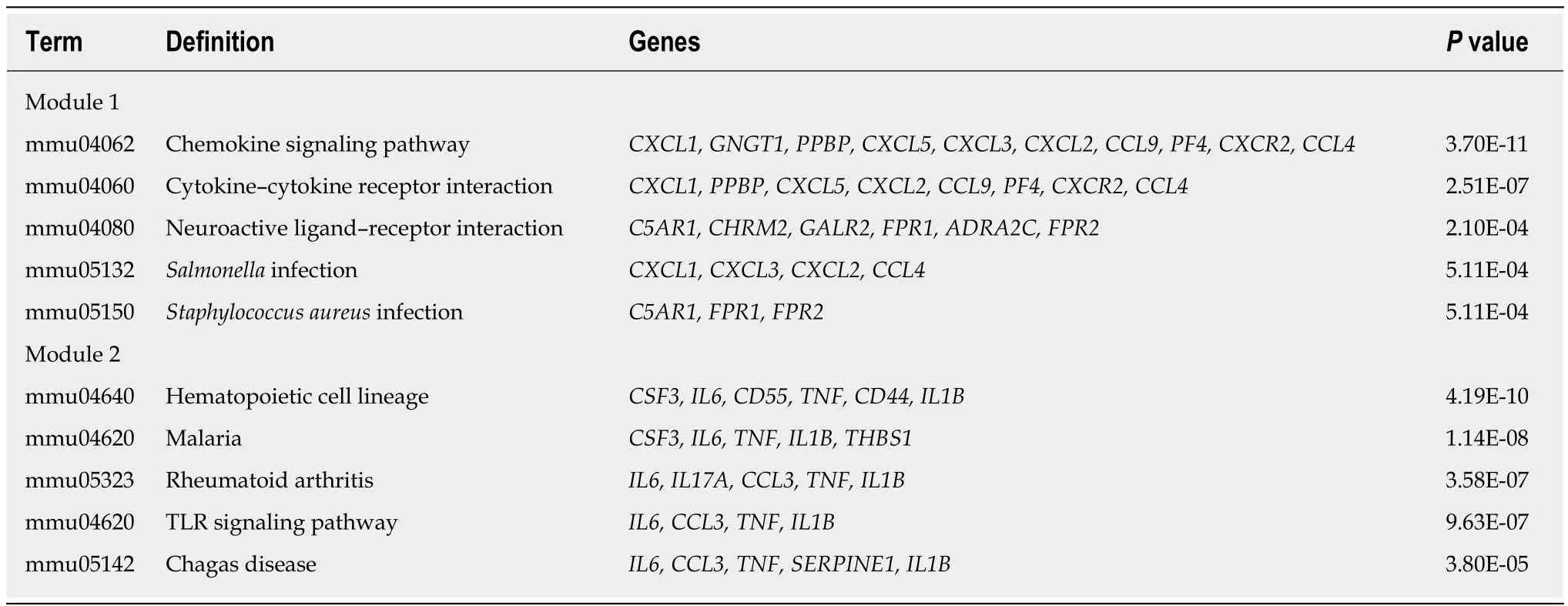
Table 3 KEGG analysis of two clusters (top five)
Based on the KEGG pathway, these hub genes were markedly enriched in TLR signaling pathway. TLRs are cellular transmembrane receptors and pattern recognition receptors that recognize pathogen-associated molecular patterns in congenital immune system. Gaining function of TLRs, TLR2, TLR3, and TLR4 have vital roles during acute wound repair[23]. Among these receptors, TLR4 is the most studied in tissue healing. The endogenous ligand MyD88 produced during the inflammatory process was identified by TLR4, which can further induce NF-κB activation and downstream regulator (i.e. IL-1, IL-6, TNF, CXCL1, and CXCL2)expression[24]. To further elucidate the critical roles of TLR4, MyD88, and NF-κB in the pathogenesis of adhesion formation, western blotting was applied to measure the protein expression in the two groups. The results showed that the proteins mentioned above were highly expressed in the PPA group. Hence, we initially speculated that the regulatory chain of “TLR4/MyD88/NF-κB/inflammatory cytokines” may serve key functions on postoperative adhesion formation. Our findings may provide preliminary evidence about TLR4/MyD88/NF-κB signal transduction in the molecular mechanism of postoperative adhesion formation. To the best of our knowledge, only one study reported that the TLR4/MyD88/NF-κB signaling pathway is a potential pathway in preventing peritoneal inflammation in peritoneal dialysis[25]. Further functional and gene knockout studies are warranted to elucidate the exact effects on the transcriptional expression of genes regulated by NF-κB axis activation.
Taken together, according to bioinformatics analysis, a series of adhesion-related hub genes and a regulatory pathway were identified. To further verify the underlying molecular mechanism in adhesion formation, experiment validation was conductedin vivo. The regulatory evidence chain of TLR4/MyD88/NF-κB/inflammatory cytokines/peritoneal adhesion was involved in the pathogenesis of PPA. These findings may provide initial sights into the underlying mechanisms of peritoneal adhesion formation.
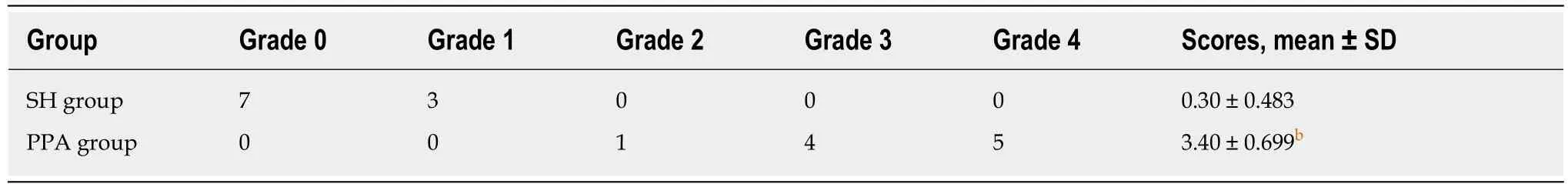
Table 4 Grading scores of peritoneal adhesions, n = 10
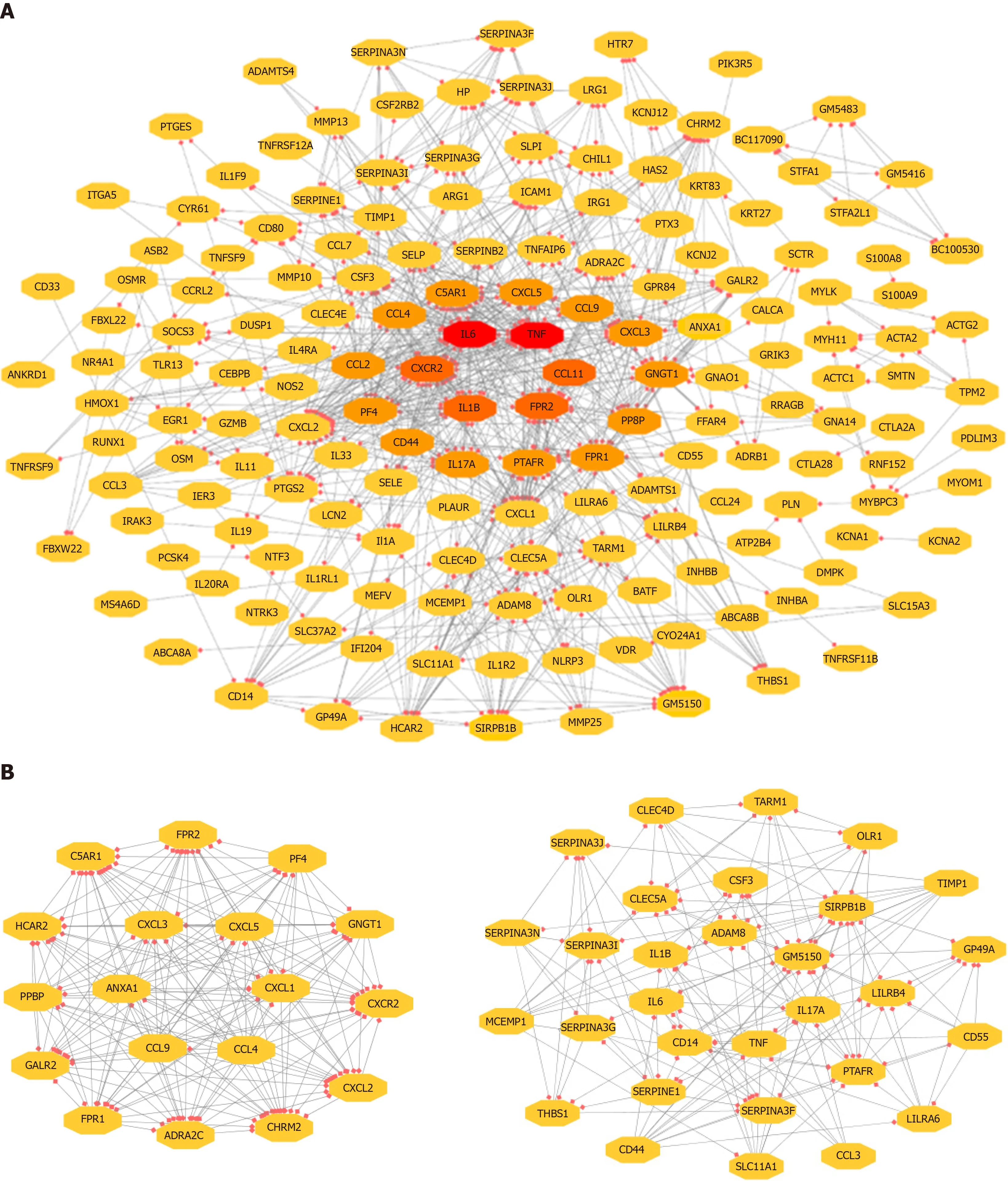
Figure 3 Visualization of the PPl network of identified DEGs.
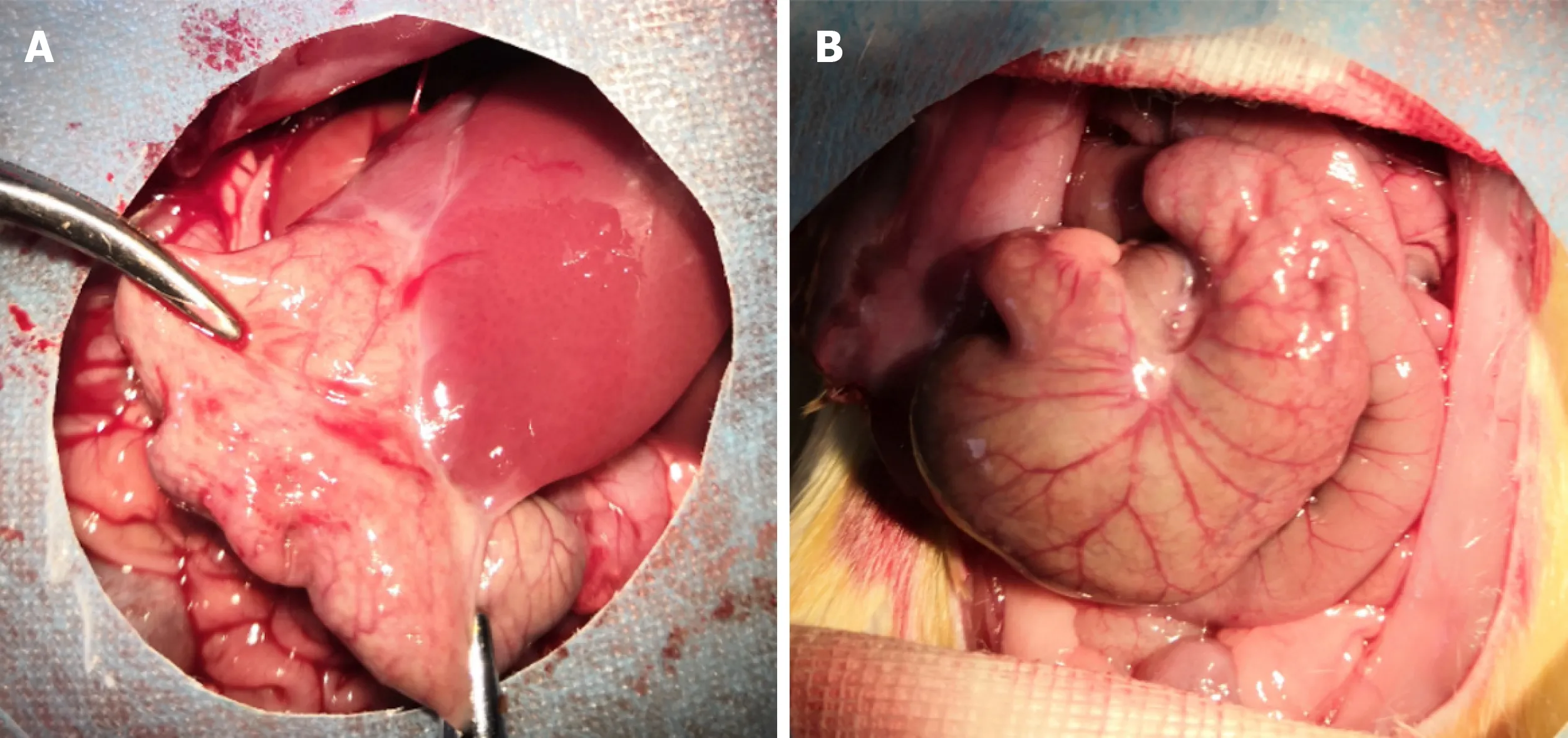
Figure 4 Representative images of PPA in each group.
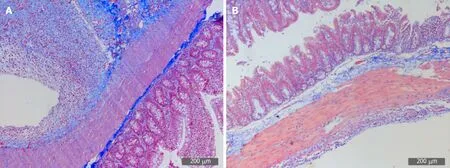
Figure 5 Representative images of Masson staining of adhesive tissues (200×).
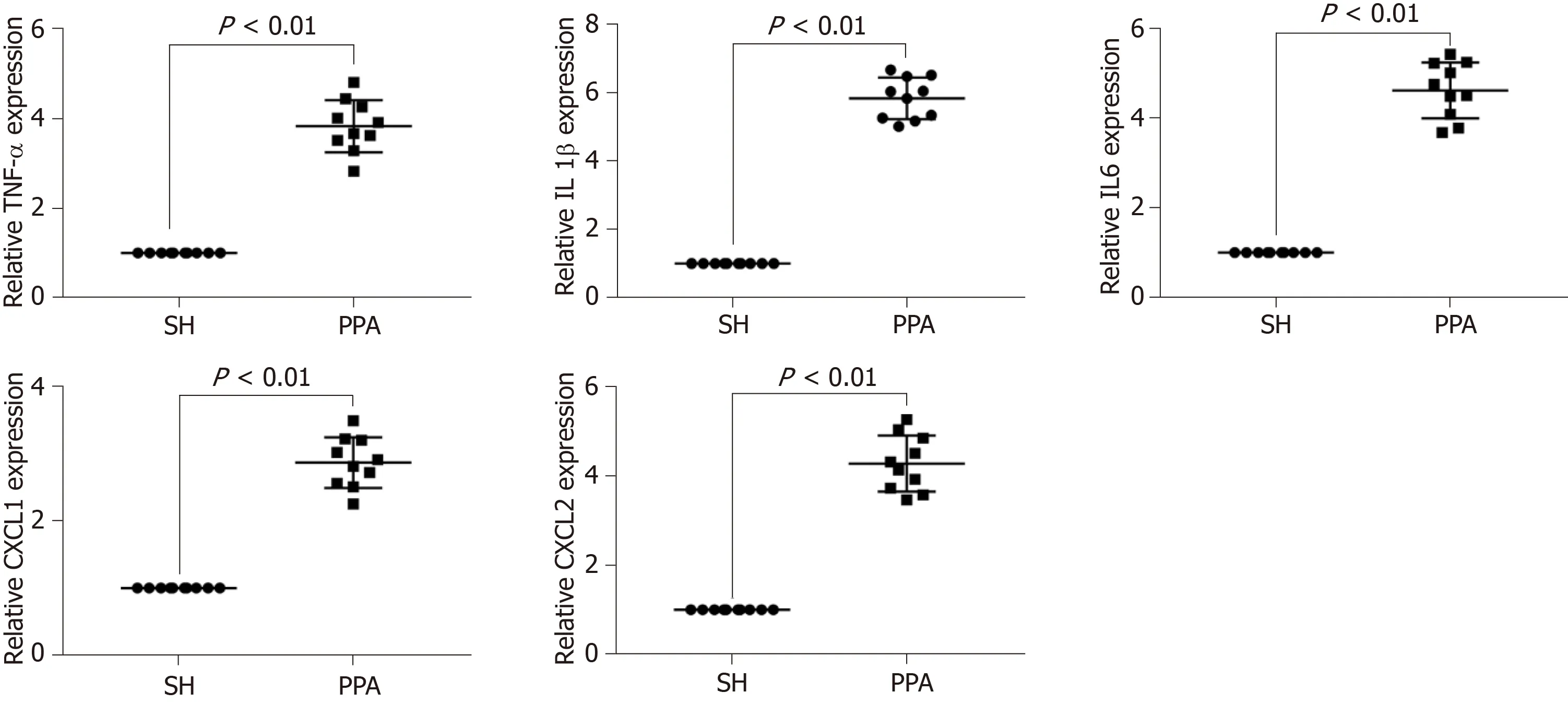
Figure 6 Representative mRNA expression level by qPCR.
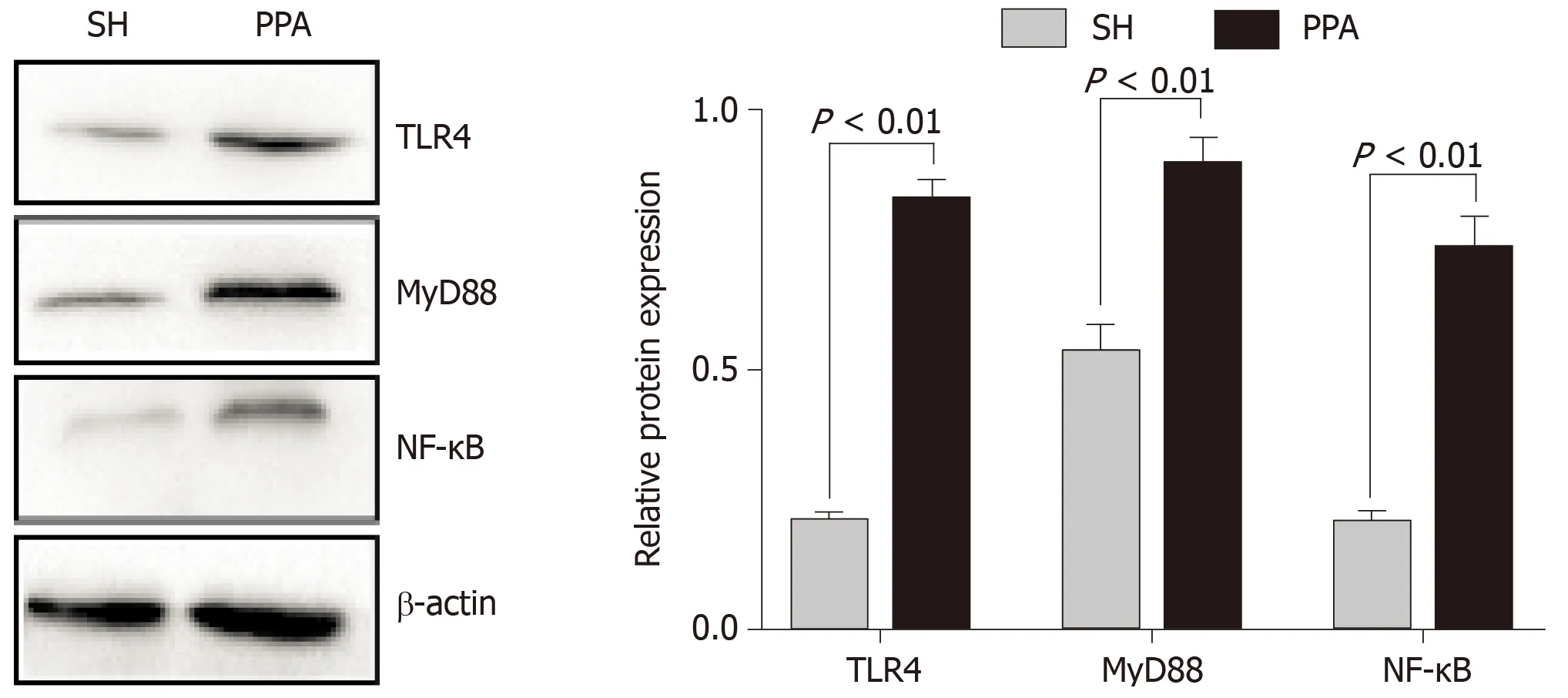
Figure 7 Relative protein levels by western blot analysis.
ARTICLE HIGHLIGHTS
Research background
Postoperative peritoneal adhesion (PPA), characterized with abdominal pain, female infertility,and even bowel obstruction after surgery, has always been a major concern. The occurrence and formation of adhesion are from complex biological processes. However, the molecular mechanisms of the basis of microarray data profile, followed by PPA formation, are largely unknown.
Research motivation
The occurrence and development of PPA is a complex biological process, during which many genes and pathways are involved in the pathogenesis of adhesion formation. As such, we developed microarray analysis combined with experimental methods to understand the underlying mechanisms of PPA at the transcriptomic and molecular levels.
Research objectives
The aim of this study was to uncover the molecular mechanisms of PPA formation after surgery using bioinformatics analysis, and to validate the results using rodent adhesion models.
Research methods
The gene expression profile was retrieved from the Gene Expression Omnibus database for our analysis. A panel of key altered genes and related pathways involved in adhesion formation were identified using bioinformatics analysis methods. And the microarray results were verified by performing quantitative PCR and western blottingin vivopreliminarily.
Research results
In total, 446 expressed genes were altered in peritoneal adhesion. We found that several hub genes (e.g., TNF, IL-1β, IL-6, CXCL1, CXCL2) were marked as significant biomarkers associated with PPA. Functional analysis suggested that these genes were enriched in the Toll-like receptor signaling pathway. According to the Kyoto Encyclopedia of Genes and Genomes pathway and published studies, TLR4, MyD88, and NF-κB played essential roles in Toll-like signaling transduction. Here, we gained a regulatory evidence chain of TLR4/MyD88/NF-κB/inflammatory cytokines/peritoneal adhesion involved in the pathogenesis of PPA. The results of the microarray analysis were consistent with the animal experiments.
Research conclusions
Our findings provide initial evidence about the regulatory evidence chain of TLR4/MyD88/NF-κB/inflammatory cytokines/peritoneal adhesion in the pathogenesis of PPA. Future studies are required to validate the results.
Research perspectives
These findings may extend our understanding of the molecular mechanisms of PPA. Further functional and gene knockout studies are warranted to elucidate the exact effects on the transcriptional expression of genes regulated by NF-κB axis activation.
杂志排行
World Journal of Gastrointestinal Oncology的其它文章
- Precision medicine for gastrointestinal cancer: Recent progress and future perspective
- Digestive tract reconstruction options after laparoscopic gastrectomy for gastric cancer
- lnterpretation of the development of neoadjuvant therapy for gastric cancer based on the vicissitudes of the NCCN guidelines
- Abnormal CD44 activation of hepatocytes with nonalcoholic fatty accumulation in rat hepatocarcinogenesis
- Laparoscopic dissection of the hepatic node: The trans lesser omentum approach
- Multi-institutional retrospective analysis of FOLFlRl in patients with advanced biliary tract cancers
