哺乳动物体细胞核移植表观遗传重编程研究进展
2019-12-24杨旭琼吴珍芳李紫聪
杨旭琼,吴珍芳,李紫聪
哺乳动物体细胞核移植表观遗传重编程研究进展
杨旭琼,吴珍芳,李紫聪
华南农业大学动物科学学院,国家生猪种业工程技术研究中心,广州 510642
体细胞核移植(somatic cell nuclear transfer, SCNT)是唯一能赋予体细胞基因组全能性的生殖工程技术,对动物种质资源保存、畜牧业发展和生物医学研究等具有重大意义。尽管该技术已经取得了许多研究进展,但哺乳动物克隆胚胎的发育效率依然很低,严重限制其在畜牧业和生物医学上的应用。导致克隆胚胎发育效率低的主要原因是体细胞重编程错误或重编程不完全,主要表现为:印记基因表达异常、DNA甲基化异常,组蛋白修饰异常等。本文简要介绍了体细胞核移植技术,系统总结了哺乳动物克隆胚胎发育效率低的主要影响因素,以期为提升体细胞克隆效率相关研究与实践提供理论参考。
体细胞核移植;;DNA甲基化;组蛋白修饰
体细胞核移植(somatic cell nuclear transfer, SCNT)又称体细胞克隆,指利用显微操作技术,以去核卵母细胞为受体,单细胞核为供体,将体细胞核移入成熟的去核卵母细胞中,激活形成克隆胚胎,进而培育出基因型与供体体细胞相同的克隆动物(图1)。体细胞核移植技术是当代生命科学研究和应用的关键技术之一,是生命科学研究高水平发展的体现,在农业动物生产、药物生产、再生医学和保护宝贵遗传资源等方面具有广泛的应用价值。然而,克隆胚胎的发育效率远远低于体外受精胚的发育效率,哺乳动物的克隆胚胎发育效率只有1%~5%[1],严重限制了克隆技术的发展。导致体细胞克隆效率低下的主要原因是体细胞重编程错误或重编程不完全[2,3]。鉴于此,研究人员希望通过一些有效的技术手段来提高克隆胚胎的发育能力,如采用不同的细胞系作为供体细胞,同时优化克隆操作的融合参数[4~7]、敲除印记基因(X-inactive specific transcript)[8]、RNA干扰技术(RNA interference, RNAi)抑制基因的异常表达[9]、注射Kdm4b或Kdm5b去甲基化酶[10,11]等。这些研究虽然取得了一定的效果,但大多数研究结果表明克隆效率并没有大幅度的提高,且克隆动物后代存在诸多异常。因此,如何克服克隆效率低下和解决克隆动物异常成为体细胞克隆研究的热点。
体细胞克隆的供体细胞是高度分化的体细胞,具有高度特异的DNA修饰和组蛋白修饰,以此来维持体细胞的细胞特性。由于克隆胚胎的发育依赖于体细胞的细胞核,所以当供体细胞核被放置到成熟的去核卵母细胞中时,供体细胞核必须经过重编程,抹去分化状态的细胞记忆,激活对早期胚胎发育具有重要作用的基因,如多能性基因、抑制与分化相关的基因,从而使得体细胞获得发育的全能性[12]。由于重编程发生在一个相对较短的时间框架内,若克隆胚胎的发育与正常的胚胎发育不一致,克隆胚胎的发育状态会出现一系列的错误。越来越多的数据表明,错误的重编程模式会使克隆动物出现表观遗传修饰的偏差。如X染色体失活[13]、印记基因与非印记基因的表达[3,12~14]、DNA甲基化[15~17]和染色体修饰[18]等。本文对体细胞核移植技术的发展以及影响克隆胚胎发育效率低的主要原因进行了总结,以期为未来提高哺乳动物克隆发育效率的研究提供参考。
1 体细胞核移植技术
1.1 体细胞核移植技术的发展历程
SCNT技术实现了体细胞的全能性。早在1952年,英国遗传学家Briggs和King将青蛙()胚胎卵裂球细胞的细胞核移植到去核的卵母细胞中,以此来研究胚胎干细胞的细胞核是否发生分化。这是首次利用两栖动物的胚胎干细胞实现胚胎细胞核移植技术,但是当时并未成功克隆出青蛙[19]。1962年,英国生物学家Gurdon首次在两栖动物上利用SCNT技术成功地将分化的青蛙体细胞克隆出小蝌蚪[20]。直到1997年第一头克隆羊“多莉”(Dolly)诞生[21],这是世界上第一个克隆成功的哺乳动物。随后,奶牛()[22]、小鼠()[23]、山羊()[24]、猪()[25,26]、兔子()[27]、猫()[28]、骡子()[29]、马()[30]、大鼠()[31]、猎犬(Canislupus familiaris)[32]和骆驼()[33]等成功克隆的20多种哺乳动物相继问世。2017年,我国成功克隆出世界上第一批体细胞克隆猴()[34]。
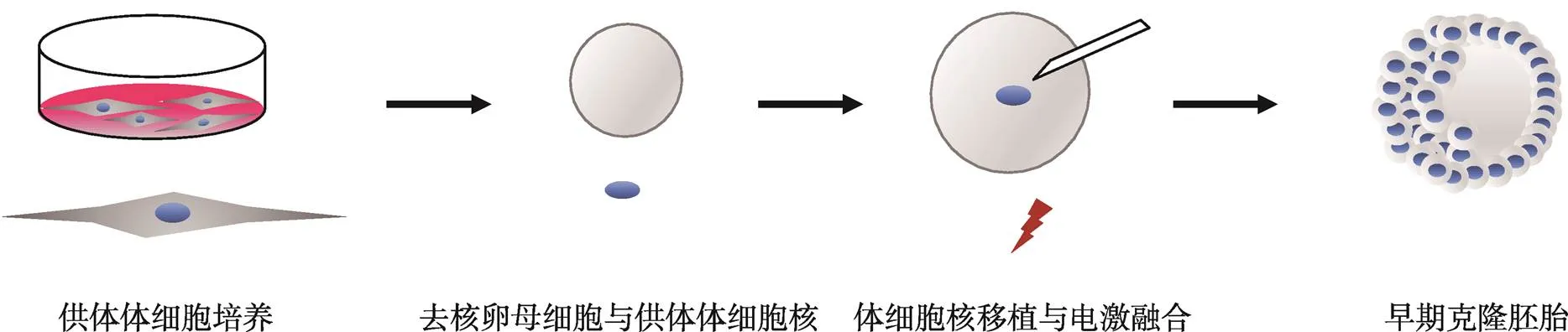
图1 体细胞核移植的流程
1.2 体细胞核移植技术的应用和存在问题
SCNT技术能够培育优良畜种,如选育高品质家畜和扩大繁殖高性能产量个体;此外还可以培育抗病物种、制备异种器官移植供体动物、制备人类疾病动物模型和转基因动物生物反应器等。除了克隆动物外,SCNT在干细胞生物学和人类疾病治疗等方面具有巨大的潜力。受精卵发育到囊胚期分化形成内细胞团(inner cell mass, ICM)和滋养层细胞(trophoblast, TE),其中ICM可分离培养出胚胎干细胞(embryonic stem cells, ESCs)。克隆胚胎发育到囊胚期,ICM也可分离培养出多能干细胞(pluripotent stem cells, PSCs)或者称核移植胚胎干细胞(nuclear transfer embryonic stem cells, ntESCs)[35]。
在生物医学疾病治疗方面,治疗性克隆能够通过培养人的ntESCs,建立并保存每个个体自身的ntESCs,用于组织和器官替代疗法。患者的ntESCs和患者本身具有相同的基因组,可避免排斥反应等不适问题[36,37]。2001年,Wakayama等[38]在成年小鼠体细胞克隆胚胎中分离出了具备多能性特征的核移植ESCs,这也是第一例ntESCs,为人ntESCs的研究提供了实验基础。Rideout等[39]通过同源重组的方法实现了突变等位基因的遗传固定,并获得ntESCs细胞系用作治疗免疫缺陷小鼠。2007年,Byrne等[40]成功获得猴子的ntESCs。2013年,Tachibana等[35]获得了第一例人的ntESCs。随后,更多的实验室相继报道获得了健康成年人[41]、糖尿病[37]及老年性黄斑变性病[36]人体细胞来源的ntESCs。众所周知,ntESCs在人类中的研究能为组织和器官功能失调或受损的患者提供干细胞新来源。这种干细胞可以更新和替换损坏了的细胞或组织,可为上百万的患者减缓病情。在临床上用患有线粒体疾病患者的卵细胞核移植到另一个健康去核卵细胞中,从而阻断线粒体疾病的下一代遗传。2017年张进等[42]利用Leigh氏综合征携带者卵细胞纺锤体移植获得了健康婴儿,这是第一例线粒体替代婴儿。当然,线粒体疾病的替代治疗存在人们关心的伦理问题,这也是阻碍其临床广泛推广的主要原因。
目前,SCNT技术已经成熟,但是依然存在一些问题,严重限制其在生产实际中的应用和发展。总的来说,哺乳动物的克隆效率都比较低下,主要表现在:克隆胚胎体外发育效率低,如猴SCNT单个卵母细胞的孵化效率仅为0.7%[40];体内发育至出生效率低,例如猪的出生率大约在0.5%~1%左右[43,44]。在克隆胚胎中,由TE分化形成的胎盘经常存在异常状态[45],胎盘异常几乎是所有克隆哺乳动物的一个共有特征,例如胎盘增生、胎盘血管缺陷、脐带畸形[46]等。此外,克隆动物的健康状况也存在一定的异常,包括肥胖、免疫及呼吸缺陷和早期死亡等[45,47,48]。由于SCNT技术受到卵母细胞、供体细胞的质量以及代孕母体等个体差异的影响,因此很难从统计学的数据分析上确定影响因素[49]。
2 表观遗传重编程对体细胞核移植胚胎发育效率的影响
生物体的大多数细胞具有相同的遗传物质,SCNT重编程主要通过表观遗传重编程来实现。在克隆胚胎发育早期,存在体细胞标记和细胞类型特异性分化记忆。无论是体细胞标记还是细胞特异性分化记忆都可能导致特定的重编程错误,致使在克隆胚胎发育过程中出现各种异常。若要使其正常发育,克隆胚胎应该以某种方式克服这两个表观遗传障碍。因此,当供体细胞核与去核的卵母细胞融合后,供体细胞核需要对核内已有的表观遗传修饰进行重编程,即擦除供体细胞表观遗传模式,激活与胚胎发育相关的基因,抑制与细胞分化相关的基因,重新获得发育的全能性。当胚胎附植于子宫后,胚胎从全能状态再分化,用于组织生成及器官发生[50]。而在克隆胚胎重编程的过程中,由于体细胞的表观遗传修饰去除不完全,未能建立起正确的表观遗传修饰来调控胚胎的正常发育,致使其出现各种异常。表观遗传重编程主要包括基因组印记、X染色体失活、DNA甲基化和组蛋白修饰等(表1)。
2.1 抑制印记基因Xist表达可显著提高体细胞核移植胚胎发育效率
XY型哺乳动物,其X和Y染色体是由同源常染色体进化而来。由于雄性和雌性X染色体上的基因数目不同,两者之间存在大规模的遗传失衡,为平衡这种剂量差异,在雌性胚胎发育过程中会选择失活其中一条X染色体[51]。基因是X染色体上顺式调控X染色体失活的印记基因,其转录产物是在X染色体失活中心(X-chromosome inactivation, XCI)开始转录的长链非编码RNA(long non-coding RNA, lncRNA),转录产物lncRNA通过包围整条X染色体使得X染色体失活[52]。
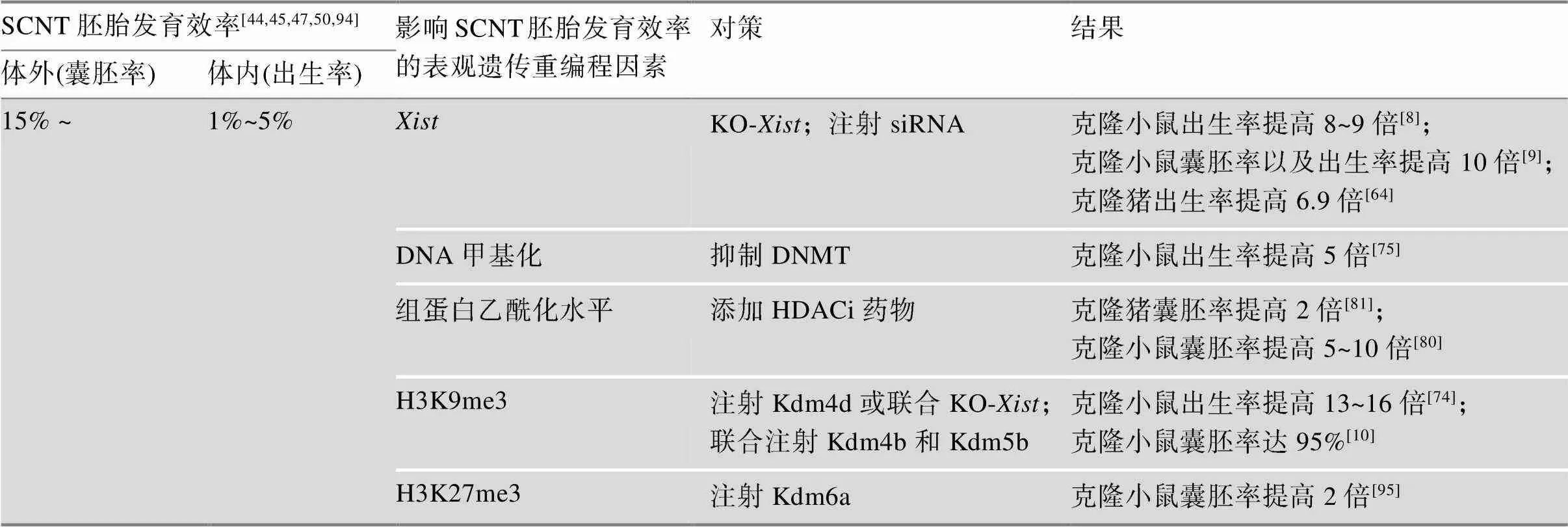
表1 小鼠和猪SCNT胚胎发育效率的表观遗传重编程影响因素及对策
哺乳动物X染色体存在两种形式的失活方式:印记失活和随机失活。XCI在胚胎发育早期建立,受胚胎发育调控,且调控方式与物种种类密切相关[53]。雌性小鼠胚胎发育过程中,在胚胎2~4细胞期基因从父源X染色体上开始转录启动表达,致使父方来源X染色体失活,此印记表达模式,在胚胎外组织中始终维持[54]。当胚胎发育到囊胚期时,这种印记形式的X染色体失活会在ICM中经历再活化,直至胚胎着床期,ICM中会随机失活一条X染色体[55]。但是,人类胚胎发育中表达模式并没有像小鼠一样,而是在胚胎发育后期随机失活[56]。
动物克隆胚胎发育过程中存在许多表达异常的基因,是其中之一[57]。印记基因的表达与表观遗传修饰乙酰化、甲基化密切相关,包括组蛋白H3和H4低乙酰化、H3-lysine 4(H3K4)低甲基化、H3-lysine 9 (H3K9)甲基化和多梳沉默复合体(polycomb repressive complex 2, PRC2)依赖的H3-lysine 27 (H3K27)甲基化等[58,59]。比较克隆胚胎和受精胚胎的转录产物,发现无论雌性或雄性小鼠克隆胚胎中基因都异常表达,其X染色体连锁基因都受到持续地特异性抑制,导致染色体水平的基因大面积下调[8]。同样,Fukuda等[60]发现小鼠克隆胚胎中异常表达,X染色体异常失活。研究表明,在小鼠克隆胚胎桑葚胚期开始异常高表达,结果导致了染色体水平的大面积基因下调,利用基因缺陷型供体细胞用于克隆实验,可显著提高小鼠克隆效率,克隆效率提高到8~9倍[8]。在雄性小鼠克隆胚胎中注入抗的小干扰RNA (siRNA),也观察到了类似的效果,同时也表明克隆胚胎植入前的异常表达严重影响克隆胚胎的发育能力[9]。敲除供体细胞的基因或干扰克隆胚胎中的基因显著的提高了小鼠的克隆效率,这说明纠正基因的错误表达对克隆胚胎的发育效率有显著作用。
对猪而言,发育异常的克隆胎儿同样存在的异常表达,且这种异常始于桑椹胚期[61]。通过RNAi的方式,在克隆胚胎1-细胞期注射siRNA,结果表明猪克隆胚的表达异常升高,小幅度提高了猪克隆胚胎发育效率[57]。一方面,由于克隆所用供体细胞是猪肾髓质部细胞,该细胞易老化,易发生癌变,致使表达异常升高;另一方面,因为siRNA作用时间太短,当猪克隆胚胎发育到桑椹胚时,siRNA已经失去它的干扰作用,所以通过注射siRNA提高猪克隆胚胎的发育效率似乎并不可行。此外,通过RNAi的方式干扰基因,对提高猪孤雌胚胎发育效率也有显著效果[62]。Yang等[63]利用TALEN技术突变猪供体细胞的基因,结果表明X染色体部分再活化,并没有提高猪克隆效率。Ruan等[64]利用TALEN技术在猪供体细胞基因第一外显子重复序列前以插入大片段的方式破坏重复序列,进而失活基因,大幅度提高了猪的克隆发育效率。但是,出生猪只数较少,共移植530个猪克隆胚胎,获得健康克隆胎儿11只。
2.2 DNA去甲基化水平影响体细胞核移植胚胎发育效率
哺乳动物DNA甲基化是指在DNA甲基化转移酶(DNA methyltransferase, DNMT)的帮助下,将DNA分子中S-腺苷蛋氨酸的甲基转移至胞嘧啶残基的第5位碳原子上的过程[65~67]。DNA甲基化由DNMT建立和维持,相反地,TET蛋白酶(ten-eleven translocation, TET)可以催化5-mC (5-methylcytosine, 5-mC)转化为5-羟甲基胞嘧啶(5-hydroxymethylcytosine, 5-hmC),进而启动DNA去甲基化程序[68,69]。哺乳动物早期胚胎发育,基因组中的DNA甲基化修饰会发生重编程过程,广泛进行去甲基化,以此在囊胚期达到最低水平。牛、鼠、猪等动物受精后,父本和母本经历不同的去甲基化方式,前者主动去甲基化,后者被动去甲基化[16,70]。DNA去甲基化是细胞多能性建立和维持的关键步骤,是重编程的第一步,同时也是核移植后早期胚胎发育正常启动和维持的重要环节[71],对表观遗传修饰起到关键作用。体细胞基因组的CpG岛大多数处于高度甲基化状态,全面去甲基化则是SCNT重编程的必须步骤[72]。相比正常的体外受精胚胎,克隆胚胎基因组去甲基化也发生在卵裂时期,但是克隆胚胎去甲基化不完全,其基因组甲基化水平更接近体细胞的状态[50]。Inoue等[73]研究发现,在小鼠受精胚胎中,除印记基因外,新合成的DNA大多未被甲基化。然而,Matoba等[74]研究发现在小鼠克隆胚胎囊胚期,某些基因的启动子部位具有高水平的DNA甲基化。Gao等[75]研究表明异常的DNA再甲基化阻碍了合子基因组激活,是影响SCNT胚胎发育的重要表观遗传障碍。DNMT的抑制能够克服DNA再甲基化的缺陷,同时提高植入后SCNT胚胎发育效率及克隆效率,抑制DNMT和过表达组蛋白赖氨酸去甲基化酶(K-demethylases, Kdms)相结合的方法可以进一步提高克隆效率。以上研究表明,DNA甲基化程度是重要的表观遗传障碍之一。
2.3 组蛋白修饰可改善体细胞核移植胚胎发育效率
组蛋白脱乙酰酶(histone deacetylase, HDAC)可调节组蛋白的乙酰化水平,从而实现对基因表达的表观遗传调控[76]。组蛋白脱乙酰酶抑制剂(histone deacetylase inhibitor, HDACi)通过其功能基团与HDAC的Zn2+形成螯合物,抑制HDAC的活性,增加细胞内组蛋白的乙酰化程度,从而提高靶基因的表达水平[76]。HDACi在动物克隆胚胎发育中被广泛用来改善不同物种胚胎发育的重编程[77]。早在2006年Kishigami等[78]和Rybouchkin等[79]发现HDACi能使小鼠克隆胚胎效率从1%提高到6%。曲古抑菌素A (trichostatin A, TSA)是一种有效的HDACi。Inoue等[80]通过添加TSA药物处理小鼠克隆胚胎,显著提高了克隆胚胎2-细胞期后的发育效率,从而将克隆效率提高了5~10倍,但是TSA对克隆胚胎中异常表达的基因数量以及表达模式没有影响。同样,HDACi药物治疗使得猪[31,81,82]、牛[83~85]的克隆胚胎发育效率均有所提高,但其对SCNT重编程的机制仍不清楚。此外,也有研究表明用HDACi处理猪克隆胚胎,H3K14、H4K5和H4K8等赖氨酸残基出现乙酰化现象[86,87]。
哺乳动物的卵母细胞和精子本身处于转录沉默,受精后,受精卵则恢复转录,该过程称为合子基因组激活(zygotic genome activation, ZGA)。不同物种的ZGA时间不同,小鼠和人的ZGA时间分别在胚胎2-细胞期和胚胎8-细胞期。当ZGA启动时,受精胚胎中母系储存的RNA迅速降解,并被新合成的合子RNA取代。同样地,克隆胚胎的ZGA也存在类似机制,且克隆胚胎早期发育过程中出现发育停滞的时间与ZGA时间高度相关。有研究表明,在小鼠克隆胚胎中大约有1000个基因组区域或基因未能在ZGA时间内激活[11]。有趣的是,在重编程抵抗区(reprogramming-resistant regions, RRRs)富含转录抑制标记物H3K9me3,这说明了供体细胞中组蛋白H3K9me3可能是阻止克隆胚胎ZGA的屏障。Matoba等[11]通过注射H3K9me3特异性去甲基酶Kdm4d的mRNAs不仅克服了ZGA缺陷,而且解决了植入前胚胎发育停滞的问题,使得幼崽出生率提高了8%以上。
Wang等[88]揭示了异染色质组蛋白修饰H3K9me3在配子细胞以及受精后和早期胚胎发育过程中的重编程与其在逆转座子沉默中的作用及调控机制。相关研究表明,供体细胞和2-细胞期克隆胚胎中在某些区域都富含异染色质组蛋白H3K9me3标记[89],且在小鼠克隆胚胎2-细胞期,存在一些区域没有去甲基化[10]。这一观察结果也证实了供体细胞中H3K9me3是SCNT重编程的表观遗传屏障[11,36,44]。研究表明在小鼠克隆胚胎2-细胞期和4-细胞期注射Kdm4b和Kdm5b去甲基化酶,针对组蛋白H3K9me3和H3K4me3去甲基化,显著提高了囊胚发育率,且从克隆胚胎中成功分离培养出ntESCs[10]。Matoba等[74]采用敲除(KO-)供体细胞与Kdm4d- mRNA注射相结合的方法,以支持细胞作为供体细胞克隆小鼠,使得克隆效率显著提高到24%。尽管如此,小鼠克隆效率依然低于体外受精发育效率。最近研究表明,通过注射Kdm4b也可以提高猪[64]、牛[90]以及猴[34]的克隆效率。以上结果说明,H3K9me3去甲基化是克隆胚胎正常发育过程中重编程所必需的组蛋白修饰,同时也是克隆胚胎成功重编程的限制因素之一[91]。此外,母源印记H3K27me3组蛋白修饰同样是影响克隆胚胎重编程的重要因素。研究发现,受精胚胎调控印记基因的母源H3K27me3结构域并未在克隆胚胎中建立[92,93],致使H3K27me3依赖性印记基因大部分失去其印记状态,成为双等位基因表达[74]。Inoue等[93]表示印记基因也受母源H3K27me3的调控,由于供体细胞的位点缺少H3K27me3标记,克隆胚胎中H3K27me3的重编程不完全,进而导致克隆胚胎异常激活。因此,为解决供体细胞中H3K27me3的缺失问题,在供体细胞母源等位基因中靶向沉积H3K27me3可能是一个必要的策略。
3 结语与展望
SCNT的成功是生命科学领域的一次重大突破,其在优良种畜扩繁、濒危物种保护、克隆性治疗等方面具有广阔的应用前景。然而,运用克隆技术成功克隆出青蛙距今已有50余年,克隆胚胎发育至成体的成功率仍保持在一个很低水平。尽管,自Dolly羊诞生20年来,科学家致力于SCNT操作过程中影响克隆胚胎发育效率的各种条件和参数的研究,但克隆效率并未得到显著提高。克隆效率低的根本原因是供体细胞核的表观重编程异常[50]。对此,人们需要对重编程过程中染色质和表观基因组的变化进行系统和详细的分析。随着测序技术的更新换代,转录组测序及相关的表观遗传学研究,使得对SCNT的重编程研究成为可能[96]。从技术上来说,获取足够的克隆样本用于此类分析仍然具有较高的难度,但近些年的相关研究证明,利用早期胚胎进行此类研究具有一定的可行性[57,97,98]。SCNT可将分化的体细胞重编程为全能性胚胎,但在克隆胚胎早期发育过程中,大多数克隆胚胎会出现停滞现象,其潜在的分子机制尚未明了。科研人员对提高克隆效率的研究,使得表观遗传障碍与其特定的重编程错误两者之间的关系变得更加清晰,从而更加准确地理解在细胞分化和克隆胚胎植入过程中,表观遗传调节机制的作用。此外,通过比较分析不同重编程系统之间的异同,来探究克隆胚胎的重编程机制也是一种可行的方案。例如,H3K9me3组蛋白修饰、染色质组装因子(CAF1)蛋白质复合物、异染色质蛋白1 (HP1)是诱导多能干细胞(induced pluripotent stem cells, iPSCs)重编程的障碍[99~101],而iPSCs重编程与SCNT的重编程机制类似。对此,在今后的研究中,进一步探究这些重编程障碍是否也在SCNT重编程中起作用,可以作为体细胞核移植潜在的研究方向。
总之,供体核的表观重编程异常修复依然是体细胞核移植研究及发展的重点。利用新型技术,如高通量测序[102,103]、CRISPR/Cas9[104,105]等,将更加快速准确地解析体细胞表观重编程机制,从而大幅度提高克隆效率,降低克隆动物异常表型的发生率,最终将SCNT技术应用于更多领域。
[1] Sung LY, Gao S, Shen H, Yu H, Song Y, Smith SL, Chang CC, Inoue K, Kuo L, Lian J, Li A, Tian XC, Tuck DP, Weissman SM, Yang X, Cheng T. Differentiated cells are more efficient than adult stem cells for cloning by somatic cell nuclear transfer., 2006, 38(11): 1323–1328.
[2] Matoba S, Zhang Y. Somatic cell nuclear transfer reprogramming: mechanisms and applications., 2018, 23(4): 471–485.
[3] Rideout WM 3rd, Eggan K, Jaenisch R. Nuclear cloning and epigenetic reprogramming of the genome., 2001, 293(5532): 1093–1098.
[4] Dinnyés A, Dai Y, Jiang S, Yang X. High developmental rates of vitrified bovine oocytes following parthenogenetic activation, in vitro fertilization, and somatic cell nuclear transfer., 2000, 63(2): 513–518.
[5] Kato Y, Tani T, Tsunoda Y. Cloning of calves from various somatic cell types of male and female adult, newborn and fetal cows., 2000, 120(2): 231–237.
[6] Lee GS, Hyun SH, Kim HS, Kim DY, Lee SH, Lim JM, Lee ES, Kang SK, Lee BC, Hwang WS. Improvement of a porcine somatic cell nuclear transfer technique by optimizing donor cell and recipient oocyte preparations., 2003, 59(9): 1949–1957.
[7] Wilmut I, Schnieke AE, Mcwhir J, Kind AJ, Campbell KHS. Viable offspring derived from fetal and adult mammalian cells., 2007, 9(1): 3–7.
[8] Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii T, Sawai K, Otte AP, Tian XC, Yang X, Ishino F, Abe K, Ogura A. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer., 2010, 330(6003): 496–499.
[9] Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N, Nakamura T, Abe K, Nakano T, Ishino F, Ogura A. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos., 2011, 108(51): 20621–20626.
[10] Liu WQ, Liu XY, Wang CF, Gao YW, Gao R, Kou XC, Zhao YH, Li JY, Wu Y, Xiu WC, Wang S, Yin JQ, Liu W, Cai T, Wang H, Zhang Y, Gao SR. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing., 2016, 2: 16010.
[11] Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation., 2014, 159(4): 884–895.
[12] Humpherys D, Eggan K, Akutsu H, Friedman A, Hochedlinger K, Yanagimachi R, Lander ES, Golub TR, Jaenisch R. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei., 2002, 99(20): 12889– 12894.
[13] Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A, Dai Y, Levine H, Pereira LV, Yang X. Aberrant patterns of X chromosome inactivation in bovine clones., 2002, 31(2): 216–220.
[14] Niemann H, Wrenzycki C, Lucas-Hahn A, Brambrink T, Kues WA, Carnwath JW. Gene expression patterns in bovine in vitro-produced and nuclear transfer-derived embryos and their implications for early development., 2002, 4(1): 29–38.
[15] Bourc'his D, Le Bourhis D, Patin D, Niveleau A, Comizzoli P, Renard JP, Viegas-Péquignot E. Delayed and incomplete reprogramming of chromosome methylation patterns in bovine cloned embryos., 2001, 11(19): 1542–1546.
[16] Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos., 2001, 98(24): 13734–13738.
[17] Kang YK, Koo DB, Park JS, Choi YH, Chung AS, Lee KK, Han YM. Aberrant methylation of donor genome in cloned bovine embryos., 2001, 28(2): 173– 177.
[18] Santos F, Zakhartchenko V, Stojkovic M, Peters A, Jenuwein T, Wolf E, Reik W, Dean W. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos., 2003, 13(13): 1116–1121.
[19] Briggs R, King TJ. Transplantation of living nuclei from blastula cells into enucleated frogs' eggs., 1952, 38(5): 455–463.
[20] Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles., 1962, 10: 622–640.
[21] Wilmut I, Schnieke AE, Mcwhir J, Kind AJ, Campbell KHS. Viable offspring derived from fetal and adult mammalian cells., 1997, 385(6619): 810–813.
[22] Kato Y, Tani T, Sotomaru Y, Kurokawa K, Kato J, Doguchi H, Yasue H, Tsunoda Y. Eight calves cloned from somatic cells of a single adult., 1998, 282(5396): 2095–2098.
[23] Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei., 1998, 394(6691): 369–374.
[24] Baguisi A, Behboodi E, Melican DT, Pollock JS, Destrempes MM, Cammuso C, Williams JL, Nims SD, Porter CA, Midura P, Palacios MJ, Ayres SL, Denniston RS, Hayes ML, Ziomek CA, Meade HM, Godke RA, Gavin WG, Overström EW, Echelard Y. Production of goats by somatic cell nuclear transfer., 1999, 17(5): 456–461.
[25] Polejaeva IA, Chen SH, Vaught TD, Page RL, Mullins J, Ball S, Dai Y, Boone J, Walker S, Ayares DL, Colman A, Campbell KH. Cloned pigs produced by nuclear transfer from adult somatic cells., 2000, 407(6800): 86–90.
[26] Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, Awata T, Hanada H, Perry ACF. Pig cloning by microinjection of fetal fibroblast nuclei., 2000, 289(5482): 1188–1190.
[27] Chesné P, Adenot PG, Viglietta C, Baratte M, Boulanger L, Renard JP. Cloned rabbits produced by nuclear transfer from adult somatic cells., 2002, 20(4): 366–369.
[28] Shin T, Kraemer D, Pryor J, Liu L, Rugila J, Howe L, Buck S, Murphy K, Lyons L, Westhusin M. A cat cloned by nuclear transplantation., 2002, 415(6874): 859.
[29] Woods GL, White KL, Vanderwall DK, Li GP, Aston KI, Bunch TD, Meerdo LN, Pate BJ. A mule cloned from fetal cells by nuclear transfer., 2003, 301(5636): 1063.
[30] Galli C, Lagutina I, Crotti G, Colleoni S, Turini P, Ponderato N, Duchi R, Lazzari G. Pregnancy: a cloned horse born to its dam twin., 2003, 424(6949): 635.
[31] Zhou Q, Renard JP, Le Friec G, Brochard V, Beaujean N, Cherifi Y, Fraichard A, Cozzi J. Generation of fertile cloned rats by regulating oocyte activation., 2003, 302(5648): 1179.
[32] Lee BC, Kim MK, Jang G, Oh HJ, Yuda F, Kim HJ, Hossein MS, Kim JJ, Kang SK, Schatten G, Hwang WS. Dogs cloned from adult somatic cells., 2005, 436(7051): 641.
[33] Wani NA, Wernery U, Hassan FA, Wernery R, Skidmore JA. Production of the first cloned camel by somatic cell nuclear transfer., 2010, 82(2): 373–379.
[34] Liu Z, Cai Y, Wang Y, Nie Y, Zhang C, Xu Y, Zhang X, Lu Y, Wang Z, Poo M, Sun Q. Cloning of macaque monkeys by somatic cell nuclear transfer., 2018, 172(4): 881–887.e7.
[35] Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, Sritanaudomchai H, Masterson K, Larson J, Eaton D, Sadler-Fredd K, Battaglia D, Lee D, Wu D, Jensen J, Patton P, Gokhale S, Stouffer RL, Wolf D, Mitalipov S. Human embryonic stem cells derived by somatic cell nuclear transfer., 2013, 153(6): 1228–1238.
[36] Chung YG, Matoba S, Liu Y, Eum JH, Lu F, Jiang W, Lee JE, Sepilian V, Cha KY, Lee DR, Zhang Y. Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells., 2015, 17(6): 758–766.
[37] Yamada M, Johannesson B, Sagi I, Burnett LC, Kort DH, Prosser RW, Paull D, Nestor MW, Freeby M, Greenberg E, Goland RS, Leibel RL, Solomon SL, Benvenisty N, Sauer MV, Egli D. Human oocytes reprogram adult somatic nuclei of a type 1 diabetic to diploid pluripotent stem cells., 2014, 510(7506): 533–536.
[38] Wakayama T, Tabar V, Rodriguez I, Perry AC, Studer L, Mombaerts P. Differentiation of embryonic stem cell lines generated from adult somatic cells by nuclear transfer., 2001, 292(5517): 740–743.
[39] Rideout WM 3rd, Hochedlinger K, Kyba M, Daley GQ, Jaenisch R. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy., 2002, 109(1): 17–27.
[40] Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Wolf DP, Mitalipov SM. Producing primate embryonic stem cells by somatic cell nuclear transfer., 2007, 450(7169): 497–502.
[41] Chung YG, Eum JH, Lee JE, Shim SH, Sepilian V, Hong SW, Lee Y, Treff NR, Choi YH, Kimbrel EA, Dittman RE, Lanza R, Lee DR. Human somatic cell nuclear transfer using adult cells., 2014, 14(6): 777–780.
[42] Zhang J, Liu H, Luo S, Lu Z, Chávez-Badiola A, Liu Z, Yang M, Merhi Z, Silber SJ, Munné S, Konstantinidis M, Wells D, Tang JJ, Huang T. Live birth derived from oocyte spindle transfer to prevent mitochondrial disease., 2017, 34(4): 361–368.
[43] Lai L, Prather RS. Production of cloned pigs by using somatic cells as donors., 2003, 5(4): 233–241.
[44] Liu Y, Li J, Løvendahl P, Schmidt M, Larsen K, Callesen H. In vitro manipulation techniques of porcine embryos: a meta-analysis related to transfers, pregnancies and piglets., 2015, 27(3): 429–439.
[45] Ogura A, Inoue K, Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer., 2013, 368(1609): 20110329.
[46] Ao Z, Liu DW, Cai GY, Wu ZF, Li ZC. Placental developmental defects in cloned mammalian animals., 2016, 38(5): 402–410.敖政, 刘德武, 蔡更元, 吴珍芳, 李紫聪. 克隆哺乳动物的胎盘发育缺陷. 遗传, 2016, 38(5): 402–410.
[47] Loi P, Iuso D, Czernik M, Ogura A. A New, Dynamic era for somatic cell nuclear transfer?, 2016, 34(10): 791–797.
[48] Ao Z, Liu D, Zhao C, Yue Z, Shi J, Zhou R, Cai G, Zheng E, Li Z, Wu Z. Birth weight, umbilical and placental traits in relation to neonatal loss in cloned pigs., 2017, 57: 94–101.
[49] Oback B. Climbing mount efficiency--small steps, not giant leaps towards higher cloning success in farm animals., 2008, 43(s2): 407–416.
[50] Yang X, Smith SL, Tian XC, Lewin HA, Renard JP, Wakayama T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning., 2007, 39(3): 295–302.
[51] Graves JAM. Sex chromosome specialization and degeneration in mammals., 2006, 124(5): 901–914.
[52] Sahakyan A, Yang Y, Plath K. The role of Xist in X-chromosome dosage compensation., 2018, 28(12): 999–1013.
[53] Furlan G, Rougeulle C. Function and evolution of the long noncoding RNA circuitry orchestrating X-chromosome inactivation in mammals., 2016, 7(5): 702–722.
[54] Shin J, Bossenz M, Chung Y, Ma H, Byron M, Taniguchi-Ishigaki N, Zhu X, Jiao B, Hall LL, Green MR, Jones SN, Hermans-Borgmeyer I, Lawrence JB, Bach I. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice., 2010, 467(7318): 977–981.
[55] Payer B. Developmental regulation of X-chromosome inactivation., 2016, 56: 88–99.
[56] Moreira De Mello JC, De Araújo ES, Stabellini R, Fraga AM, De Souza JES, Sumita DR, Camargo AA, Pereira LV. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome., 2010, 5(6): e10947.
[57] Zeng F, Huang ZH, Yuan YJ, Shi JS, Cai GY, Liu DW, Wu ZF, Li ZC. Effects of RNAi-mediated knockdown of Xist on the developmental efficiency of cloned male porcine embryos., 2016, 62(6): 591–597.
[58] Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing., 2002, 298(5595): 1039–1043.
[59] Nora EP, Heard E. Chromatin structure and nuclear organization dynamics during X-chromosome inactivation., 2010, 75: 333–344.
[60] Fukuda A, Cao F, Morita S, Yamada K, Jincho Y, Tane S, Sotomaru Y, Kono T. Identification of inappropriately reprogrammed genes by large-scale transcriptome analysis of individual cloned mouse blastocysts., 2010, 5(6): e11274.
[61] Yuan L, Wang AF, Yao CG, Huang YY, Duan FF, Lv QY, Wang DX, Ouyang HS, Li ZJ, Lai LX. Aberrant expression of Xist in aborted porcine fetuses derived from somatic cell nuclear transfer embryos., 2014, 15(12): 21631–21643.
[62] Chen XY, Zhu ZW, Yu FX, Huang J, Jia RX, Pan JZ. Effect of shRNA-mediated Xist knockdown on the quality of porcine parthenogenetic embryos., 2019, 248(1): 140–148.
[63] Yang Y, Wu D, Liu D, Shi J, Zhou R, He X, Quan J, Cai G, Zheng E, Wu Z, Li Z. Mutation of the XIST gene upregulates expression of X-linked genes but decreases the developmental rates of cloned male porcine embryos., 2017, 84(6): 525–534.
[64] Ruan D, Peng J, Wang X, Ouyang Z, Zou Q, Yang Y, Chen F, Ge W, Wu H, Liu Z, Zhao Y, Zhao B, Zhang Q, Lai C, Fan N, Zhou Z, Liu Q, Li N, Jin Q, Shi H, Xie J, Song H, Yang X, Chen J, Wang K, Li X, Lai L. XIST derepression in active X chromosome hinders pig somatic cell nuclear transfer., 2018, 10(2): 494–508.
[65] Sulewska A, Niklinska W, Kozlowski M, Minarowski L, Naumnik W, Niklinski J, Dabrowska K, Chyczewski L. DNA methylation in states of cell physiology and pathology., 2007, 45(3): 149–158.
[66] Guo L, Li H, Han ZM. Effect of DNA methylation and histone modification during the de-velopment of cloned animals., 2010, 32(8): 762–768.郭磊, 李慧, 韩之明. DNA甲基化和组蛋白修饰在克隆动物发育过程中的作用. 遗传, 2010, 32(8): 762–768.
[67] Song HW, An TZ, Piao SH, Wang CS. Mammalian DNA methylation and its roles during the induced re-programming of somatic cells., 2014, 36(5): 431–438.宋红卫, 安铁洙, 朴善花, 王春生. 哺乳动物DNA甲基化及其在体细胞诱导重编程中的作用. 遗传, 2014, 36(5): 431–438.
[68] Deng DJ. DNA methylation and demethylation: current status and future per-spective., 2014, 36(5): 403–410.邓大君. DNA甲基化和去甲基化的研究现状及思考. 遗传, 2014, 36(5): 403–410.
[69] Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond., 2017, 18(9): 517–534.
[70] Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome., 2000, 403(6769): 501–502.
[71] Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei., 2004, 6(10): 984–990.
[72] Zhang Y, Charlton J, Karnik R, Beerman I, Smith ZD, Gu H, Boyle P, Mi X, Clement K, Pop R, Gnirke A, Rossi DJ, Meissner A. Targets and genomic constraints of ectopic Dnmt3b expression., 2018, 7: pii: e40757.
[73] Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos., 2011, 334(6053): 194.
[74] Matoba S, Wang H, Jiang L, Lu F, Iwabuchi KA, Wu X, Inoue K, Yang L, Press W, Lee JT, Ogura A, Shen L, Zhang Y. Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development., 2018, 23(3): 343– 354.e345.
[75] Gao R, Wang C, Gao Y, Xiu W, Chen J, Kou X, Zhao Y, Liao Y, Bai D, Qiao Z, Yang L, Wang M, Zang R, Liu X, Jia Y, Li Y, Zhang Y, Yin J, Wang H, Wan X, Liu W, Zhang Y, Gao S. Inhibition of aberrant DNA re-methylation improves post-implantation development of somatic cell nuclear transfer embryos., 2018, 23(3): 426–435.e425.
[76] Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer., 2002, 1(4): 287–299.
[77] Ji HL, Lu SS, Pan DK. Epigenetic reprogramming by somatic cell nuclear transfer: questions and potential solutions., 2014, 36(12): 1211–1218.纪慧丽, 卢晟盛, 潘登科. 体细胞核移植后表观遗传重编程的异常及其修复. 遗传, 2014, 36(12): 1211–1218.
[78] Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NV, Wakayama S, Bui HT, Wakayama T. Significant improvement of mouse cloning technique by treatment with trichostatin a after somatic nuclear transfer., 2006, 340(1): 183–189.
[79] Rybouchkin A, Kato Y, Tsunoda Y. Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer., 2006, 74(6): 1083–1089.
[80] Inoue K, Oikawa M, Kamimura S, Ogonuki N, NakamuraT, Nakano T, Abe K, Ogura A. Trichostatin a specifically improves the aberrant expression of transcription factor genes in embryos produced by somatic cell nuclear transfer., 2015, 5: 10127.
[81] Bohrer RC, Duggavathi R, Bordignon V. Inhibition of histone deacetylases enhances DNA damage repair in SCNT embryos., 2014, 13(13): 2138–2148.
[82] Jin JX, Kang JD, Li S, Jin L, Zhu HY, Guo Q, Gao QS, Yan CG, Yin XJ. PXD101 significantly improves nuclear reprogramming and the in vitro developmental competence of porcine SCNT embryos., 2015, 456(1): 156–161.
[83] Akagi S, Matsukawa K, Mizutani E, Fukunari K, Kaneda M, Watanabe S, Takahashi S. Treatment with a histone deacetylase inhibitor after nuclear transfer improves the preimplantation development of cloned bovine embryos., 2011, 57(1): 120–126.
[84] Li X, Ao X, Bai L, Li D, Liu X, Wei Z, Bou S, Li G. VPA selectively regulates pluripotency gene expression on donor cell and improve SCNT embryo development., 2018, 54(7): 496–504.
[85] Song BS, Yoon SB, Sim BW, Kim YH, Cha JJ, Choi SA, Jeong KJ, Kim JS, Huh JW, Lee SR, Kim SH, Kim SU, Chang KT. Valproic acid enhances early development of bovine somatic cell nuclear transfer embryos by alleviating endoplasmic reticulum stress., 2014, 26(3): 432–440.
[86] Zhao J, Hao Y, Ross JW, Spate LD, Walters EM, Samuel MS, Rieke A, Murphy CN, Prather RS. Histone deacetylase inhibitors improve in vitro and in vivo developmental competence of somatic cell nuclear transfer porcine embryos., 2010, 12(1): 75–83.
[87] Martinez-Diaz MA, Che L, Albornoz M, Seneda MM, Collis D, Coutinho AR, El-Beirouthi N, Laurin D, Zhao X, Bordignon V. Pre-and postimplantation development of swine-cloned embryos derived from fibroblasts and bone marrow cells after inhibition of histone deacetylases., 2010, 12(1): 85–94.
[88] Wang C, Liu X, Gao Y, Yang L, Li C, Liu W, Chen C, Kou X, Zhao Y, Chen J, Wang Y, Le R, Wang H, Duan T, Zhang Y, Gao S. Reprogramming of H3K9me3-dependentheterochromatin during mammalian embryo development., 2018, 20(5): 620–631.
[89] Djekidel MN, Inoue A, Matoba S, Suzuki T, Zhang CX, Lu FL, Jiang L, Zhang Y. Reprogramming of chromatin accessibility in somatic cell nuclear transfer is DNA replication independent., 2018, 23(7): 1939– 1947.
[90] Liu X, Wang Y, Gao Y, Su J, Zhang J, Xing X, Zhou C, Yao K, An Q, Zhang Y. H3K9 demethylase KDM4E is an epigenetic regulator for bovine embryonic development and a defective factor for nuclear reprogramming., 2018, 145(4), pii: dev158261.
[91] Hang XW, Cheng XR, Wang N, Zhang YW, Liao C, Jin LH, Lei L. Histone variant H3.3 and its functions in reprogramming., 2018, 40(3): 186– 196.黄星卫, 程香荣, 王楠, 张雨薇, 廖辰, 金连弘, 雷蕾. 组蛋白H3变体H3.3及其在细胞重编程中的作用. 遗传, 2018, 40(3): 186–196.
[92] Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. Maternal H3K27me3 controls DNA methylation-independent imprinting., 2017, 547(7664): 419–424.
[93] Inoue A, Jiang L, Lu F, Zhang Y. Genomic imprinting of Xist by maternal H3K27me3., 2017, 31(19): 1927–1932.
[94] Keefer CL. Lessons learned from nuclear transfer (cloning)., 2008, 69(1): 48–54.
[95] Bai GY, Song SH, Zhang YW, Huang X, Huang XW, Sun RZ, Lei L. Kdm6a overexpression improves the development of cloned mouse embryos., 2018, 26(1): 24–32.
[96] K L, Chen YJ, Gao SR. Historical review of reprogramming and pluripotent stem cell research in China., 2018, 40(10): 825–840.康岚, 陈嘉瑜, 高绍荣. 中国细胞重编程和多能干细胞研究进展. 遗传, 2018, 40(10): 825–840.
[97] Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, Zhang Y. Establishing chromatin regulatory landscape during mouse preimplantation development., 2016, 165(6): 1375–1388.
[98] Ke Y, Xu Y, Chen X, Feng S, Liu Z, Sun Y, Yao X, Li F, Zhu W, Gao L, Chen H, Du Z, Xie W, Xu X, Huang X, Liu J. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis., 2017, 170(2): 367–381.e320.
[99] Cheloufi S, Elling U, Hopfgartner B, Jung YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Euong Ang C, Tenen D, Wesche DJ, Abazova N, Hogue M, Tasdemir N, Brumbaugh J, Rathert P, Jude J, Ferrari F, Blanco A, Fellner M, Wenzel D, Zinner M, Vidal SE, Bell O, Stadtfeld M, Chang HY, Almouzni G, Lowe SW, Rinn J, Wernig M, Aravin A, Shi Y, Park PJ, Penninger JM, Zuber J, Hochedlinger K. The histone chaperone CAF-1 safeguards somatic cell identity., 2015, 528(7581): 218–224.
[100] Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome., 2012, 151(5): 994–1004.
[101] Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, Mckee R, Huang CY, Patel S, Lopez D, Mishra N, Pellegrini M, Carey M, Garcia BA, Plath K. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency., 2013, 15(7): 872–882.
[102] Okae H, Matoba S, Nagashima T, Mizutani E, Inoue K, Ogonuki N, Chiba H, Funayama R, Tanaka S, Yaegashi N, Nakayama K, Sasaki H, Ogura A, Arima T. RNA sequencing-based identification of aberrant imprinting in cloned mice., 2014, 23(4): 992– 1001.
[103] Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, Gould J, Liu S, Lin S, Berube P, Lee L, Chen J, Brumbaugh J, Rigollet P, Hochedlinger K, Jaenisch R, Regev A, Lander ES. Optimal-transport asnalysis of single-cell gene expression identifies developmental trajectories in reprogramming., 2019, 176(4): 928–943.e922.
[104] Fan ZQ, Yang M, Regouski M, Polejaeva IA. Gene knockouts in goats using CRISPR/Cas9 system and somatic cell nuclear transfer., 2019, 1874: 373–390.
[105] Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes., 2014, 32(4): 347–355.
Advances in epigenetic reprogramming of somatic cells nuclear transfer in mammals
Xuqiong Yang, Zhenfang Wu, Zicong Li
Somatic cell nuclear transfer (SCNT) is the only reproductive engineering technique that can confer genomic totipotency on somatic cell. SCNT is of great significance for animal germplasm conservation, animal husbandry development, and biomedical research. Although many research advances have been made in this technology, the developmental rate of SCNT mammalian embryos is very low, which seriously limits the application of SCNT in animal husbandry and biomedicine. The primary reason for the low efficiency of cloned embryos is somatic cell reprogramming errors or incomplete reprogramming. These errors or incompleteness present as the abnormal expression of imprinted gene, abnormal DNA methylation, and abnormal histone modification. In this review, we summarize the main factors that influence the low development efficiency of mammalian cloned embryos to provide theoretical reference for the research and practice of improving somatic cell cloning efficiency.
somatic cell nuclear transfer (SCNT);; DNA methylation; histone modification
2019-07-03;
2019-10-07
国家自然科学基金面上项目(编号:31772554)资助[Supported by the National Natural Science Foundation of China(No. 31772554)
杨旭琼,硕士研究生,专业方向:动物遗传育种与繁殖。E-mail: 1814639793@qq.com
李紫聪,教授,博士生导师,研究方向:动物遗传育种与繁殖。E-mail: lizicongcong@163.com
10.16288/j.yczz.19-193
2019/11/19 13:16:00
URI: http://kns.cnki.net/kcms/detail/11.1913.r.20191118.1633.002.html
(责任编委: 高绍荣)
