Identification and Binding Mechanism Evaluation of Novel ACE Inhibitory Peptides from Ovalbumin
2019-12-04YUZhipengWUSijiaZHAOWenzhuDINGLongLIUJingbo
YU Zhipeng, WU Sijia, ZHAO Wenzhu,*, DING Long, LIU Jingbo,*
(1. National & Local Joint Engineering Research Center of Storage, Processing and Safety Control Technology for Fresh Agricultural and Aquatic Products, College of Food Science and Engineering, Bohai University, Jinzhou 121013, China;2. Lab of Nutrition and Functional Food, Jilin University, Changchun 130062, China)
Abstract: The present study aimed to characterize potent angiotensin converting enzyme (ACE) inhibitory peptides from ovalbumin. Ovalbumin was in silico cleaved by gastrointestinal proteases. Subsequently, the toxicity, solubility, and absorption, distribution, metabolism, and excretion (ADME) properties of the tripeptides were predicted, and out of these,tripeptides strongly bound to ACE were selected and their activity was confirmed. As a result, a novel ACE inhibitory tripeptide CIK with IC50 value of (161 ± 0.06) μmol/L was identified. CIK-ACE complex was stabilized by 5 conventional hydrogen bonds, 2 carbon hydrogen bonds, and 4 salt bridges. The best docking poses of CIK could bind with the amino acid residues of ACE Gln281, His353, Ala354, Glu376, Val380, His383, Glu384, Lys511, His513, Tyr523, and Phe527. The best pharmacophore model was generated consisting of three hydrogen bond acceptors, one hydrophobic region, and one positive ionizable center, and four features were matched with the tripeptide CIK.
Keywords: angiotensin converting enzyme inhibitory peptides; ovalbumin; multistep virtual screening; Cys-Ile-Lys;pharmacophore model
Angiotensin converting enzyme (ACE) as a key enzyme in the modulation of blood pressure plays a key role in the renin-angiotension-aldosterone system (RAAS)[1]. ACE transforms an inactive form of angiotensin I (ANG I; Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-His-Leu) to vasoconstrictor angiotensin II (ANG II; Asp-Arg-Val-Tyr-Ile-His-Pro-Phe)[2], leading to the degradation of bradykinin in the kallikrein-kimin system[3]. Thus, ACE is a protein target for therapeutic hypertension, and there are many drugs that have been developed against ACE, such as Captopril,Enalapril, and Lisinopril. Although the chemically synthetic ACE inhibitors are effective, they cause potential health hazards[4-6]. Therefore, natural ACE inhibitors have recently received increasing attention. Peptides derived from natural food protein sources have been shown significant ACE inhibitory activity, such as hydrolysates from Bambara bean protein[7], tomato waste protein[8], rice bran protein[9], and egg white protein[10]. Thus, ovalbumin is major protein fractions in the total egg proteins and account for about 55% of the total proteins[11]. Ovalbumin is a good source of bioactive peptides. Many studies focused on enzymatic hydrolysates of ovalbumin and its ACE inhibitory activity[12-14]. Therein,gastrointestinal (GI) digestion is a common method to release bioactive peptides from natural foods. In vitro GI digestion of bioactive peptides is rapid, inexpensive, and safe[15].Furthermore, it has been used to generate bioactive peptides which can resist in vivo degradation of gastrointestinal enzymes and perform physiological functions.
However, the isolation and purification of bioactive peptides from protein hydrolysate is generally timeconsuming and expensive[16]. The process can be simplified and accelerated by in silico GI digestion and multistep virtual screening method[1]. Many studies have proven the availability of the in silico screening method, which can be regarded as a valid alternative to classical enzymatic hydrolysis method[17-19]. Some studies suggested that the smaller molecular weight of a peptide, through the cell membrane and cross the intestinal barrier easier[20], and also suggested that the amino acid sequence of peptide is generally short chain to interact with ACE active sites[4]. Tripeptides with reasonable molecular weight have been shown to have significant ACE inhibitory activity, such as MCS[21], LKP[22],VWP, VNP[23], and LHV[24].
Based on the above rationale, the main objective of the present study was to characterize novel ACE inhibitory tripeptides from ovalbumin using in silico GI digestion and multistep virtual screening method. Furthermore, the CDOOKER program in discovery studio 2017 was used to explore the molecular mechanism of inhibition of ACE by the tripeptide. Finally, pharmacophore model was constructed to analyze the key chemical features of ACE inhibitory peptides.
1 Materials and Methods
1.1 Materials and reagents
ACE (protease from rabbit lung), hippuryl-histidylleucine (HHL), and hippuric acid were purchased from Sigma-Aldrich Co. (St. Louis, Missouri, USA). Acetonitrile,methanol and trifluoroacetic acid (TFA) were purchased from Fisher Scientific Co. (Waltham, MA, USA) and were of chromatographic grade. All the other reagents and chemicals used were analytical grade. All synthetic peptides used in this study were provided by Shanghai Top Peptide Biological Technology Corporation (Shanghai, China).
1.2 Instruments and equipment
Discovery studio 2017 R2 client were purchased from Neotrident Technology Ltd. (Beijing, China); Reversed phase high-performance liquid chromatography (RP-HPLC)were purchased from Shimadzu (China) Co. Ltd., Beijing Branch (Beijing, China); HH-501 thermostat water bath were purchased from Changzhou Guoyu Instrument manufacturing Co. Ltd. (Changzhou, Jiangsu, China).
1.3 Method
1.3.1 Proteolysis simulation of ovalbumin
Three typical proteases of Pepsin (EC 3.4.23.1), Trypsin(EC 3.4.21.4), and Chymotrypsin (EC 3.4.21.1) were chosen and used in present study. The program ExPASy PeptideCutter (http://web.expasy.org/peptide_cutter/)[11]was used to predict the peptides sequence released from ovalbumin (Accession of NCBI: 0705172A)[25]. Number of peptides were obtained and compared with known ACE-inhibitory peptides in BIOPEP-UWM (http://www.uwm.edu.pl/biochemia/index.php/en/biopep)[26]. Subsequently,unknown tripeptides were selected and their toxicity,solubility, and absorption, distribution, metabolism, and excretion (ADME) properties were predicted.
1.3.2 Prediction of toxicity, solubility, and ADME properties
The tool ToxinPred (http://crdd.osdd.net/raghava//toxinpred/) was utilized to predict the toxicity of tripeptides according to the important physico-chemical properties[27].The peptide property calculator (http://www.innovagen.com/)was used to estimate the solubility[28]. The ADME properties,such as Human Intestinal Absorption (HIA), Caco-2 permeability, and metabolism parameters predictions were performed using the web server admetSAR (http://lmmd.ecust.edu.cn/admetsar1/)[29].
1.3.3 Molecular docking and analysis of the interactions
The crystal structure of ACE (1O86)-Lisinopril chosen from Protein Data Bank (PDB) was used as the target to screen peptides strongly bond with ACE[6]. The structure of ACE was prepared by removing water and adding up the hydrogen atoms[30]. The binding sites were identified based on the Lisinopril, the current ligand of ACE (1O86). The structures of peptides were drawn by Discovery Studio (DS)2017 Client software. CDOCKER protocol of DS 2017 was used for molecular docking. The docking were carried out with coordinates x: 41.207 3, y: 33.943 1 and z: 46.520 1 with a radius of 9. The best pose was output based on the CDOCKER score.
1.3.4 ACE inhibitory activity assay
The assay of in vitro ACE inhibitory activity was performed by RP-HPLC method by Yu et al[31]. The experiment was repeated three times.
1.3.5 Construction and validation of pharmacophore model
Six ACE inhibitory peptides with lower IC50values included tripeptides MCS (IC50value of 0.29 μmol/L)[21],MKP (IC50value of 0.30 μmol/L)[4], LKP (IC50value of 0.32 μmol/L)[22], IVR (IC50value of 0.81 μmol/L)[32], LRP(IC50value of 1.20 μmol/L)[33], and VIP (IC50value of 1.69 μmol/L)[34](shown in Fig. 1) were used as training set. The six tripeptides were generated used Discovery Studio 2017 R2 Client, and then the Principal value of the six tripeptides was set at 2. The MaxOmitFeat value of the peptides was set at 0[35]. The pharmacophore with qualitative common features was generated by the pharmacophores module of Discovery Studio 2017 R2 Client. Maximum pharmacophore hypotheses were set at 10. The minimum interfeature distance was set to 2.97 Å. All the other parameters were kept the defaults. Furthermore, the pharmacophore models were validated using Ligand Profiler program of Discovery Studio 2017 R2 Client. Fifteen ACE inhibitory peptides (i.e., VWP, VNP, LHV, MAW, VMP,TLS, ASL, CIK, GVR, SLR, VVR, KYK, NPR, AER, and AVF) were used as decoy set. Most selective model for ACE and rigid fitting method between the tripeptides of decoy set and pharmacophore was used in this study.
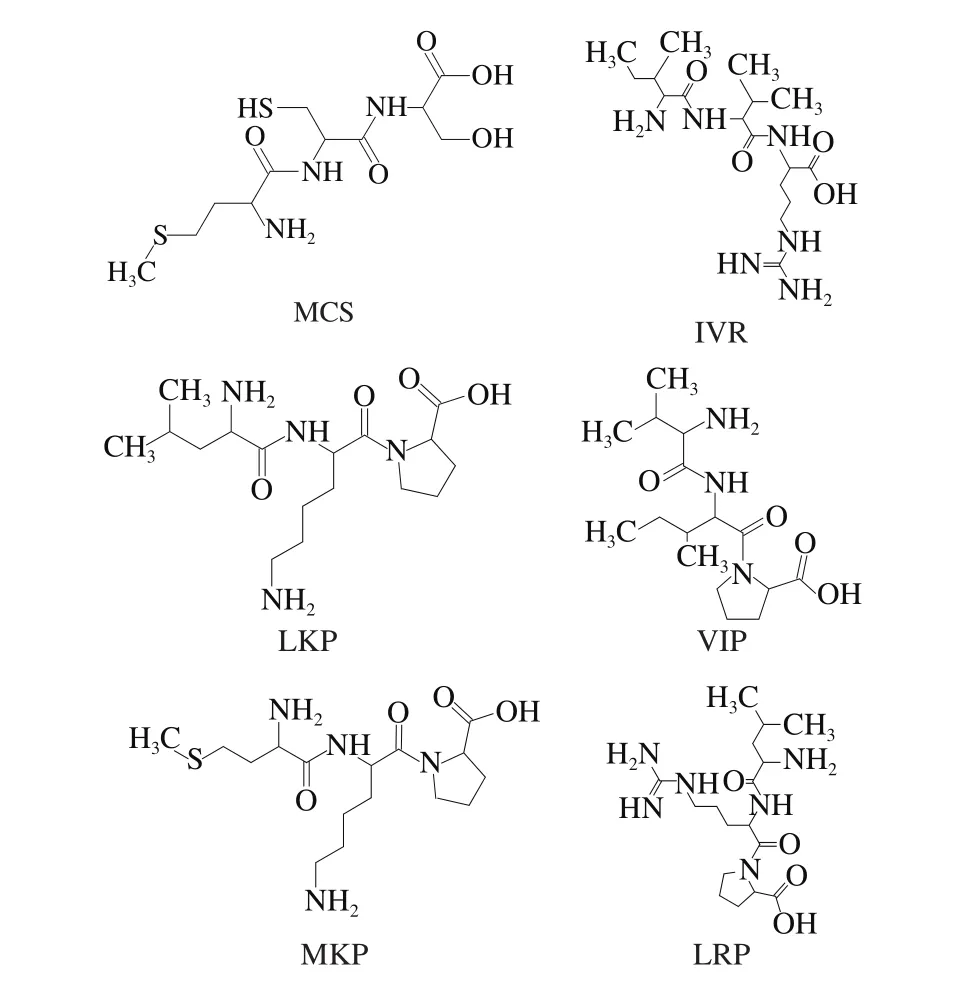
Fig. 1 Chemical structures of five ACE inhibitory peptides as training set for pharmacophore construction
1.4 Statistical analysis
2 Results and Analysis
2.1 Toxicity, solubility, and ADME properties predictions of tripeptides
Following combined gastrointestinal digestion, a total of 104 peptide sequences were generated, and 11 unknown tripeptides (shown in Table 1) were selected for prediction of their the toxicity, solubility, and ADMET properties. The results showed that all of them were non-toxic along with all the important physicochemical properties. Tripeptides GAK, VVR, DIL, ASR, PEY, LEL, TEW, and CIK showed good water-solubility while others showed poor watersolubility. Water-solubility has an influence on the degree of absorbing of bioactive peptides. Dissolution is a limiting factor for the performance of physiological functions[36].Tripeptides with good water solubility may present high biological availability. Thus, tripeptides GAK, VVR, DIL,ASR, PEY, LEL, TEW, and CIK were selected to perform prediction of ADME properties. The absorption factor of tripeptides GAK, VVR, DIL, ASR, PEY, LEL, TEW,and CIK can be summarized in terms of HIA and Caco-2 permeability. Peptides PEY, CIK, and TEW were labeled as HIA+, and the peptides GAK, VVR, DIL, ASR, and LEL were labeled as HIA-, which indicated that the HIA% of PEY, CIK, and TEW were higher than 30%, on the contrary,the HIA% of GAK, VVR, DIL, ASR, and LEL were lower than 30% (Table 1). HIA could help predicting the intestinal absorption of a peptide[37], which is a key to screen potent ACE inhibitor peptide. All of the tripeptides were labeled as moderate-poor permeability (Caco2-), which indicated that the Caco-2 permeability values (Papp) of the tripeptides were lower than or equal to 8 × 10-6cm/s[38]. Moreover, all of the tripeptides were neither substrate nor inhibitor, which means that CYP450 could not recognize those tripeptides. There are less drug-drug interactions in the course of metabolism.Consequently, peptides CIK, PEY, and TEW with better pharmacokinetic and pharmacodynamics properties passed the ADME filter and were chosen for further study.

Table 1 In silico calculation results of toxicity, solubility, ADME properties and docking score of tripeptides from ovalbumin
2.2 Virtual screening for ACE inhibitory tripeptides
In order to screen out the potential ACE inhibitory tripeptides and explore the molecular mechanism of ACE interactions with the tripeptides, peptides PEY, CIK and TEW satisfying the criteria in the previous simulation filter were subjected for molecular docking by using CDOOCKER program, a flexible docking tool of Discovery Studio 2017 software. The 3D structure of ACE (PDB ID: 1O86) in complex with Lisinopril and derived from human was chosen as the target.
The best docking poses of tripeptides CIK, PEY, and TEW were shown in Fig. 2, and value of CDOCKERINTERACTION-ENERGY was -97.398, -96.014 2, and-85.242 9 kcal/mol, respectively. A lower ‘CDOCKERINTERACTION-ENERGY’ score denotes a more favorable binding. As shown in Fig. 2a, the atoms HE22 and HZ3 of the residues Gln281 and Lys511 of ACE formed conventional hydrogen bonds with the atoms O32 and O54 of CIK generating lengths 2.85 Å and 2.27 Å, respectively. The residues His353 (NE2), Ala354 (O), and Glu384 (OE2) of ACE also formed conventional hydrogen bonds with the atoms H3, H4, and H11 of CIK at a distance of 2.98, 2.98 Å,and 2.27 Å, respectively. The hydrogen atom (H6) of CIK formed carbon hydrogen bond with NE2 of the residue His353 (2.72 Å). Also, another carbon hydrogen bond is observed between the residue His513 (HE1) and the oxygen atom (O55) of CIK, which formed a distance at 2.43 Å.Four salt bridges were observed in the complex, the first one involves the oxygen atom (OE2) of residue Glu162 with the hydrogen atom (H52) of CIK (2.43 Å); the second one involves the oxygen atom (OD1) of the residue Asp377 with the hydrogen atom (H50) of CIK (1.92 Å); the third one involves the oxygen atom (OE2) of the residue Glu384 with the hydrogen atom (H2) of CIK (2.10 Å); and the fourth one involves the hydrogen atom (HZ1) of the residue Lys511 with the oxygen atom (O55) of CIK (1.90 Å). Moreover,the residue Glu376 (OE2) formed an attractive charge with CIK, and the residue His383 formed a Pi-Sulfur interaction with CIK. Additionally, some hydrophobic interactions were also involved with the residues Val380, His383, Tyr523, and Phe527 of ACE. As shown in Fig. 2b, PEY-ACE complex is stabilized by 2 conventional hydrogen bonds, 4 carbon hydrogen bonds, and 2 salt bridges. One of the conventional hydrogen bond is between hydrogen atom (H33) of PEY with nitrogen atom (NE2) of the residue His353 (2.64 Å),and the other involves the hydrogen atom (HH) of the residue Tyr520 with oxygen atom (O28) of PEY (2.06 Å). The first carbon hydrogen bond involves the interactions between the residue His353 (HE1) and PEY (O29) at a distance of 2.67 Å;the second involves the interactions between the residue His387 (HD2) and PEY (O52) at a distance of 3.08 Å;the third involves the interactions between PEY (H6) and the residue Asp415 (OD2) at a distance of 2.79 Å; and the fourth involves the interactions between the residue His513(HE1) and PEY (O29) at a distance of 2.57 Å. One of the salt bridges is between PEY (H3) with Asp415 (OD2) at a distance of 2.55 Å, and the other is between Lys511 (HE1)with PEY (O29) at a distance of 1.94 Å. And a hydrophobic interaction between the residue Val518 and PEY was observed. Additionally, His383, Arg522, and Zn701 of ACE are involved in some electrostatic interactions.
In the docked complex, the atoms H10 and O29 of TEW formed conventional hydrogen bonds with ACE at the oxygen atom (OE2) of Glu162 and the hydrogen atom (HH)of Tyr520, each at a distance 1.98 Å and 2.65 Å, respectively(shown in Fig. 2c). The hydrogen atom (H33) of TEW formed 2 conventional hydrogen bonds with ACE at the residues Ala354 (O) and Glu384 (OE2), each at a distance 2.95 Å and 2.60 Å, respectively. The oxygen atoms (O56) and hydrogen atom (H35) of TEW formed carbon hydrogen bonds with His387 (HD2) and Tyr523 (hydroxyl group) of ACE each at a distance 3.10 Å and 2.52 Å, respectively. The hydrogen atom (HE1) of the residue His513 of ACE formed two carbon hydrogen bonds with O29 and O31 of TEW with distance 2.57 Å and 2.70 Å. Furthermore, one salt bridge between nitrogen atom (NZ1) of the residue Lys511 of ACE with oxygen atom (O29) of TEW (1.89 Å) were observed.Additionally, the residues Glu162, Glu376, and Zn701 of ACE established 3 attractive charge interactions with TEW.
The structure of ACE active site composed of three pockets, such as S1, S2, and S1’. S1 pocket includes Ala354,Glu384, and Tyr523 residues and S2 pocket includes Gln281,His353, Lys511, His513, and Tyr520 residues, while S1’contains Glu162 residue[39-42]. Further, ACE has a zinc ion(Zn(II)) in its active site coordinated by residues His383,His387, and Glu411[39], which may fortify its significant role in ACE inhibition.
The interactions of Lisinopril and Captopril with ACE were shown in Fig. 2d and Fig. 2e, and values of CDOCKER-INTERACTION-ENERGY were -70.442 6 and-41.140 7 kcal/mol, respectively. The results have demonstrated that Lisinopril interacts with ACE at Glu162,Gln281, His353, Ala354, His383, Glu384, Lys511, His513,Tyr520, Tyr523, and Zn701, and Captopril interacts with ACE at His353, His383, and Tyr523. Therefore, the residues His353, His383, and Tyr523 of ACE might play major roles in ACE binding and should be very important screening indicators.

Fig. 2 Molecular interactions of the tripeptides Cys-Ile-Lys (CIK) (a),Pro-Glu-Tyr (PEY) (b), Thr-Glu-Trp (TEW) (c), lisinopril (d) and captopril (e) at the active site of ACE
Taken together, the tripeptide CIK performed the lowest docking score which can indicate the tripeptide had highbinding affinities with the ACE. Tripeptide CIK could bind with the active sites of ACE, i.e., Gln281, His353, Ala354,His383, Glu384, Lys511, His513, Tyr520, and Tyr523.Moreover, tripeptide CIK had the highest number of predicted hydrogen bonds. Thus, tripeptide CIK was subjected to further ACE-inhibitory activity assay.
2.3 In vitro ACE inhibitory activity of tripeptide CIK
The ACE inhibitory activity of synthetic tripeptide CIK was performed by RP-HPLC method. Tripeptide CIK showed high ACE inhibitory activity with IC50value of (161 ± 0.06) μmol/L.In recent years, many ACE inhibitory tripeptides have been prepared, and their sequences have been identified.Comparing to tripeptides AER (293.81 μmol/L)[43], KYK(280.12 μmol/L), SLR (170 μmol/L), GVR (166.48 μmol/L)[44],VVR (249.69 μmol/L), and NPR (285.4 μmol/L)[45], CIK showed a greater ACE inhibitory activity. The results proved that gastrointestinal enzymes play an effective role on the release of ACE inhibitory peptides from the ovalbumin. The results suggested thatin silicoGI digestion and multistep virtual screening approach may be a more reliable method to substitute for classical enzymatic hydrolysis method.
2.4 ACE mechanism-based inhibitory pharmacophore model construction validation
Ten chemical feature based pharmacophore models were generated by using HipHop program of Discovery Studio 2017[46]. The best pharmacophore model was assessed on the Fit values of the peptides which contain five features[47].As shown in Table 2, among the ten pharmacophores,pharmacophore-3 estimated the highest fit value of 4.999 77 for the most active peptide MCS and 2.631 36 for the least active peptide LRP. Pharmacophore-3 consists of three hydrogen bond acceptors (HBA), one hydrophobic region(Hyd), and one positive ionizable (PI) with rank value of 84.153. Consequently, pharmacophore-3 was chosen as the best model (shown in Fig. 3a). The mapping on the tripeptide CIK (shown in Fig. 3c) showed that three HBA located on the oxygen atoms O13, O32, and O54 of the carboxyl groups,and one PI located on the nitrogen atom N1 of the amine group. Four pharmacophore features of pharmacophore-3 were matched with the tripeptide CIK.
In addition, the mapping results suggested the Hyd is missing in most of lower active tripeptides, indicating that Hyd feature play a significant role in ACE inhibitory peptides screening (shown in Table 3). Thus, future studies should focus on the structural optimization of CIK based on the Hyd feature of pharmacophore-3.
Table 3 Mapping results of ten pharmacophores with tripeptides
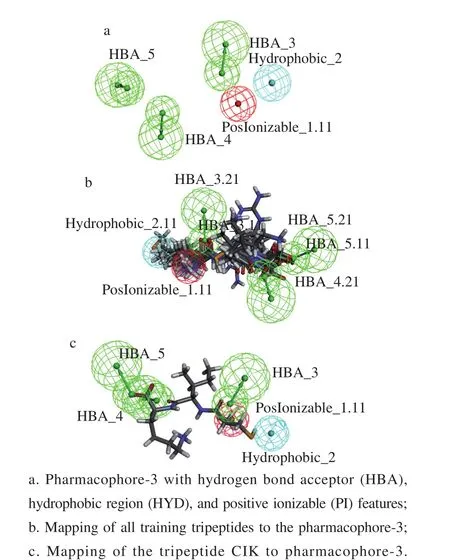
Fig. 3 Representation of the ACE pharmacophore model
3 Conclusion
In conclusion, tripeptide CIK from ovalbumin was identified using a combination of in silico digestion with gastrointestinal enzymes and hierarchical virtual screening.Peptide CIK could bind with the active site residues of ACE by 11 hydrogen bonds and showed significant ACE inhibitory activity in vitro with IC50value of (161 ± 0.06) μmol/L.Pharmacophore-3 was the best pharmacophore model consisted of the common chemical features hydrogen bond acceptor, hydrophobic region, and positive ionizable,and four pharmacophore features were matched with the tripeptide CIK. Hyd feature play a significant role in ACE inhibitory peptides screening. Future studies should focus on the structural optimization of CIK based on the Hyd feature of pharmacophore-3. This work suggested that in silico digestion and multistep virtual screening method may be a more reliable method to substitute for classical enzymatic hydrolysis method.
