冷鲜鸡肉表面四环素和磺胺耐药菌的菌群多样性分析
2019-11-11姚春霞黄柳娟周昌艳王卫国
邵 毅,姚春霞,,黄柳娟,冯 博,周昌艳,白 冰,王 华,王卫国
冷鲜鸡肉表面四环素和磺胺耐药菌的菌群多样性分析
邵 毅1,2,姚春霞1,2,※,黄柳娟1,冯 博1,周昌艳1,2,白 冰1,王 华3,4,王卫国5
(1. 上海市农业科学院农产品质量标准与检测技术研究所,上海 201403;2. 上海市设施园艺技术重点实验室,上海 201403;3. 美国俄亥俄州立大学食品科学与技术系,哥伦布 43210;4. 复旦大学生命科学院微生物学和微生物工程系,上海 200433;5. 上海国荣果业专业合作社,上海 201516)
为探索冷鲜鸡肉产品表面的抗生素耐药菌的菌群结构,利用IonS5TMXL测序平台对18个市售冷鲜鸡肉样品表面的可培养四环素耐药菌和磺胺耐药菌进行了研究。结果表明,2类耐药菌中分别注释出59个和58个已明确属名的属,相对丰度最大的3个门均为变形菌门、拟杆菌门和厚壁菌门。共享菌属中不动杆菌属、假单胞菌属、变形杆菌属、柠檬酸杆菌属、香味菌属和漫游球菌属的禽源和人源分离株的多重耐药性已被大量研究所证实;而禽源肉杆菌属等16个菌属的耐药特性还未见报道。各采样点分别鉴定出5~39个特有操作分类单元,分属3~32个属。该研究反映了冷鲜鸡肉表面2类耐药菌的污染状况,为后期冷鲜鸡肉产品表面耐药菌的迁移风险评估和控制技术研究提供参考。
微生物;细菌;抗生素;冷鲜鸡;四环素耐药菌;磺胺耐药菌;菌群多样性
0 引 言
抗生素耐药(antibiotic resistant,ART)致病菌可能通过食物链从农业生产及农产品中扩散至人类[1],耐药(antibiotic resistance,AR)基因还能通过水平基因迁移(horizontal gene transfer,HGT)从养殖动物源细菌迁移至人类共生菌中[2-3],因此,ART菌的快速增多对人类公共卫生安全造成巨大威胁[4]。耐药性的发生和迁移机制复杂,除抗生素筛选压外,AR基因池的大小[5]和ART菌的种属[6]是影响食源性耐药基因迁移潜势的关键因素。然而,目前食源性ART菌的研究主要在致病菌中展开,且依赖于有限菌种或单个菌落的培养分离[7]。在复杂的细菌生态系统中,除致病菌外,数量庞大的其他细菌的耐药数据十分有限。借助高通量二代测序技术,对潜在的ART细菌进行菌群组成研究,有助于完善ART菌的种属信息,进而全面揭示耐药机制。
为避免禽流感等动物源性疾病的人群暴发,冷鲜鸡逐渐成为禽肉市场的主要产品类型。但冷鲜鸡在屠宰和贮运过程中很可能发生微生物的污染和滋生,成为ART菌的载体[8-10]。这些ART菌的菌群组成和迁移风险尚不明确。因此,本研究通过选择性培养获得具有四环素或磺胺耐药表型的细菌,并通过IonS5TMXL平台分析其菌落结构,从理论上探讨ART菌的潜在污染来源和迁移风险,为冷鲜鸡肉产品表面有害微生物的控制和安全加工奠定基础。
1 材料与方法
1.1 试验材料与主要试剂
18份冷鲜鸡样品(表1)于2018年9月4日至2018年10月16日购于上海市6个大型超市和生鲜超市,冷鲜鸡样品的生产或包装日期为采样当天。采样时避免样品与手及人呼出的气体接触。样品用无菌袋盛装,在冰盒(约4 ℃)中于3 h内运回实验室,进行ART菌的分离。
脑心浸液肉汤(brain heart infusion broth,BHI)琼脂培养基(英国 Oxoid);四环素(tetracycline,Tet)、磺胺甲恶唑(sulfamethoxazole,Sul)、甲氧苄啶(trimethoprim,Tri)(美国Sigma);真菌抑制剂放线菌酮(cycloheximide,Cyc)(美国Amresco);生理盐水(广东环凯微生物科技有限公司);细菌基因组提取试剂盒(德国QIAGEN)。
表1 冷鲜鸡肉样品信息
1.2 仪器设备
SX-500型高压灭菌锅(日本TOMY)用于细菌培养基的制备;BAGMIXER400型均质机(法国Interscience)用于鸡肉样品的拍打均质;1300 SERIES A2型生物安全柜(美国Thermo Fisher scientific)用于细菌的分离操作;Medcenter Einrichtungen GmbH型恒温培养箱(德国Friocell)用于细菌的培养;Mini-Sub cell GT型水平核酸电泳系统(美国Bio-rad)和NanoDrop 2000c型超微量分光光度计(美国Thermo)分别用于菌体DNA提取质量的定性和定量分析;Criterion Stain Free型凝胶成像系统(美国Bio-rad)用于菌体DNA电泳结果的显示;T100型PCR仪(美国Bio-rad)用于菌体16S rDNA的聚合酶链式反应(polymerase chain reaction,PCR)扩增。
1.3 方 法
1.3.1 四环素耐药菌(tetracycline resistant bacteria,Tetr)和磺胺耐药菌(sulfamethoxazole resistant bacteria,Sulr)的分离
参考Huang等[11]的方法从冷鲜鸡样品表面分离对四环素或磺胺甲恶唑/甲氧苄啶耐药的细菌:无菌条件下将表面肌肉和皮剪碎,混匀,准确称量25 g置于无菌袋中,加入225 mL无菌生理盐水,拍打均质5 min,用移液枪吸取200L混匀的均质液均匀涂布于含有放线菌酮和四环素(Cyc+Tet)或放线菌酮和磺胺甲恶唑及甲氧苄啶(Cyc+ Sul+Tri)的BHI平板上,所有平板中Cyc的浓度均为100g/mL。为了尽量减少耐药假阳性细菌的生长,参考美国临床和实验室标准协会(Clinical and Laboratory Standards Institute,CLSI)对几种典型临床致病菌对四环素或磺胺甲恶唑/甲氧苄啶耐药折点的规定[12],将培养皿中四环素、磺胺甲恶唑和甲氧苄啶的浓度点分别定为48、228和12g/mL,即大多数致病菌相应耐药折点的3倍;同时,将本实验室前期优化的鸡肉表面抗生素耐药细菌分离培养条件((32±1)℃,48 h)[8]中的培养时间缩短为24 h。
1.3.2 DNA提取及PCR扩增
刮取培养皿上所有菌体,用2 mL生理盐水重悬混匀,取100L提取细菌总DNA。经1%琼脂糖凝胶电泳和核酸定量仪测定,确定所提细菌总DNA的质量后,PCR扩增核糖体亚基16S rDNA基因的V3-V4区。上游引物为341F:5’-CCTAYGGGRBGCASCAG-3’,下游引物为806R:5’-GGACTACNNGGGTATCTAAT-3’。扩增结束后,用2%琼脂糖凝胶电泳和核酸定量仪检测PCR产物的质量,委托北京诺禾致源生物信息科技有限公司基于IonS5TMXL测序平台,利用单端测序(Single-End)的方法,对构建的小片段文库进行高通量测序。
1.3.3 高通量测序数据分析
将IonS5TMXL下机数据导出fastq文件。根据barcode序列区分各个样本的数据,并进行嵌合体过滤,得到用于后续分析的有效数据(clean reads)。以97%的一致性(identity)对所有样本的有效标签(effective tags)进行操作分类单元(operational taxonomic unit,OTU)聚类和物种分类分析。随后,一方面用SILVA132(http://www.arb-silva.de/)的SSUrRNA数据库在界(kingdom)、门(phylum)、纲(class)、目(order)、科(family)、属(genus)和种(species)7个水平对每个OTU的序列做物种注释分析,得到对应的物种信息和基于物种的丰度分布情况;另一方面,对OTU进行丰度、-多样性计算、维恩(Venn)图和花瓣图等分析,以得到样本内物种丰富度和均匀度信息、不同样本或分组间的共有(共享菌群[13])和特有(特有菌群)OTU信息等。用EXCEL的单因素方差分析比对各样品中四环素耐药菌和磺胺耐药菌-多样性指数的差异,计算P值(P-value),当< 0.05时,差异显著。用EXCEL绘制菌落丰度柱状图。
2 结果与分析
2.1 α-多样性分析
对每个冷鲜鸡肉样品表面的Tetr菌和Sulr菌的组成分别进行多样性分析(-多样性),评估其复杂度和多样性。菌群覆盖度(good’s coverage)指数反映了测序深度,当测序深度覆盖到样品中所有的物种时,其值为1;物种数(observed species)指数和超(chao1)指数反映了单个样品中物种的数量,即群落的丰富度;香农(shannon)指数和辛普森(simpon)指数反映了群落的多样性,相同丰富度时,均匀度越高,群落的多样性越大,2个指数的值就越大。结果表明,各样品中2种ART菌的覆盖指数为0.999~1(表 2),说明几乎所有序列都被测出,本次多样性分析结果能反映样品菌群组成的真实情况。18个样品测序共获得503个OTU,平均每个样品的Tetr菌和Sulr菌中分别获得139个和156个OTU。本试验涉及的冷鲜鸡样品中,Tetr菌的OTU数、丰富度指数和多样性指数与Sulr菌相比,均无显著性差异,可能是BHI培养基对菌种的筛选偏好造成的。
2.2 菌群结构分析
在门、纲、目、科和属5个水平上对菌群结构进行分析。结果表明,在门水平上,不论是Tetr菌还是Sulr菌,相对丰度最大的3个门均为变形菌门(Proteobacteria)、拟杆菌门(Bacteroidetes)和(厚壁菌门Firmicutes),与已报道的冷鲜鸡肉产品表面细菌的主要菌门[14]类似,3个菌门之和分别占两类ART菌的99.69%~100.00%和95.73%~99.99%(图1)。
在属水平上,Tetr菌和Sulr菌中各注释出59个和58个已明确属名的属。林奕岑等[13]在肉鸡盲肠细菌多样性分析研究中鉴定出85个属的细菌,其中51个已被注释;肖英平等[14]注释了冷鲜鸡肉表面细菌的菌属在数量级上为10~102之间,其中十大菌属中除了嗜冷杆菌属(spp.)和黄杆菌属(spp.)仅在本研究的个别样品中发现外,其余包括希瓦氏属(spp.)、假单胞菌属(spp.)、不动杆菌属(spp.)、环丝菌属(spp.)、香味菌属(spp.)、魏斯氏菌属(spp.)、漫游球菌属(spp.)和肉杆菌属(spp.)在内的8个属在本研究的冷鲜鸡表面ART菌中都获得了鉴定,表明冷鲜鸡表面的细菌的耐药比率较高。
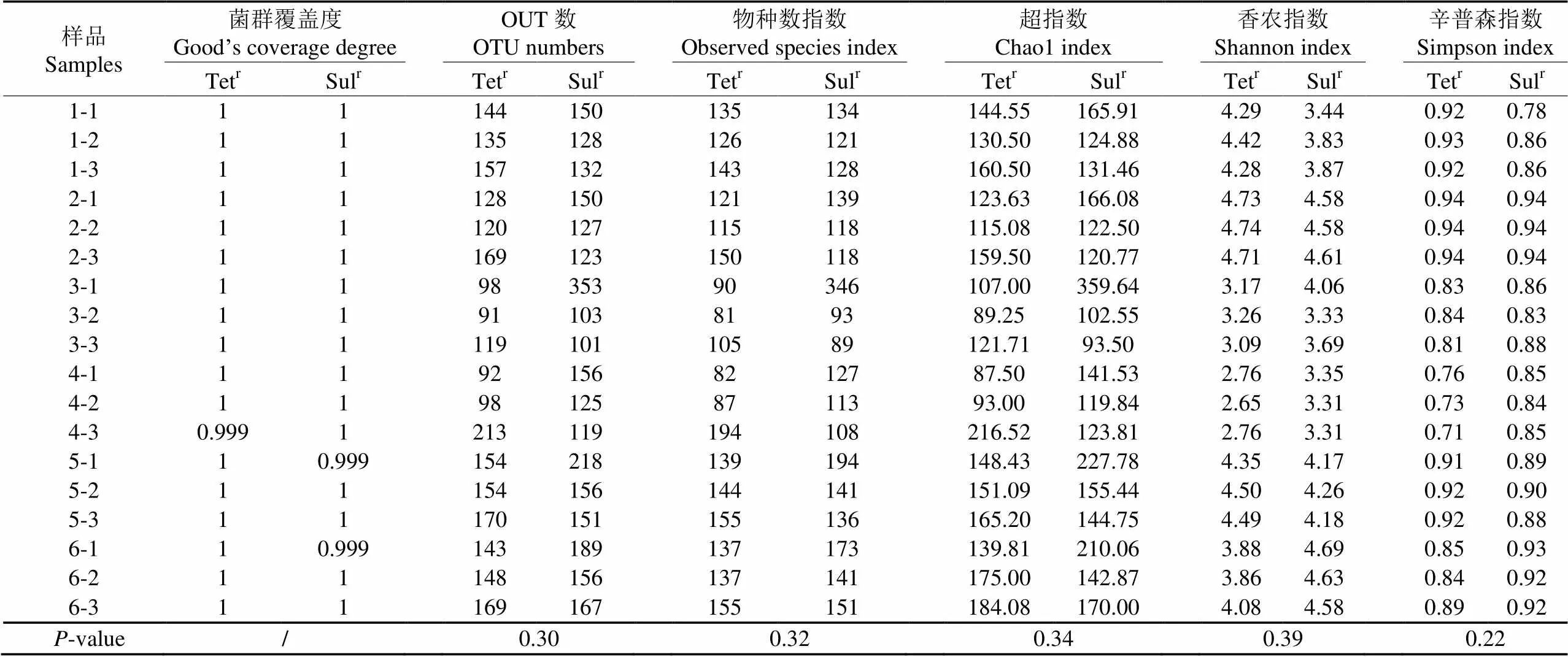
表2 样品OTU数量和α-多样性指数
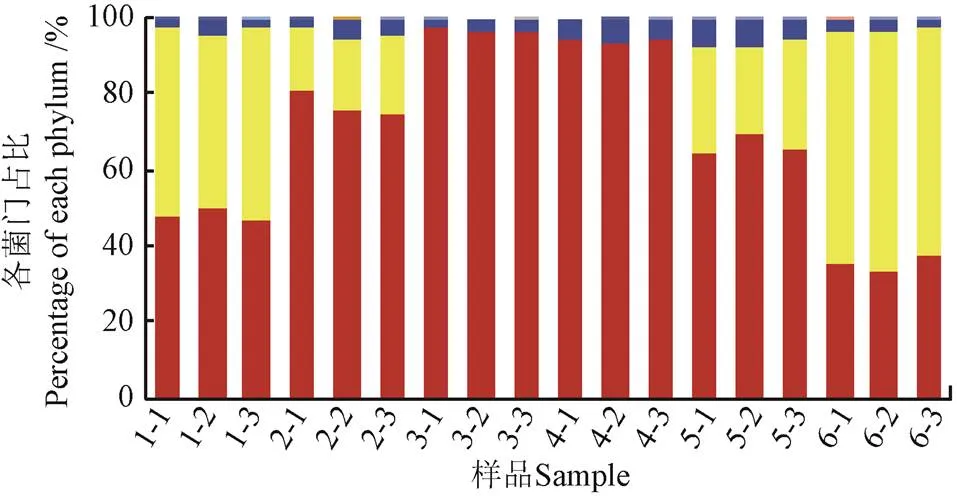
a. 四环素耐药菌
a. Tetracycline-resistant bacteria
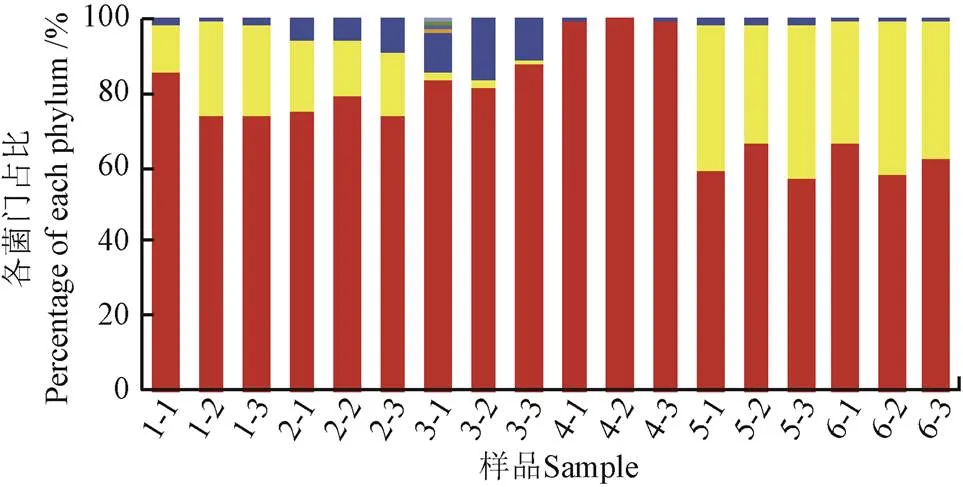
b. 磺胺耐药菌
b. Sulfamethoxazole-resistant bacteria

图1 冷鲜鸡肉表面四环素耐药菌和磺胺耐药菌在门水平上的分布
冷鲜鸡肉表面四环素耐药菌和磺胺耐药菌在属水平上的分布见图2。
图2a显示,Tetr菌和Sulr菌在属水平上的菌群结构有所不同,丰度平均值最高的Tetr菌依次为不动杆菌属、香味菌属、假单胞菌属、柠檬酸杆菌属(spp.)和稳杆菌属(spp.),而丰度最高的Sulr菌依次为假单胞菌属、香味菌属、气单孢菌属(spp.)、不动杆菌属和变形杆菌属(spp.)(图2b)。前人研究表明,肠球菌属(spp.)、葡萄球菌属(spp.)、肉杆菌属、乳球菌属(spp.)和乳杆菌属(spp.)的细菌较多地参与了AR基因的水平迁移[5-6],在本研究中也鉴定出了其中的葡萄球菌属和肉杆菌属,因此有必要在今后的研究中进一步确认相应菌属单个菌落的耐药性,并评估AR基因向人类共生菌迁移的风险。
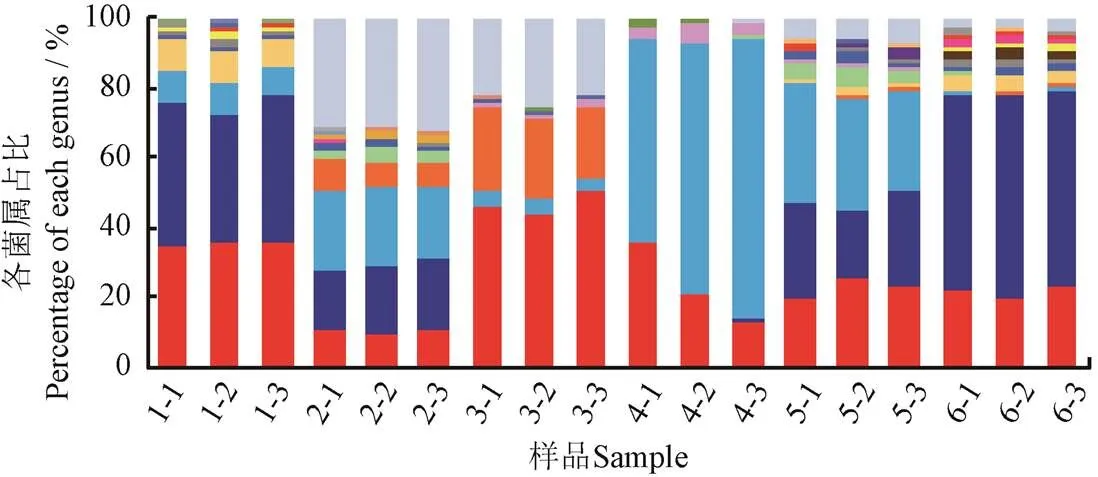
a. 四环素耐药菌
a. Tetracycline-resistant bacteria
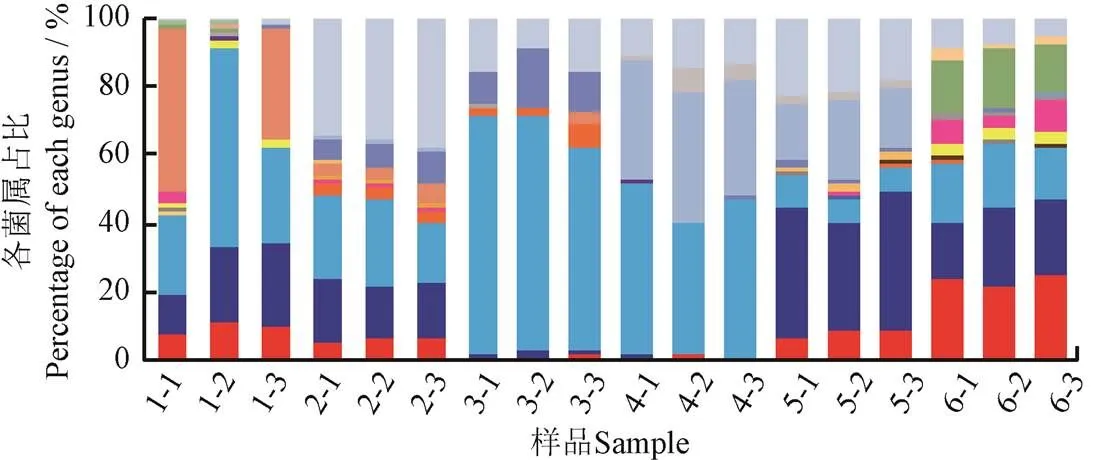
b. 磺胺耐药菌
b. Sulfamethoxazole-resistant bacteria

图2 冷鲜鸡肉表面四环素耐药菌和磺胺耐药菌在属水平上的分布
2.3 共享菌群分析
共享菌群在不同来源的冷鲜鸡样品表面均有检出,反映所有样品的菌群组成共性,推测其在其他冷鲜鸡中存在的可能性较高,造成的耐药风险也相应较高,因此予以重点分析。统计所有样品中共同存在的OTU,结果表明,在门水平上,不论是Tetr菌还是Sulr菌,共享菌群都包括变形菌门、拟杆菌门和厚壁菌门。在属水平上,Tetr菌和Sulr菌各有18个菌属(56个OUT)和19个菌属(63个OUT)的共享菌群(表3),前人报道的冷鲜鸡肉优势菌属[15]在本研究中均有发现,说明冷鲜鸡肉表面各类优势细菌均已不同程度地存在具有耐药性的种。
作为冷鲜鸡肉微生物菌群中的优势菌属,不动杆菌属、假单胞菌属和肉杆菌属是鸡肉腐败过程中的主要污染菌属[16-18];变形杆菌属普遍存在于健康肉鸡和市售生鸡产品[19]以及养鸡环境[20]中;柠檬酸杆菌属是常见的肠道非致病菌(在机体抵抗力降低时偶尔致病),冷鲜鸡[15]和腐败的扒鸡[21]中都曾分离到柠檬酸杆菌;香味菌属是罕见的机会致病菌,目前的感染报道主要发生在中国;而漫游球菌属和魏斯氏菌属在禽类中的研究报道尚不多。值得注意的是,不动杆菌属、假单胞菌属、肉杆菌属和魏斯氏菌属等菌属的细菌在高盐的鸡肉调理品中仍存在[22],说明其耐环境胁迫的能力较强。8类优势菌属中,不动杆菌属、假单胞菌属和变形杆菌属的禽源分离株已多次被证实具有不同程度的耐药性,甚至是多重耐药性[23-25]。尽管鸡源柠檬酸杆菌属、香味菌属和漫游球菌属的耐药性研究较少,但有限的数据也已表明,禽源柠檬酸杆菌对氟喹诺酮类、氨基糖苷类、氯霉素类、四环素类等均产生了较高的多重耐药性[26-27];在鸡肉中分离到的香味菌属细菌(SKS05-GRD)对阿米卡星、庆大霉素和卡那霉素的耐药性是质粒介导的[28],存在耐药基因的水平迁移风险;一个携带四环素、卡那霉素、头孢氨苄AR基因的质粒,存在于河流漫游球菌()、干燥棒状杆菌()、大肠杆菌()等多种细菌中,可能存在不同种属细菌间的AR基因水平迁移事件[29]。此外,肉杆菌属和魏斯氏菌属的耐药特性还未见报道。另一方面,近十几年的人类临床数据表明,不动杆菌属细菌在中国的检出率、多重耐药率和广泛耐药菌比率均呈上升趋势,其中的鲍曼不动杆菌()对多种抗生素的耐药率均超过60%[30],成为中国目前最主要的“超级细菌”[31];广泛存在于自然界的条件致病菌假单胞菌属细菌的分离阳性率也明显增加,已成为医院感染的重要致病菌之一[32];人类临床分离的香味菌属对大多数可用抗生素具有高度抗性[33]。因此,应特别注意这些在鸡肉和人类中都能分离到的细菌,其污染来源、耐药性的发生和迁移机制、以及是否存在AR基因在人源与动物源同类细菌之间迁移的可能性等问题值得深入研究。
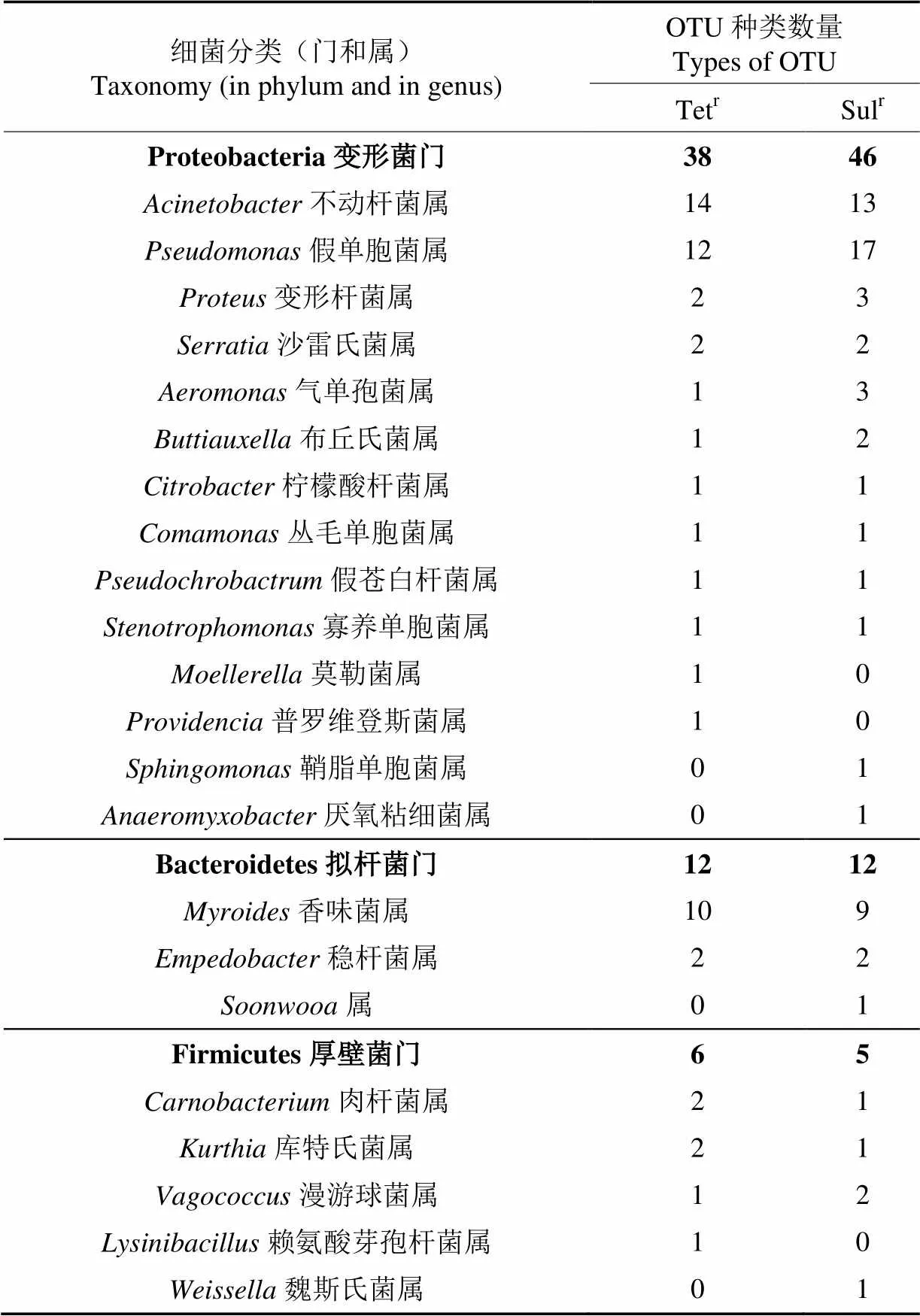
表3 冷鲜鸡肉中四环素耐药菌和磺胺耐药菌的共享菌群
针对鸡肉中的非优势菌属的研究尚未深入展开,其耐药特性仅有几例报道。如人类临床分离到的多株沙雷氏菌属(spp.)细菌含有AR基因,其赋予对-内酰胺、氨基糖苷类、喹诺酮类、大环内酯类和多肽抗微生物剂的抗性[34],而一株火鸡来源的液化沙雷菌()被证实对四环素、氨苄青霉素、链霉素、庆大霉素、红霉素和青霉素产生了六重耐药[35];鸡源豚鼠气单胞菌()可对12种抗生素产生不同程度的耐药性,对头孢噻肟、呋喃妥因、氯霉素和环丙沙星较为敏感[36];院感重要机会致病菌嗜麦芽寡养单胞菌()的耐药性强[37],而在养殖业,在鸡蛋表面[38]和家禽屠宰场空气中[39]均分离到了寡养单胞菌属(spp.)细菌。除此以外,从本研究涉及的冷鲜鸡肉样品中发现的共享Tetr菌和Sulr菌中的布丘氏菌属(spp.)、丛毛单胞菌属(spp.)、假苍白杆菌属(spp.)、莫勒菌属(spp.)、普罗维登斯菌属(spp.)、鞘脂单胞菌属(spp.)、厌氧粘杆菌属(spp.)、稳杆菌属(spp.)、spp.属、库特氏菌属(spp.)和赖氨酸芽孢杆菌属(spp.)等菌属的耐药特性、耐药菌株还未见报道。
2.4 特有菌群
本研究中,将仅在某一个冷鲜鸡样品表面检出的菌群定义为特有菌群,该指标可指示单个样品从养殖、屠宰加工直至贮藏运输过程中的潜在污染情况。来自6个采样点的鸡肉样品中均存在特有菌群,已鉴定到属水平的特有OTU有5~39种,分属3~32个菌属,污染ART菌的复杂度差异较大。其中,尽管来自3号和4号超市的冷鲜鸡样品是预包装产品,但分别具有较多种类的特有Sulr菌和Tetr菌(表4),预测其原料或生产加工过程可能存在ART菌污染源,后续可利用细菌多样性分析技术,通过比对冷鲜鸡肉成品中的特有菌群及其对应的生产链各环节采集的样品的菌群,进行鸡肉表面污染细菌的溯源和验证。
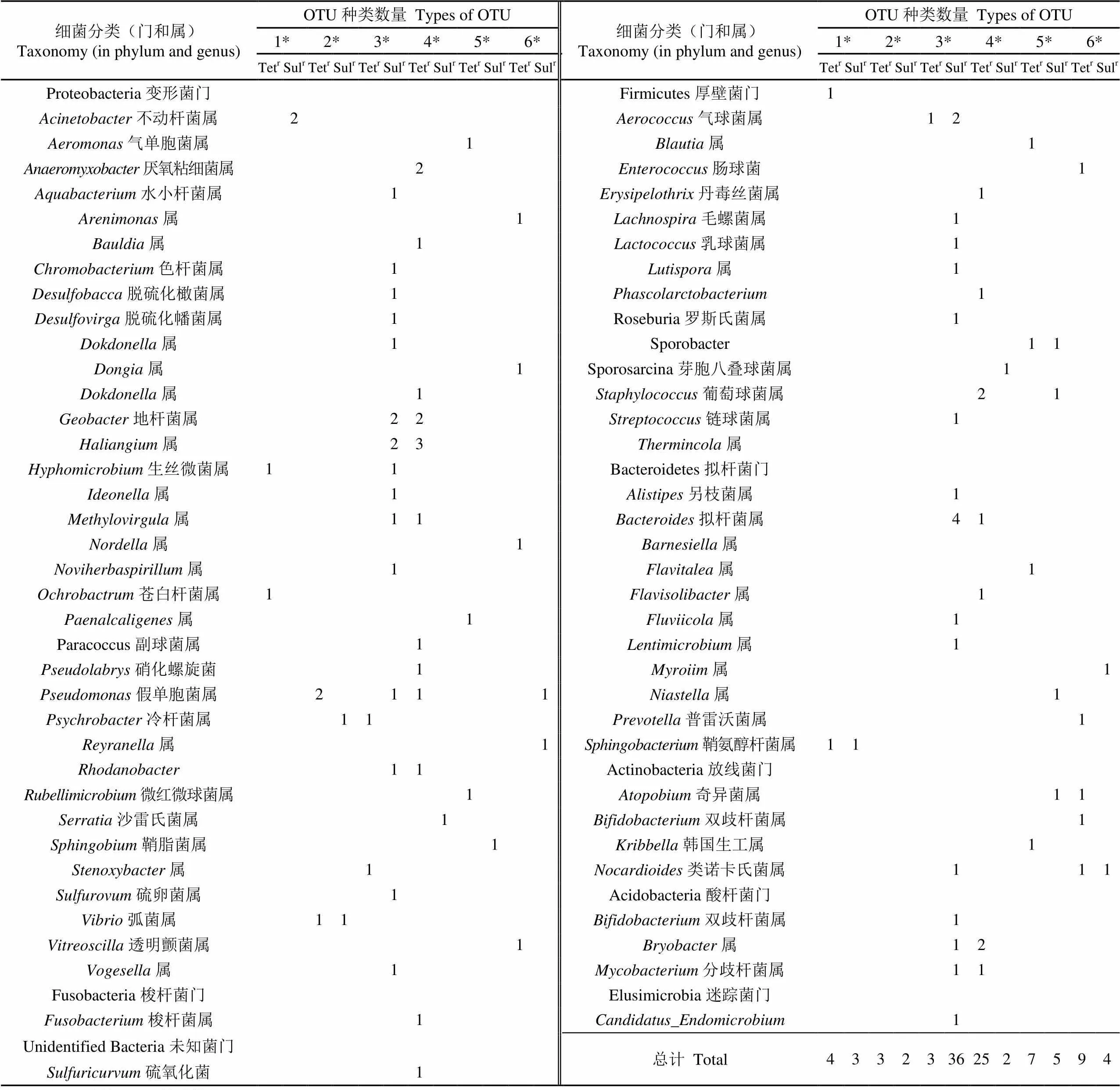
表4 冷鲜鸡肉中四环素耐药菌和磺胺耐药菌的特有菌群
注:1*~6*为第1~6号采样点。
Note: 1*-6*stand for sample No. 1 to 6.
3 结 论
1)利用二代测序技术分析冷鲜鸡肉表面可培养的四环素耐药菌(tetracycline resistant bacteria,Tetr)和磺胺耐药菌(sulfamethoxazole resistant bacteria,Sulr)的菌群多样性,在2类ART菌中各注释出59个和58个已明确属名的属,与前人关于冷鲜鸡肉表面细菌多样性分析的研究数据相比,占比较高,预示冷鲜鸡表面多类细菌已具备抗生素耐药性。
2)共享菌群的分析结果进一步说明,冷鲜鸡肉表面各类优势细菌均已不同程度地存在具有耐药性的种。前人研究已多次证实其中不动杆菌属、假单胞菌属、变形杆菌属、柠檬酸杆菌属、香味菌属和漫游球菌属的禽源和人源分离株具有多重耐药性,因此,应充分重视冷鲜鸡肉表面细菌的抗生素耐药性问题,并深入研究耐药基因在上述禽源和人源细菌间水平迁移的潜在风险。此外,共有菌群中的肉杆菌属、魏斯氏菌属、沙雷氏菌属、气单胞菌属、寡养单胞菌属、布丘氏菌属、丛毛单胞菌属、假苍白杆菌属、莫勒菌属、普罗维登斯菌属、鞘脂单胞菌属、厌氧粘杆菌属、稳杆菌属、属、库特氏菌属和赖氨酸芽孢杆菌属的耐药特性应进一步确认,其污染来源和迁移风险有待揭示。
由于不同的培养方法会影响细菌的分离结果,因此,单一的培养条件会限制微生物多样性分析结果的全面性。比如本研究中,尽管每个样品中Tetr菌和Sulr菌在属水平上的菌群分布有明显差异,但两者的操作分类单元(operational taxonomic unit,OTU)数量、丰富度指数和多样性指数均无显著性差异,说明菌相信息的获得收到了培养的限制。因此,冷鲜鸡肉表面耐药菌的全面揭示有赖于不依赖培养的分析技术的建立。
[1] Depoorter P, Persoons D, Uyttendaele M, et al. Assessment of human exposure to 3rd generation cephalosporin resistant(CREC) through consumption of broiler meat in Belgium[J]. International Journal of Food Microbiology, 2012, 159(1): 30-38.
[2] Wang Hua, Manuzon M, Lehman M, et al. Food commensal microbes as a potentially important avenue in transmitting antibiotic resistance genes[J]. Microbiology Letters, 2006, 254(2): 226-231.
[3] Verraes C, Van Boxstael S, Van Meervenne E, et al. Antimicrobial resistance in the food chain: A review[J]. International Journal of Environmental Research and Public Health, 2013, 10(7): 2643-2669.
[4] Centerz T J. Efforts to slacken antibiotic resistance: Labeling meat products from animals raised without antibiotics in the United States[J]. Science of The Total Environment, 2016, 563(9): 1088-1094.
[5] Wang Hua. Commensal Bacteria, Microbial Ecosystems, and Horizontal Gene Transmission: Adjusting Our Focus for Strategic Breakthroughs Against Antibiotic Resistance[M]. In Jaykus L, Wang H, Schlesinger L (ed.), Foodborn Microbes: Shaping the Host Ecosystems, 1st ed. Washington, DC: ASM Press, 2009: 267-281.
[6] Rossi F, Rizzotti L, Felis E, et al. Horizontal gene transfer among microorganisms in food: Current knowledge and future perspective[J]. Food Microbiology, 2014, 42(9): 232-243.
[7] 崔泽林,冯婷婷,周与华,等. 家养及大型养殖场鸡肠道微生物菌群及四环素耐药菌多样性的比较研究[J]. 微生物与感染,2017,12(3):146-155. Cui Zelin, Feng Tingting, Zhou Yuhua, et al. Comparison of microbiota and dissemination of tetracycline resistant bacteria between chickens from a small farmhouse and a big feedlot[J]. Journal of Microbes and Infections, 2017, 12(3): 146-155. (in Chinese with English abstract)
[8] 李姝,邵毅,周昌艳,等. 市售鸡肉及内脏中磺胺耐药菌污染特征[J]. 食品科学,2017,38(21):170-174. Li Shu, Shao Yi, Zhou Changyan, et al. Prevalence and characteristics of sulfamethoxazole resistant bacteria in retail chicken meat and giblets[J]. Food Science, 2017, 38(21): 170-174. (in Chinese with English abstract)
[9] 邵毅,李姝,黄柳娟,等. 市售鸡肉中四环素耐药菌污染特征初探[J]. 中国食品学报,2018,18(1):250-256. Shao Yi, Li Shu, Huang Liujuan, et al. A pilot study on the prevalence and characteristics of tetracycline resistant bacteria in retail chicken samples[J]. Journal of Chinese Institute of Food Science and Technology, 2018, 18(1): 250-256. (in Chinese with English abstract)
[10] 何祥祥,杨华,戴宝玲,等. 冷鲜鸡污染微生物磺胺类和喹诺酮类耐药基因分析[J]. 浙江农业学报,2017,29(4):555-559. He Xiangxiang, Yang Hua, Dai Baoling, et al. Detection of the antibiotic genes of sulfonamides and quinolones of bacteria in refrigerated chicken[J]. Acta Agriculturae Zhejiangensis, 2017, 29(4): 555-559. (in Chinese with English abstract)
[11] Huang Ying, Zhang Lu, Tiu L, et al. Characterization of antibiotic resistance in commensal bacteria from an aquaculture ecosystem[J]. Front Microbiology, 2015, 6: 914. Doi: 10.3389/fmicb.2015.00914.
[12] Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI supplement M100[M]. Pennsylvania: Clinical and Laboratory Standards Institute, 2018.
[13] 林奕岑,徐帅,倪学勤,等. 利用Illumina MiSeq测序平台分析肉鸡盲肠微生物多样性[J]. 中国农业大学学报,2016,21(12):65-73. Lin Yicen, Xu Shuai, Ni Xueqin, et al. Diversity of the cecal microbiome of broiler chicken based on Illumina MiSeq sequencing platform[J]. Journal of China Agricultural University, 2016, 21(12): 65-73. (in Chinese with English abstract)
[14] 肖英平,何祥祥,戴宝玲,等. 采样方法对冷鲜鸡表面细菌DNA提取及高通量测序结果的影响[J]. 食品科学,2017,38(24):260-264. Xiao Yingping, He Xiangxiang, Dai Baoling, et al. Effects of different sampling methods on microbial DNA extraction from chilled chicken and the high-throughput sequencing of amplification products[J]. Food Science, 2017, 38(24): 260-264. (in Chinese with English abstract)
[15] 温冬玲,成淑君,刘悦,等. 高通量测序分析不同增菌温度下冷鲜鸡肉细菌的群落多样性[J]. 食品科学,2018,39(24):156-161. Wen Dongling, Cheng Shujun, Liu Yue, et al. Analysis of bacterial community diversity of chilled chicken at different enrichment temperatures using high-throughput sequencing[J]. Food Science, 2018, 39(24): 156-161. (in Chinese with English abstract)
[16] 吴海虹,刘朏,刘芳,等. 低温贮藏对冷鲜鸡腐败菌菌群变化的影响[J]. 现代食品科技,2016,32(4):177-181. Wu Haihong, Liu Fei, Liu Fang, et al. Effect of storage temperature on changes in spoilage bacterial community in chilled chicken[J]. Modern Food Science and Technology, 2016, 32(4): 177-181. (in Chinese with English abstract)
[17] Morales P, Aguirre J, Troncoso M, et al. Phenotypic and genotypic characterization ofspp. present in spoiled poultry fillets sold in retail settings[J]. LWT-Food Science and Technology, 2016, 73: 609-614.
[18] Samapundo S, de Baenst I, Aerts M, et al. Tracking the sources ofbacteria contaminating chicken cuts during processing[J]. Food Microbiology, 2019, 81(8): 40-50.
[19] 郭玉梅,张慧贤,秦丽云,等. 肉鸡和市售生鸡产品中变形杆菌的研究[J]. 中国卫生检验杂志,2014,24(14):2009-2010. Guo Yumei, Zhang Huixian, Qin Liyun, et al. Study on proteus in chicken and raw chicken products[J]. Chinese Journal of Health Laboratory Technology, 2014, 24(14): 2009-2010. (in Chinese with English abstract)
[20] Olonitola O S, Fahrenfeld N, Pruden A. Antibiotic resistance profiles among mesophilic aerobic bacteria in Nigerian chicken litter and associated antibiotic resistance genes[J]. Poul Sci, 2015, 94(5): 867-74.
[21] 张春江,黄峰,张良,等. 扒鸡加工中主要致腐菌群落结构解析[J]. 中国食品学报,2017,17(2):227-234. Zhang Chunjiang, Huang Feng, Zhang Liang, et al. Analysis of the spoilage microbial communities of braised chicken during processing[J]. Journal of Chinese Institute of Food Science and Technology, 2017, 17(2): 227-234. (in Chinese with English abstract)
[22] 范晓攀,王娉,陈颖,等. 肉类调理食品中细菌多样性的分析[J]. 现代食品科技,2017,33(1):237-242. Fan Xiaopan, Wang Ping, Chen Ying, et al. Bacterial diversity of prepared meat products[J]. Modern Food Science & Technology, 2017, 33(1): 237-242. (in Chinese with English abstract)
[23] Liu Dong, Liu Zengshan, Hu Pan, et al. Characterization of a highly virulent and antimicrobial-resistantstrain isolated from diseased chicks in China[J]. Microbiology and Immunology, 2016, 60(8): 533-539.
[24] Ansari F, Khatoon H. Multiple antibiotic resistance among gram negative bacteria isolated from poultry[J]. Indian Journal of Experimental Biology, 1994, 32(3): 211-212.
[25] 李欣南,韩镌竹,宁宜宝. 鸡源奇异变形杆菌的分离鉴定及耐药性研究[J]. 黑龙江畜牧兽医,2015(6): 165-167. Li Xinnan, Han Juanzhu, Ning Yibao, et al. Isolation and identification of Proteus mirabilis from chicken and study of its drug resistance[J]. Heilongjiang Animal Science and Veterinary Medicine, 2015(6): 165-167. (in Chinese with English abstract)
[26] 刘星,孟韩冰,孔祥彬. 鸡肠毒综合征致病菌的自动生化鉴定及药敏分析[J]. 河南畜牧兽医:综合版,2008,29(7):7-9. Liu Xing, Meng Hanbing, Kong Xiangbin. Automatic biochemical identification and drug sensitivity analysis of chicken enterotoxic syndrome pathogens[J]. Henan Journal of Animal Husbandry and Veterinary Medicine, 2008, 29(7): 7-9. (in Chinese with English abstract)
[27] 付秀玲,苑丽,陈红英,等. 临床分离鸡致病菌超广谱-内酰胺酶的检测及药敏分析[J]. 河南畜牧兽医,2006,27(12):6-9. Fu Xiuling, Yuan Li, Chen Hongying, et al. Detection and drug sensitivity analysis of extended-spectrum β-lactamase pathogens from clinical isolates of chicken[J]. Henan Journal of Animal Husbandry and Veterinary Medicine, 2006, 27(12): 6-9. (in Chinese with English abstract)
[28] Suganthi R, Shanmuga Priya T, Saranya A, et al. Relationship between plasmid occurrence and antibiotic resistance inSKS05-GRD isolated from raw chicken meat[J]. World Journal of Microbiology and Biotechnology, 2013, 29(6): 983-990.
[29] 牛天琦. 鸡粪中多重耐药细菌的分离鉴定及介导抗性基因水平转移元件的检测[D]. 新乡:河南师范大学,2015. Niu Tianqi. Isolation Identification of Multidrug-Resistant Bacteria in Chicken Manure and the Testing of Resistance Gene Horizontal Transfer Elements[D]. Xinxiang: Henan Normal University, 2015. (in Chinese with English abstract)
[30] 张辉,张小江,徐英春,等. 2005-2014年CHINET不动杆菌属细菌耐药性监测[J]. 中国感染与化疗杂志,2016,16(4):429-436. Zhang Hui, Zhang Xiaojiang, Xu Yingchun, et al. Resistance profile ofisolates in hospitals across China: Results from CHINET Antimicrobial Resistance Surveillance Program 2005-2014[J]. Chinese Journal of Infection and Chemotherapy, 2016, 16(4): 429-436. (in Chinese with English abstract)
[31] 陈佰义,何礼贤,胡必杰,等. 中国鲍曼不动杆菌感染诊治与防控专家共识[J]. 中国医药科学,2012,2(8):3-8. Chen Baiyi, He Lixian, Hu Bijie, et al. Expert opinion on the diagnosis, treatment and prevention of Acinetobacter baumannii infection in China[J]. National Medical Journal of China, 2012, 2(8): 3-8. (in Chinese with English abstract)
[32] Guan Xiangdong, He Lixian, Hu Bijie, et al. Laboratory diagnosis, clinical management and infection control of the infections caused by extensively drug-resistant Gram-negative: A Chinese consensus statement[J]. Clin Microbiol Infect, 2016, 22 (S1): S15-S25.
[33] Hu Shaohua, Yuan Shuxing, Qu Hai, et al. Antibiotic resistance mechanisms ofsp.[J]. Journal of Zhejiang University SCIENCE B, 2016 17(3): 188-199.
[34] Sandner-Miranda L, Vinuesa P, Cravioto A, et al. The Genomic basis of intrinsic and acquired antibiotic resistance in the genus[J]. Front Microbiology, 2018, 9: 828. doi: 10.3389/fmicb.2018.00828.
[35] Kilonzo-Nthenge A, Rotich E, Nahashon S N. Evaluation of drug-resistantin retail poultry and beef[J]. Poultry Science, 2013, 92(4): 1098-107.
[36] Abu-Elala N, Abdelsalam M, Marouf S, et al. Comparative analysis of virulence genes, antibiotic resistance, andbased phylogeny of motilespecies isolates from Nile tilapia and domestic fowl[J]. Letters in Applied Microbiology, 2015, 61(5): 429-436.
[37] Sánchez María B. Antibiotic resistance in the opportunistic pathogen[J]. Front Microbiology, 2015, 6: 658. doi: 10.3389/fmicb.2015.00658.
[38] 陈力力,杨伊磊,青文哲,等. 鸡蛋壳表面细菌数量及种群多样性分析[J]. 现代食品科技,2015,31(10):74-79. Chen Lili, Yang Yilei, Qing Wenzhe, et al. Aerobic plate count and bacterial diversity on the surface of eggshells[J]. Modern Food Science and Technology, 2015, 31(10): 74-79. (in Chinese with English abstract)
[39] 戴宝玲,肖英平,孙凤来,等. 家禽定点屠宰场不同屠宰区域空气的微生物结构[J]. 食品科学,2018,39(21):219-223. Dai Baoling, Xiao Yingping, Sun Fenglai, et al. Structure of airborne microbial communities in different slaughter areas of poultry slaughterhouse[J]. Food Science, 2018, 39(21): 219-223. (in Chinese with English abstract)
Diversity of tetracycline- and sulfamethoxazole-resistant bacteria on surface of cold fresh chicken
Shao Yi1,2, Yao Chunxia1,2,※, Huang Liujuan1, Feng Bo1, Zhou Changyan1,2, Bai Bing1, Wang Hua3,4, Wang Weiguo5
(1.,,201403,; 2.,201403,; 3.,,43210,; 4.,,,200433,; 5.,201516,)
The increased use of antimicrobial-resistant (AR) bacteria in agricultural production has posed a great threat to human health, as AR pathogens could spread from agricultural product to human body via food chain. The antimicrobial resistance (ART) genes in animal- and environment-derived bacteria can migrates to human commensal bacteria through horizontal gene transfer (HGT). Increased evidences have shown the critical role of commensal bacteria in HGT of ART genes, affected by both size and species of ART bacteria. Because of the horizontal gene transfer, it is essential to understand antimicrobial resistance in bacterial community rather than in several species. Currently, there is a knowledge gap in our understanding of tendentious carriers of ART gene as most existing studies focused only on a limited pathogenic bacteria. Cold fresh chicken is consumed worldwide, and our previous work had found existence of tetracycline-resistant (Tetr) bacteria and sulfamethoxazole-resistant (Sulr) bacteria in chicken products which might harbor a pool of AR gene, offering a potential avenue for transmission of antimicrobial resistant bacteria and AR genes to human beings. Motivated by these findings, we randomly selected 18 cold fresh chicken samples from six supermarkets in Shanghai from September to October in 2018 to investigate possible existence of Tetrand Sulrbacteria, as well as the potential risk of antimicrobial resistance migration. The culturable Tetrand Sulrbacteria on the surface of the chicken samples were recovered by brain heart infusion (BHI) agar, containing 48g/mL tetracycline or 228g/mL sulfamethoxazole plus 12g/mL trimethoprim. Their diversity was analyzed by the IonS5TMXL sequencing platform. The results showed that the top three phyla with highest relative abundance of ART bacteria were Proteobacteria, Bacteroidetes and Firmicutes. In Tetrand Sulrbacteria, 59 and 58 specific genera were identified respectively. Compared with the literature report on diversity of total bacteria on surface of cold fresh chicken, the genera of the two ART bacteria found in our study accounted for a high proportion of the total microbial population, indicating that many bacteria on the surface of cold fresh chicken were resistant to the two antimicrobials. The results of shared floras found 18 genera (56 OUTs) and 19 genera (63 OUTs) in Tetrand Sulrbacteria respectively, among which the multi-drug resistance of avian and humanspp,spp.,spp.,spp. andspp. had been well established. Thus, the potential risk of horizontal transfer of AR genes from avian to human need to further investigation. However, the characteristics of antimicrobial resistance in other 16 genera of shared floras isolated from avian, includingspp.,spp.,spp.,spp.,spp.,spp.,spp.,spp.,spp.,spp.,spp.,spp.,spp.,spp.,spp. andspp., have not been reported yet. We found that the specific floras of ART bacteria differed from each sample, and 5 to 39 unique OTUs, from 3 to 32 genus, were identified in samples taken from all supermarkets. The specific ART floras data could be used to trace the origin of the ART bacteria during the processing chain. Overall, our results identified existence of ART bacteria on surface of cold fresh chicken and provided reference for further investigation into their potential risk and control. Since separation of bacteria could be affected by cultural method, our diversity analysis was limited to a single culture condition and relied on establishment of analytical techniques of culture-independent in the future.
microorganism; bacteria; antibiotics; cold fresh chicken; tetracycline-resistant bacteria; sulfamethoxazole-resistant bacteria; diversity
2019-05-09
2019-07-15
上海市农委基础研究项目(沪农科攻字(2014)第7-3-6号);上海市市级农口系统青年人才成长计划(沪农青字(2018)第1-38号);国家自然科学基金项目(31401599);上海市农业科学院学科领域建设专项(农科农助2019(10))
邵毅,副研究员,博士,主要从事农产品微生物安全研究。E-mail:shao_saas@163.com
姚春霞,副研究员,博士,主要从事农产品质量安全研究。E-mail:chunxia.yao@163.com
10.11975/j.issn.1002-6819.2019.17.036
S182;TS207.4
A
1002-6819(2019)-17-0301-08
邵 毅,姚春霞,黄柳娟,冯 博,周昌艳,白 冰,王 华,王卫国. 冷鲜鸡肉表面四环素和磺胺耐药菌的菌群多样性分析[J]. 农业工程学报,2019,35(17):301-308. doi:10.11975/j.issn.1002-6819.2019.17.036 http://www.tcsae.org
Shao Yi, Yao Chunxia, Huang Liujuan, Feng Bo, Zhou Changyan, Bai Bing, Wang Hua, Wang Weiguo. Diversity of tetracycline- and sulfamethoxazole-resistant bacteria on surface of cold fresh chicken[J]. Transactions of the Chinese Society of Agricultural Engineering (Transactions of the CSAE), 2019, 35(17): 301-308. (in Chinese with English abstract) doi:10.11975/j.issn.1002-6819.2019.17.036 http://www.tcsae.org
