国产驴蹄草的细胞地理学研究(英文)
2019-09-10王广艳周宁钱敏张婵杨永平
王广艳 周宁 钱敏 张婵 杨永平

摘 要: 為探讨国产毛茛科(Ranunculaceae)驴蹄草属(Caltha)两种植物的演化,该文利用传统染色体压片技术和流式细胞术,并结合前人染色体研究结果,对我国驴蹄草23个居群和花葶驴蹄草10个居群进行了细胞学研究。结果表明:驴蹄草是由四倍体(2n=4x=32)、六倍体(2n=6x=48)和八倍体(2n=8x=64)构成的多倍体复合群,花葶驴蹄草具有四倍体(2n=4x=32)和八倍体(2n=8x=64)两种倍性水平。驴蹄草和花葶驴蹄草均是四倍体较为常见,目前尚未见有二倍体报道。由于驴蹄草和花葶驴蹄草大部分居群采自中国青藏高原地区,可能在冰期时存在古二倍体,其适应性较弱,逐渐被其他的倍性取代,这是由于不同细胞型对环境适应性的结果。驴蹄草可能存在两条进化路线:一条是从甘肃到达云南;另一条是从西藏到达云南。前期分子系统学研究显示花葶驴蹄草与驴蹄草的亲缘关系较近,该研究结果中花葶驴蹄草染色体比驴蹄草要小,花葶驴蹄草可能比驴蹄草相对进化。目前花葶驴蹄草只有10个居群,还需进一步增加居群量来解析其演化路线。
关键词: 细胞地理, 驴蹄草, 花葶驴蹄草, 多倍化
Polyploidy, the duplication of entire sets of chromosomes, is a key process in the evolution and diversification of vascular plants (Hegarty et al., 2013; Otto & Whitton, 2000). Previous studies have found that polyploids are better to adapt to stress or novel niches than their diploid progenitors (Ehrendorfer, 1980; Grant, 1981; Levin, 2004; Morton, 1993; Otto & Whitton, 2000; Stebbins, 1985). Furthermore, intraspecific variation in ploidy level is frequently observed in angiosperms (Kolárˇ et al., 2015; Wood et al., 2009). It is known that polyploidization is one of the few speciation processes that may operate in sympatry, due to the possible immediate emergence of reproductive isolation between individuals with different ploidy levels (Husband & Sabara, 2003). Therefore, the geographic distribution of cytotypes could provide valuable information about the origin and maintenance of different ploidy levels (Baack, 2004; Kolárˇ et al., 2009; Rieseberg & Willis, 2007; Segraves et al., 1999).
The perennial herb Caltha palustris grows from 600-4 000 m in mountain regions, valleys, marshlands, forests, streams, and on grassy slopes in the north temperate region (Wang et al., 2001). After C. palustris was first described by Linnaeus (1753), great variability of some morphological characters was described in this species, such as plant size, leaf shape and size, leaf margins, flowers, mature follicles, rooting at nodes, tepal number and color, and seed color and symmetry (Smit, 1973; Kumar & Singhal, 2008). It is previously shown that morphological diversity is a product of environmental conditions (Blagojevic et al., 2013). The current study primarily focused on cytotype distribution in the C. palustris complex, which includes tetraploids (Wang et al., 2013; Yang, 2002; Yuan & Yang, 2006), hexaploids (Parfenov & Dmitrieva, 1985; Wang et al., 2013; Yang, 2002; Yuan & Yang, 2006), and octoploids (Wang et al., 2013; Yang, 2002; Yuan & Yang, 2006) (x=8, Langlet, 1927). Furthermore, molecular phylogenetic evidence also shows that C. scaposa is sister to C. palustris (with 100% bootstrap support) (Cheng & Xie, 2014; Schuettpelz & Hoot, 2004). Caltha scaposa is endemic to SinoHimalaya, and grows from 2 800 to 4 100 m in wet parts of alpine meadows and valleys. Only two cytotypes have been detected: tetraploids (Wang et al., 2013) and octoploids (Wang et al., 2013; Yuan & Yang, 2006) (x=8, Langlet, 1927). The existence of different cytotypes in C. palustris and C. scaposa may indicate strong spatial segregation. As a result of inche differentiation (Ehrendorfer, 1980; Lewis, 1980), reproductive exclusion (Levin, 1975; VanDijk & BakxSchotman, 1997), and historical factors (AnCˇev, 2006), these distinct cytotypes may experience differential reproductive success and occurrence of particular evolutionary constraints or demographic stochasticity (MunozPajares et al., 2017).
By conducting a novel analysis of previous cytotype distribution data, we herein present a cytogeographical study of C. palustris and C. scaposa in China. Our aims in this study were as follows: (1) To assess the geographic distribution of different cytotypes in C. palustris and C. scaposa to propose a scenario of dispersal events; (2) To determine the major driving force of speciation in C. palustris and C. scaposa.
1 Materials and Methods
1.1 Taxon sampling
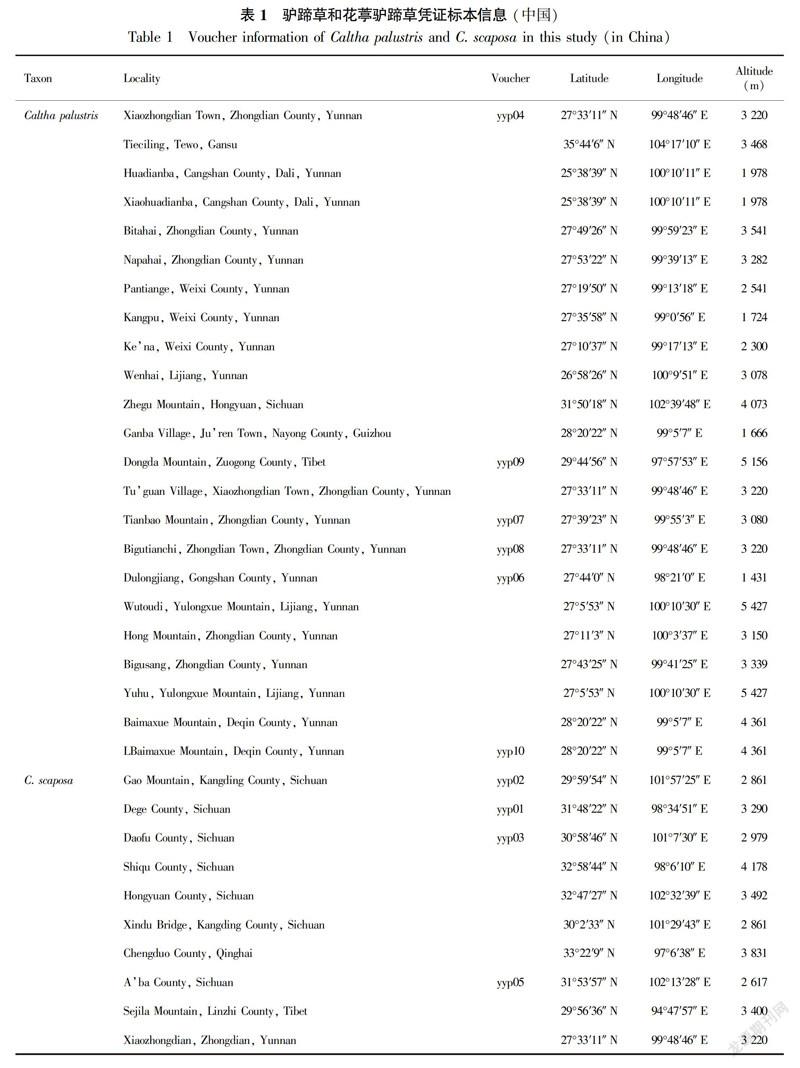
In this study, we sampled six C. palustris accessions and four C. scaposa accessions (Table 1). In total, 15-20 plants from each population were sampled. Geographical coordinates were recorded in the field using a GPS instrument. Living plants were cultivated in a greenhouse, and voucher specimens were deposited in the herbarium of Kunming Institute of Botany, Chinese Academy of Sciences. We performed cytogeographical analysis using these accessions and previously reported data (Yang, 2002; Yuan & Yang, 2006; Table 2).
1.2 Chromosome number
Root tips were collected from each individual and pretreated with a solution of 0.002 mol·L1 8hydroxyquinoline at 20-21 ℃ for 4-5 h. After fixation for 50 min in Carnoy’s solution (ethanol∶acetic acid=3∶1) at 4 ℃, the root tips were dissociated in a mixture of 1 N HCl and 45% acetic acid (1∶1) at 60 ℃ for 30 s, stained with 1% acetic orcein for 2-3 h and squashed on a glass slide (Wang et al., 2013). Chromosome numbers were determined for each accession from at least 50 cells of at least two seedlings by mitotic observations. Mitotic interphase nuclei and prophase chromosomes preparations followed Tanaka (1971, 1977, 1987), and the designation of the centromeric position followed Levan et al. (1964). Karyotype asymmetry was classified according to Stebbins (1971).
1.3 Flow cytometry and DNA ploidy level determination
Propidium iodide flow cytometry (FCM) analysis was performed using fresh leaf samples from our greenhouse. Approximately 0.5 cm2 of leaf material was finely diced using a new razor blade in a Petri dish that contained 1 500-2 000 μL of WPB nuclear solution buffer (0.2 mol·L1 TrisHCl, 4 mmol·L1 MgCl·6H2O, 2 mmol·L1 EDTA Na2·2H2O, 86 mmol·L1 NaCl, 10 mmol·L1 Na2S2O5, 1% PVP10, 1% [v/v] Triton X100, pH 7.5) (Tian et al., 2011). The nuclear suspension was then filtered through disposable filters (30 μm) to remove cell debris, and stained with 150 μL propidium iodide (50 μg·mL1; including RNAse [500 μg·mL1]) for 10 min. Samples were analyzed on a CyFlow Space (Partec, Münster, Germany) flow cytometer equipped with a blue laser operating at 488 nm. At least 5 000 nuclei were measured for each sample. FlowMax ver. 2.82 was used to analyze the resulting histograms. By comparison with a known ploidy level (4x; yyp04), we estimated the ploidy levels of other samples based on the histograms. The ploidy level of each sample was calculated as described by Tian et al., 2011:
Ploidy level of sample=(mean of sample peak/mean of standard peak) × ploidy level of the standard species.
2 Results and Analysis
2.1 Chromosome counts and DNA ploidy level determination
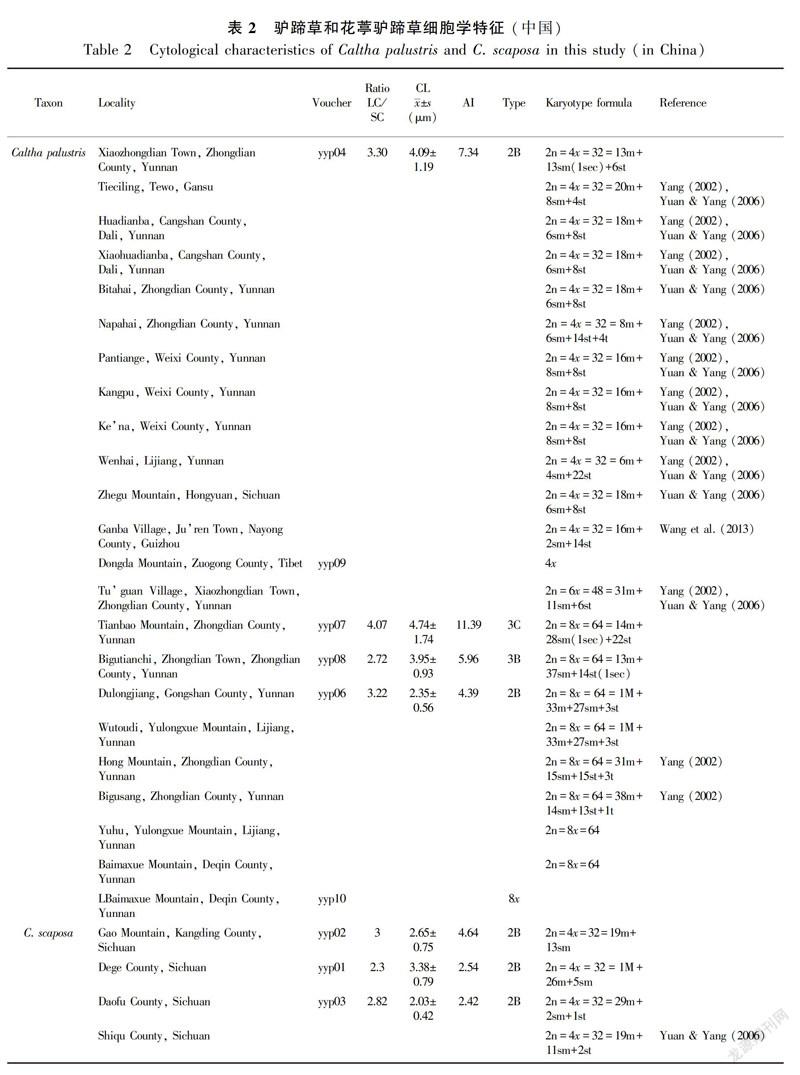
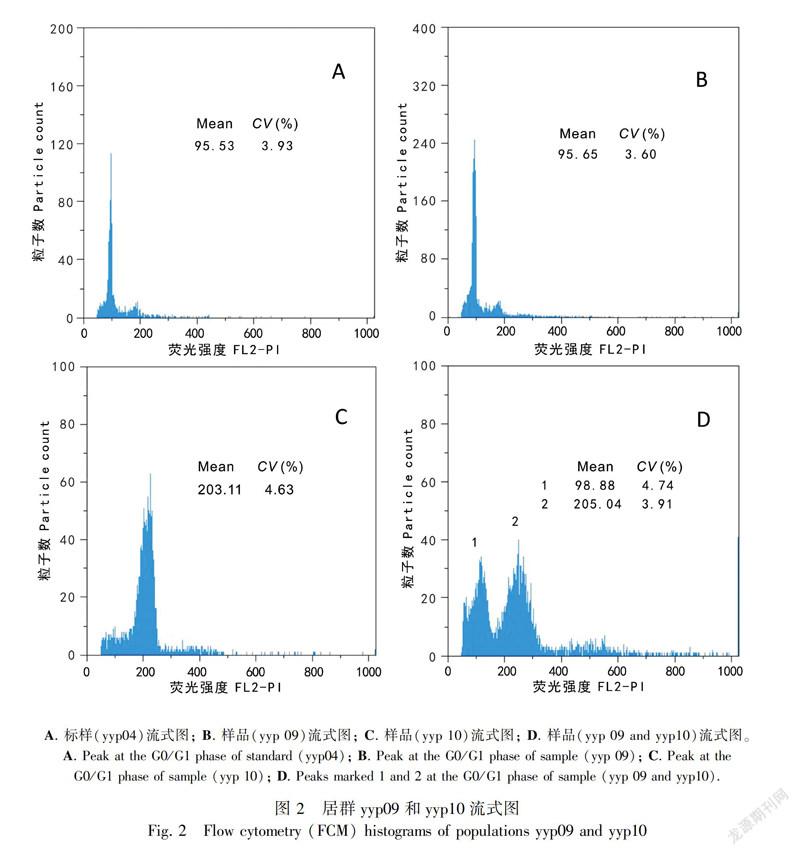
In this study, ploidy levels included 13 4x C. palustris, one 6x C. palustris, nine 8x C. palustris, seven 4xC. scaposa, and three 8x C. scaposa accessions (Table 2); These specimens were collected from Gansu (one accession), Yunnan (16 accessions), Sichuan (four accessions), Tibet (one accession), Guizhou (one accession), and Qinghai (one accession). Metaphase chromosomes of eight accessions are shown in Fig. 1. We successfully estimated the ploidy levels of two C. palustris accessions (yyp09 and yyp10) by FCM at 4x and 8x (Fig. 2).
2.2 Cytogeography
The ploidy distribution of C. palustris and C. scaposa was revealed based on currently available data. All C. palustris accessions were singleploidy, although our sample was very limited in some accessions; however, secondary constriction chromosomes were observed in three C. palustris accessions (Table 2). The tetraploid cytotype was more common than the other cytotypes (hexaploids and octoploids). Moreover, the tetraploid karyotype also exhibited obvious variation among accessions. The samples from Diqing Tibetan Autonomous Prefecture (Yunnan) included tetraploid, hexaploid, and octoploid cytotypes. Two cytotypes (tetraploids and octoploids) were found in Lijiang (Yunnan). Only one cytotype existed in Tewo (Gansu), Dali (Yunnan), Gongshan (Yunnan), Hongyuan (Sichuan), Nayong (Guizhou), and Zuogong (Tibet). All C. scaposa accessions were singleploidy, and the tetraploid cytotype was also common. Two cytotypes (tetraploids and octoploids) existed in Sichuan, and one cytotype each in Tibet, Qinghai, and Yunnan. In addition, only one contact areas between different cytotypes were detected. In the Xiaozhongdian accession, a region of overlap between the ranges of 4x C. palustris and 8x C. scaposa was observed.
3 Discussion
FCM offers a rapid and precise method for identifying taxa of different ploidy levels, enabling researchers to map the finescale distribution of ploidies within individual populations (Suda et al., 2004). FCM has been used in ploidy analysis, e.g., in Ranunculus (Ranunculaceae) (Cires et al., 2010) and C. leptosepala s.l. (Wefferling et al., 2017). In our study, ploidy levels of two accessions (yyp09 and yyp10) were estimated by FCM. The current study revealed that C. palustris may be viewed as a polyploid complex, which presents clear patterns of cytotype distribution. Polyploidy is a prevalent phenomenon in the chromosomal evolution of extant species and genera (Otto & Whitton, 2000), and it may have contributed to the origin of flowering plants (De Bodt et al., 2005). As a result, plant scientists have recognized that polyploid lineages may have complex relationships with each other and their diploid ancestors, making application of species concepts problematic (Soltis et al., 2007, 2009).
C. palustris polyploid complex showed a varied cytotype distribution. No diploids and few hexaploids were found in this study, but tetraploid and octoploid cytotypes were common and widespread. Similarly, in C. scaposa, tetraploids and octoploids were common, whereas diploids and hexaploids were not found. Such distribution patterns are often explained by cytotype adaptive differences to the underlying heterogeneity of environmental factors (Lewis, 1980). All accessions except for the Guizhou and Gansu accessions were from extreme habitats, like alpine mountains in the QinghaiTibetan Plateau. Polyploidy is common in plants from cold climates with harsh and stressful environments (Grant, 1981; Lve & Lve, 1949, 1967). Therefore, a relatively high frequency of polyploidy was observed in this species. Ancestral diploids may be present in this region during glacial periods and colonized most regions at the end of the glaciation cycles. However, other ploidy levels could gradually replace diploids, because of their increased fitness in changing environment (Cui et al., 2008).
The chromosome counts observed in the C. palustris complex indicate that ploidy changes may be important in its evolution. Chromosome counts often show obvious differences in different accessions within a particular species. Our analysis showed that the Hengduan Mountains could be better viewed as a polyploid complex of diploids, tetraploids, and hexaploids. Symmetrical karyotypes are widely accepted to be more primitive than asymmetrical ones (Stebbins, 1971). In our combined data (Table 1), the accessions from Zhongdian (Yunnan) with different types (3B, 3C), AI (11.39, 7.34, 5.96), and the secondary constrictions showed the highest asymmetric tendencies. Therefore, we speculate that two possible evolutionary trends may exist: one from Gansu to Yunnan, and the other from Tibet to Yunnan of China.
Molecular phylogeny have shown that C. scaposa is closely related to C. palustris (Cheng & Xie, 2014; Schuettpelz & Hoot, 2004). C. scaposa cytotype distribution was determined from only ten populations; therefore, C. scaposa cytogeography could not be comprehensively analyzed. Moreover, the chromosome size of this species was smaller than that of C. palustris. The size of the chromosome is also a feature subject to evolutionary change, the direction of chromosome evolution could have a decrease trend in chromosome size (Martel et al., 2004). Therefore, smaller chromosomes may be a relatively derived evolutionary character. Consequently, in the future, additional samples need to be analyzed to better elucidate C. scaposa cytogeography.
References:
ANCˇEV M, 2006. Polyploidy and hybridization in Bulgarian (Brassicaceae) distribution and evolutionary role [J]. Phytol Balcan, 12: 357-366.
BAACK EJ, 2004. Cytotype segregation on regional and microgeographic scales in snow buttercups (Ranunculus adoneus, Ranunculaceae) [J]. Amer J Bot, 91: 1783-1788.
BLAGOJEVIC J, JOVANOVIC V, ADNADEVIC T, et al., 2013. Chromosome status of marsh marigold Caltha palustris L. (Ranunculaceae) from Serbia [J]. Genetika, 45: 793-798.
CHENG J, XIE L, 2014. Molecular phylogeny and historical biogeography of Caltha (Ranunculaceae) based on analyses of multiple nuclear and plastid sequences [J]. J Syst Evol, 52: 51-67.
CIRES E, CUESTA C, NGELES M, et al., 2010. Intraspecific genome size variation and morphological differentiation of Ranunculus parnassifolius (Ranunculaceae), an AlpinePyreneanCantabrian polyploid group [J]. Biol J Linn Soc, 101: 251-271.
CUI XK, AO CQ, ZHANG Q, et al., 2008. Diploid and tetraploid distribution of Allium przewalskianum Regel. (Liliaceae) in the QinghaiTibetan Plateau and adjacent regions [J]. Caryologia, 61(2): 192-200.
DE BODT S, MAERE S, VAN DE PEER Y, 2005. Genome duplication and the origin of angiosperms [J]. Trends Ecol Evol, 20: 591-597.
EHRENDORFER F, 1980. Polyploidy and distribution [M]// LEWIS WH. Polyploidy: Biological Relevance: 45-60.
GRANT V, 1981. Plant speciation [M]. 2nd. New York: Columbia University Press.
HEGARTY MJ, COATE J, SHERMANBROYLES S, et al., 2013. Lessons from natural and artificial polyploids in higher plants [J]. Cyt Gen Res, 140: 204-225.
HUSBAND BC, SABARA HA, 2003. Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium (Onagraceae) [J]. New Phytol, 161: 703-713.
KOLR F, PISOVA S, ZAVESKA E, et al., 2015. The origin of unique diversity in deglaciated areas traces of Pleistocene processes in northEuropean endemics from the Galium pusillum polyploid complex (Rubiaceae) [J]. Molec Ecol, 24: 1311-1344.
KOLAR F, TECH M, TRAVNICEK P, et al., 2009. Towards resolving the Knautia arvensis agg. (Dipsacaceae) puzzle: primary and secondary contact zones and ploidy segregation at landscape and microgeographic scales [J]. Ann Bot, 103: 963-974.
KUMAR P, SINGHAL VK, 2008. Cytology of Caltha palustris L. (Ranunculaceae) from cold regions of western Himalayas [J]. Cytologia, 73: 137-143.
LANGLET O, 1927. Beitrge zur zytologie der Ranunculazeen [J]. Svensk Bot Tidskr, 21: 1-17.
LEVAN A, FEDGA K, SANDBERG A, 1964. Nomenclature for centromeric position on chromosomes [J]. Hereditas, 52: 201-220.
LEVIN DA, 2004. The ecological transition in speciation [J]. New phytol, 161: 91-96.
LEVIN DA, 1975. Minority cytotype exclusion in local plant populations [J]. Taxon, 1: 35-43.
LEWIS W, 1980. Polyploidy in species population [M]// LEWIS WH. Polyploidy, biological relevance. New York: Plenum Press: 103-144.
LINNAEUS C, 1753. Caltha [M]// LINNAEUS C. Species plantarum: 558.
LVE A, LVE D, 1949. The geobotanical significance of polyploidy I. Polyploidy and latitude [J]. Port Acta Biol Ser, A: 273-352.
LVE A, LVE D, 1967. Polyploidy and altitude, Mt. Washington [J]. Biol Zent (Supple), 86: 307-312.
MARTEL E, PONCET V, LAMY F, et al., 2004. Chromosome evolution of Pennisetum species (Poaceae): Implications of ITS phylogeny [J]. Plant Syst Evol, 249: 139-149.
MORTON J, 1993. Chromosome numbers and polyploidy in the flora of Cameroons Mountain [J]. Opera Bot, 121: 159-172.
MUNOZPAJARES AJ, PERFECTTI F, LOUREIRO J, et al., 2017. Niche differences may explain the geographic distribution of cytotypes in Erysimum mediohispanicum [J]. Plant Biol, 20(s1): 139-147.
OTTO SP, WHITTON J, 2000. Polyploid incidence and evolution [J]. Ann Rev Genet, 34: 401-437.
PARFENOV VI, DMITRIEVA SA, 1985. Kariologicheskaja differenciacija u vidov flory Belorussii iee rol, v formoobrazovanii [J]. Dokl Akad Nauk Byelorussk SSR (Minsk), 29: 544-557.
RIESEBERG LH, WILLIS JH, 2007. Plant speciation [J]. Science, 317: 910-914.
SCHUETTPELZ E, HOOT SB, 2004. Phylogeny and biogeography of Caltha (Ranunculaceae) based on chloroplast and nuclear DNA sequences [J]. Am J Bot, 91: 247-253.
SEGRAVES KA, THOMPSON JN, SOLTIS PS, et al., 1999. Multiple origins of polyploidy and the geographic structure of Heuchera grossulariifolia [J]. Molec Ecol, 8: 253-262.
SMIT GS, 1973. A revision of Caltha (Ranunculacaea) [J]. Blumea, 21: 119-150.
SOLTIS DE, SOLTIS PS, SCHEMSKE DW, et al., 2007. Autopolyploidy in angiosperms: Have we grossly underestimated the number of species? [J] Taxon, 56: 13-30.
SOLTIS DE, ALBERT VA, LEEBENSMACK J, et al., 2009. Polyploidy and angiosperm diversification [J]. Am J Bot, 96(1): 336-348.
STEBBINS G, 1971. Chromosomal evolution in higher plants [M]. London: Edward Arnold.
STEBBINS GL, 1985. Polyploidy, hybridization, and the invasion of new habitats [J]. Ann Missouri Bot Gard, 72: 824-832.
SUDA J, MALCOVA R, ABAZID D, et al., 2004. Cytotype distribution in Empetrum (Ericaceae) at various spatial scales in the Czech Republic [J]. Folia Geobot, 39: 169-171.
TANAKA R, 1971. Types of resting nuclei in Orchidaceae [J]. Bot Mag Tokyo, 84: 118-122.
TANAKA R, 1977. Recent karyotype studies [M]// OGAWA K et al. Tokyo: Asakura Publisher: 293-326.
TANAKA R, 1987. The karyotype theory and wide crossing as an example in Orchidaceae [M]// HONG DY. Plant chromosome research 1987, proceedings of the SinoJapanese symposium on plant chromosomes. Hiroshima: 1-10.
TIAN XM, ZHOU XY, GONG N, 2011. Applications of flow cytometry in plant researchanalysis of nuclear DNA content and ploidy level in plant cells [J]. Chin Agric Sci Bull, 27: 21-27.
VANDIJK P, BAKXSCHOTMAN T, 1997. Chloroplast DNA phylogeography and cytotype geography in autopolyploid Plantago media [J]. Molec Ecol, 6: 345-352.
WANG GY, MENG Y, YANG YP, 2013. Karyological analyses of 33 species of the tribe Ophiopogoneae (Liliaceae) from Southwest China [J]. J Plant Res, 126: 597-604.
WANG LY, REN C, YANG QE, 2013. Cytology of two species in the genus Caltha (Ranunculaceae) from China [J]. J Trop Subtrop Bot, 21(1): 21-28. [王龍远, 任琛, 杨亲二, 等, 2013. 国产毛茛科驴蹄草属两种植物的细胞学研究 [J]. 热带亚热带植物学报, 21(1): 21-28.]
WANG WT, FU DZ, LI LQ, et al., 2001. Ranunculaceae, Flora Reipublicae Popularis Sinicae [M]. Beijing: Science Press: 133-438. [王文采, 傅德志, 李良千, 等, 2001. 毛茛科, 中国植物志 [M]. 北京: 科学出版社: 133-438. ]
WEFFERLING KM, CASTRO S, LOUREIRO J, et al., 2017. Cytogeography of the subalpine marsh marigold polyploidy complex (Caltha leptosepala s. l., Ranunculaceae) [J]. Am J Bot, 104(2): 271-285.
WOOD TE, TAKEBAYASHI N, BARKER MS, et al., 2009. The frequency of polyploid speciation in vascular plants [J]. Proc Natl Acad Sci USA, 106: 13875-13879.
YANG Q, 2002. Cytology of the tribe Trollieae and of the tribe Cimicifugeae in the Ranunculaceae a comparative study [J]. Acta Phytotax Sin, 40: 52-65.
YUAN Q, YANG QE, 2006. Cytology, palynology, and taxonomy of Asteropyrum and four other genera of Ranunculaceae [J]. Bot J Linn Soc, 152: 15-26.
YUAN Q, YANG QE, 2006. Tribal relationships of Beesia, Eranthis and seven other genera of Ranunculaceae evidence from cytological characters [J]. Bot J Linn Soc, 150: 267-289.
