海藻希瓦氏菌感染对半滑舌鳎肠道菌群结构及相关功能基因表达的影响
2019-09-10张燕玉韩卓然孙敬锋吕爱军胡秀彩刘军锋
张燕玉 韩卓然 孙敬锋 吕爱军 胡秀彩 刘军锋


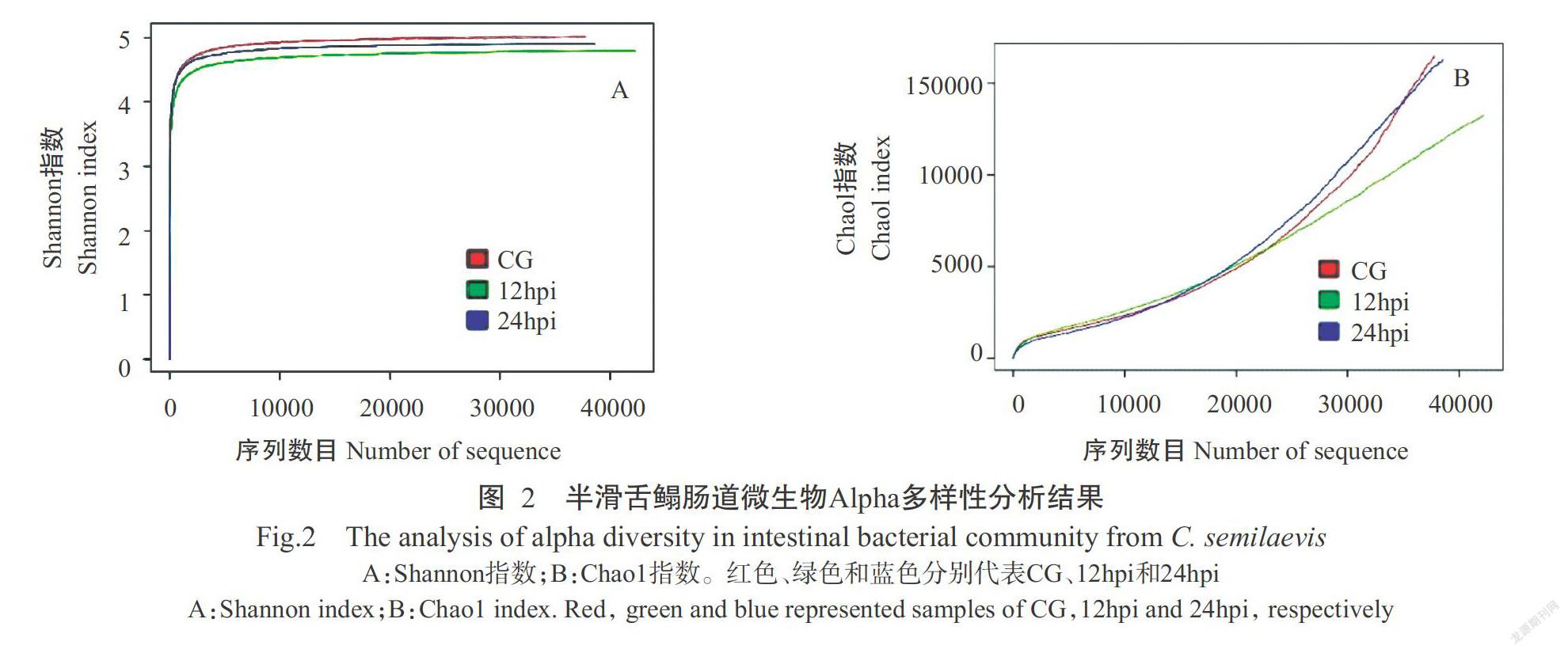



摘要:【目的】明確海藻希瓦氏菌(Shewanella algae)感染对半滑舌鳎(Cynoglossus semilaevis)肠道菌群结构及相关功能基因表达的影响,揭示肠道菌群和肠道组织相关功能基因在疾病发生及免疫应答过程中的作用机制。【方法】以致病性海藻希瓦氏菌人工感染半滑舌鳎后,采用16S rDNA高通量测序技术探究其肠道菌群组成结构的变化情况,并利用实时荧光定量PCR检测分析半滑舌鳎肠道组织中参与疾病发生和免疫应答相关功能基因的表达规律。【结果】共测序获得118657条有效序列,按97%的序列相似度聚类后得到6732个OTUs。Alpha多样性分析结果显示,Shannon指数和Chao1指数以感染前(CG)的健康半滑舌鳎最高,在感染后12 h(12hpi)最低;感染海藻希瓦氏菌前后半滑舌鳎肠道优势菌门无明显变化,但不同类群的相对丰度发生变化。在属水平上,Elizabethkingia、曼噬甲壳菌属(Chitinophaga)、Brevinema、苯基杆菌属(Phenylobacterium)、假单胞菌属(Pseudomonas)、乳杆菌属(Lactobacillus)、Marivita和雷尔氏菌属(Ralstonia)的相对丰度在CG半滑舌鳎肠道菌群组成中占比最高,希瓦氏菌属(Shewanella)、Petrimonas、Proteiniphilum和Aminobacterium的相对丰度在12hpi的占比最高,食酸菌属(Acidovorax)、芽孢杆菌属(Bacillus)和弧菌属(Vibrio)的相对丰度在感染后24 h(24hpi)的占比最高。半滑舌鳎肠道组织相关功能基因的表达变化表现为:果糖二磷酸醛缩酶A基因(ALDOA)的相对表达量在24hpi时显著高于CG(P<0.05,下同);磷脂酶B1基因(PLB1)、热休克蛋白70 kD蛋白1A基因(HSPA1A)、组氨酸三聚体核苷结合蛋白1基因(HINT1)和γ谷氨酰转移酶1基因(GGT1)的相对表达量显著高于CG和24hpi;海藻糖酶基因(TREH)的相对表达量在12hpi时显著低于CG和24hpi。【结论】半滑舌鳎感染海藻希瓦氏菌后其肠道菌群多样性降低、菌群结构发生变化,肠道组织中免疫功能相关基因(HSPA1A和HINT1)及代谢功能相关酶类基因(ALDOA、PLB1、GGT1和TREH)呈差异表达,说明海藻希瓦氏菌感染引起半滑舌鳎肠道微生态紊乱,且肠道组织中免疫功能相关基因和代谢功能相关酶类基因分别参与机体的免疫应答及疾病发生过程。
关键词: 半滑舌鳎;海藻希瓦氏菌;肠道菌群;16S rDNA高通量测序;实时荧光定量PCR
中图分类号: S941.42 文献标志码: A 文章编号:2095-1191(2019)10-2300-08
Effects of infection with Shewanella algae on the microbial communities and expression of related functional genes in the intestine of Cynoglossus semilaevis
ZHANG Yan-yu1, HAN Zhuo-ran1, SUN Jing-feng1*, LYU Ai-jun1,
HU Xiu-cai1, LIU Jun-feng2
(1College of Fisheries, Tianjin Agricultural University/Tianjin Key Lab of Aqua-Ecology and Aquaculture, Tianjin 300384; 2Tianjin Yushengtang Biotechnology Co., Ltd., Tianjin 300404, China)
Abstract:【Objective】The purpose of this study was to investigate the changes of intestinal microbial communities and expression of related functional genes in the intestine of Cynoglossus semilaevis after artificial infection with the pathogenic Shewanella algae, and reveal the role of intestinal microbial flora and related functional genes in the process of disease occurrence and intestinal immune response in the host. 【Method】After C. semilaevis were artificially infected with the pathogenic S. algae,16S rDNA high-throughput sequencing technique was used to study the changes of intestinal microbial communities, and real-time fluorescence quantitative PCR technology was used to study the expression pattern of functional genes involved in the process of disease occurrence and immune response in the intestinal tissues. 【Result】A total of 118657 effective tags were obtained and assigned to 6732 OTUs based on a 97% sequence similarity level. The results of alpha diversity analysis showed that the indexes of Shannon and Chao1 were the highest in the control group (CG), and the lowest in the group of 12 h post-injection(12hpi). At phylum level,although the identified dominant intestinal bacteria were consistent between the control and infection groups, the relative abundance of the bacterial taxa varied. At genus level, the relative abundances of Elizabethkingia, Chitinophaga, Brevinema, Phenylobacterium, Pseudomonas, Lactobacillus, Marivita, andalstonia were the highest in the intestinal microbiota of CG; the relative abundances of Shewanella, Petrimonas, Proteiniphilum, and Aminobacterium in 12hpi group were more than those in other groups. Acidovorax, Bacillus, and Vibrio had the highest relative abundance in the group of 24 h post-injection (24hpi). The expression patterns of related functional genes in the intestine of C. semilaevis were as follows:the relative expression of fructose drphosphate aldolase A gene ALDOA in 24hpi group was significantly higher than that in CG(P<0.05, the same below); the relative expression of phospholipase b1 gene(PLB1), heat shock protein 70 kD protein 1A gene(HSPA1A),histidine trimer nucleoside binding protein 1 gene(HINT1) and gamma glutamyltransferase 1 gene(GGT1)in 12hpi group was significantly higher than those in CG and 24hpi groups; the relative expression of trehalase gene(TREH) in 12hpi group was significantly lower than those in CG and 24hpi groups.【Conclusion】The structure of the microflora changes with the diversity of intestinal microflora of C. semilaevis decreasing after infection with S. algae. The immune function related genes(HSPA1A and HINT1) and metabolism-related enzyme genes (ALDOA, PLB1,GGT1 and TREH) are diffe-rentially expressed. It indicates that infection with S. algae results in the disturbance of intestinal microecology of C. semilaevis, and the immune function related genes and metabolism-related enzyme genes are involved in the process of immune response against bacterial infection and disease occurrence.
Key words: Cynoglossus semilaevis;Shewanella algae; intestinal microflora; 16S rDNA high-through sequencing technique; real-time fluorescence quantitative PCR
0 引言
【研究意义】半滑舌鳎(Cynoglossus semilaevis)因具有产量高、肉质鲜美等优点,已发展成为海水渔业中最具养殖价值的鱼类品种之一。近年来,随着渔业工厂化养殖技术的不断成熟,半滑舌鳎规模化养殖密度越来越高,导致养殖水质恶化、疾病频发,给养殖户带来极大的经济损失。侵染半滑舌鳎的常见病原菌有副溶血弧菌(Vibrio parahemolyticus)(胡璇等,2014)、创伤弧菌(V. vulnificus)(高桂生等,2016)及杀鲑气单胞菌(Aeromonas salmonicida)(周红霞等,2017)等。至今,越来越多研究表明肠道菌群结构与水产养殖动物的健康密切相关(Pérez et al.,2010)。肠道微生物群作为一个复杂的生态系统,通过参与免疫应答、营养吸收及疾病防御,对促进宿主健康至关重要(Lin et al.,2014)。鱼类肠道作为机体重要的组织器官,不仅承担着消化吸收功能,还在免疫防御过程中发挥重要作用(Wang et al.,2018)。因此,研究半滑舌鳎感染病原菌后其肠道菌群结构及相关功能基因表达的变化规律,可为揭示肠道菌群和肠道组织相关功能基因在疾病发生及免疫应答过程中的作用机制提供参考依据。【前人研究进展】肠道微生物对于宿主健康至关重要,针对水产动物的相关研究主要集中在机体健康状态下的肠道菌群组成结构和多样性分析(Giatsis et al.,2015;Narrowe et al.,2015;Han et al.,2018)。最近的研究表明,环境因素和养殖条件对水产动物肠道菌群组成结构有显著影响(Talwar et al.,2018)。不同外界因素如生活环境(Cornejo-Granados et al.,2017)、饮食组成(Duan et al.,2017;He et al.,2017)、养殖水质(Suo et al.,2017)及生长阶段(Cornejo-Granados et al.,2018)等均可导致南美白对虾(Litopenaeus vannamei)肠道菌群组成差异。在鱼类上,年龄(Lauzon et al.,2010)、饲料成分(Desai et al.,2012)、食性(Ye et al.,2014)及性别(Li et al.,2016b)等因素均会影响肠道菌群组成结构及其种类丰度。除这些因素外,病原微生物感染及疾病发生对水生动物的肠道微生态系统也有重要影响。白斑综合征病毒(WSSV)感染可改变中华绒螯蟹(Eriocheir sinensis)和南美白对虾的肠道微生物种类及其相对丰度(Ding et al.,2017;Wang et al.,2019);弧菌可引起斑节对虾(Penaeus monodon)和南美白对虾肠道细菌动态变化(Rungrassamee et al.,2014),并改变南美白对虾肠道微生物群落结构(He et al.,2017);南美白对虾急性肝胰腺坏死病的发生常伴随着宿主肠道微生物群落变化,导致与疾病有关的特异性细菌出现并增殖(Chen et al.,2017);溶藻弧菌(V. alginolyticus)感染三疣梭子蟹(Portunus trituberculatus)可引起肠道微生物不同类群的丰度产生变化(Xia et al.,2018);在鱼类方面,从患疮疖病的圆口铜鱼(Coreius guichenoti)和嗜水气单胞菌(A. hydrophila)感染的斑马鱼(Danio rerio)中均发现肠道微生物群落组成及其多样性发生变化(Li et al.,2016a;Yang et al.,2017b)。鱼类疾病的发生发展和病原生物感染均伴随着肠道菌群结构变化,肠道作为黏膜免疫应答的重要器官,在抗感染免疫防御过程中发挥重要作用(Wang et al.,2018),如在黏孢子蟲(Enteromyxum leei)感染黑鲷(Sparus aurata)(Davey et al.,2011)和溶藻弧菌感染半滑舌鳎(Yang et al.,2017a)的肠道组织中某些免疫相关基因发生差异表达。目前,关于鱼类抗感染免疫应答的研究主要集中于脾脏和头肾等系统性免疫器官,而针对肠道中功能基因参与免疫应答及疾病发生过程的相关研究鲜见报道。【本研究切入点】本课题组前期从以肠道炎症为特征的患病半滑舌鳎中分离获得致病性海藻希瓦氏菌(Shewanella algae),且证实肠道是其产生致病作用主要的靶器官(Han et al.,2017b),但目前对海藻希瓦氏菌感染引起的半滑舌鳎肠道菌群动态变化及疾病发生和肠道免疫应答反应中的相关功能基因尚缺乏深入了解。【拟解决的关键问题】通过16S rDNA高通量测序技术探究人工感染海藻希瓦氏菌后半滑舌鳎肠道菌群结构的变化情况,并利用实时荧光定量PCR检测分析半滑舌鳎肠道组织中参与疾病发生和免疫应答相关功能基因的表达规律,旨在揭示肠道菌群和肠道组织相关功能基因在疾病发生及免疫应答过程中的作用机制。
1 材料与方法
1. 1 试验材料
试验用半滑舌鳎购自天津市海发珍品实业发展有限公司,健康无病,体长29±2 cm/尾,体重130±5 g/尾。养殖水温(23±1)℃,pH 7.5,盐度22‰,溶解氧含量5.5~6.0 mg/L。每日喂食2次,所喂饲料为普通商业饲料;早晚各换水1次,防止粪便及剩余饲料败坏养殖水质。暂养1周,待鱼体状况稳定后进行感染试验;感染前随机选取5尾用于病原菌检测。
1. 2 细菌培养及感染试验
海藻希瓦氏菌CSG-15株分离自患病半滑舌鳎肝脏组织,由天津市水产生态及养殖重点实验室保存提供。将CSG-15株接种至无菌2216E液体培养基中,30 ℃培养18 h后,5000 r/min离心3 min,收集菌体,再用灭菌的0.9%生理盐水悬浮菌体以获得1.0×108 CFU/mL的菌悬液。将半滑舌鳎随机均分为3组,每组9尾鱼,分为3个平行。其中2组按200 μL/尾的剂量腹腔注射菌悬液,另一组注射等量无菌生理盐水。分别于感染前(CG)及感染后12 h(12hpi)和24 h(24hpi)收集半滑舌鳎肠道组织及粪便样本,将每组样本分别混合,-80 ℃保存备用,其中,粪便样本用于肠道微生物基因组DNA提取,肠道组织用于RNA提取。
1. 3 肠道微生物基因组DNA提取
按照QIAamp Fast DNA试剂盒(德国QIAGEN公司)说明提取粪便样品中微生物基因组DNA,以0.8%琼脂糖凝胶电泳检测DNA片段的完整性及大小,采用微量分光光度计检测DNA纯度。
1. 4 PCR扩增及测序
以稀释后的基因组DNA(1 ng/µL)为模版,用包含Barcode特异性序列的引物对16S rDNA基因V4区进行扩增(Guo et al.,2016)。正、反向引物分别为515F:5'-GTGCCAGCMGCCGCGGTAA-3'和806R:5'-GGACTACHVGGGTWTCTAAT-3'(Xiong et al.,2015)。按照Han等(2018)的方法,回收PCR产物并构建文库,利用Qubit和实时荧光定量PCR对构建好的文库进行定量,使用HiSeq 2500 PE250进行高通量测序分析。
1. 5 数据处理
根据Barcode序列和PCR扩增引物序列,从下机数据中拆分出各样品数据,并将每个样品的序列拼接为原始Tags数据(Raw Tags),质控后得到有效序列(Effective Tags)。利用UPARSE按97%的序列相似度将有效序列聚类成OTUs(Operational taxonomic units),同时对OTUs代表序列进行物种注释(Edgar,2013);以Qiime v1.7.0计算Chao1、Shannon和Simpson等指数,并计算Unifrac距离及构建UPGMA聚类分析树;采用R软件分析Alpha和Beta多样性指数及组间差异。
1. 6 肠道组织总RNA提取及cDNA文库构建
根据RNAiso Plus总RNA提取试剂盒(TaKaRa公司)说明提取半滑舌鳎肠道组织总RNA,使用1.0%琼脂糖凝胶电泳和微量分光光度计检测RNA的完整性、浓度和质量。采用PrimeScriptTM RT reagent Kit with gDNA Eraser试剂盒(TaKaRa公司)对RNA样本进行反转录,获得的cDNA冷冻保存,用于实时荧光定量PCR。
1. 7 实时荧光定量PCR
利用实时荧光定量PCR对热休克蛋白70 kD蛋白1A(HSPA1A)、组氨酸三聚体核苷结合蛋白1(HINT1)、果糖二磷酸醛缩酶A(ALDOA)、磷脂酶B1(PLB1)、海藻糖酶(TREH)和γ谷氨酰转移酶1(GGT1)的基因表达情况进行检测,以β-actin为内参基因。通过NCBI Primer BLAST设计引物(表1),委托生工生物工程(上海)股份有限公司合成。实时荧光定量PCR反应体系20.0 μL:SYBR® Premix Ex TaqTM Ⅱ 10.0 μL,cDNA模板2.0 μL,上、下游引物(10 μmol/L)各0.4 μL,灭菌蒸馏水7.2 μL。扩增程序:95 ℃预变性3 min;95 ℃ 7 s,57 ℃ 10 s,72 ℃ 15 s,进行45个循环。每次循环结束采集荧光信号,然后进行熔解曲线的采集与数据分析,采用2-△△Ct法计算各目的基因的相对表达量。每个基因扩增设3次重复,利用Duncanʼs新复极差法进行多重比较。
2 结果与分析
2. 1 半滑舌鳎肠道微生物多样性分析结果
质控后共测序获得有效序列118657条,平均每个样本为39552条。其中,12hpi时获得有效序列数目最多(45346条),CG获得有效序列数目最少(39724条)。各组有效序列按97%的序列相似度进行聚类,结果3组样本共获得6732个OTUs。CG、12hpi和24hpi对应的OTUs数目分别为2506、2819和2265个,其中3组样本的共有OTUs为225个(图1)。Alpha多样性分析结果表明,Shannon指数和Chao1指数在CG半滑舌鳎中最高,在12hpi半滑舌鳎中最低(图2)。
2. 2 半滑舌鳎肠道微生物组成结构分析结果
在门水平上共鉴定出38个菌门,其中,12hpi和24hpi半滑舌鳎肠道微生物组成均以变形菌门(Proteobacteria)、拟杆菌门(Bacteroidetes)和厚壁菌门(Firmicutes)占绝对优势,三者的相对丰度之和超过90%;CG半滑舌鳎肠道微生物组成以变形菌门、拟杆菌门、厚壁菌门和螺旋体门(Spirochaetes)为优势菌门,其相对丰度之和也在90%以上(图3-A)。变形菌门和厚壁菌门的相对丰度在12hpi半滑舌鳎肠道微生物组成中占比最高,分别为48.28%和22.08%;而拟杆菌门的相对丰度在CG半滑舌鳎肠道微生物组成中占比最高,为7.85%。
在属水平上共鉴定出578个菌属,相对丰度较高的前15个优势菌属分别为希瓦氏菌属(Shewanella)、Elizabethkingia、曼噬甲壳菌属(Chitinophaga)、Petrimonas、Proteiniphilum、Brevinema、苯基杆菌属(Phenylobacterium)、Aminobacterium、食酸菌属(Aci-dovorax)、芽孢杆菌属(Bacillus)、假单胞菌属(Pseudomonas)、乳杆菌属(Lactobacillus)、Marivita、弧菌属(Vibrio)和雷尔氏菌属(Ralstonia)(图3-B)。其中,Elizabethkingia、曼噬甲壳菌属、Brevinema、苯基杆菌属、假单胞菌属、乳杆菌属、Marivita和雷尔氏菌属的相对丰度在CG半滑舌鳎肠道微生物组成中占比最高,希瓦氏菌属、Petrimonas、Proteiniphilum和Aminobacterium的相对丰度在12hpi半滑舌鳎肠道微生物组成中占比最高,食酸菌属、芽孢桿菌属和弧菌属的相对丰度在24hpi半滑舌鳎肠道微生物组成中占比最高。
2. 3 半滑舌鳎肠道微生物Beta多样性分析结果
基于Unifrac距离构建UPGMA聚类分析树,结果(图4)显示,CG和12hpi的半滑舌鳎肠道微生物样本先聚为一支,二者与24hpi半滑舌鳎肠道微生物样本的Unifrac距离相对较远,说明海藻希瓦氏菌感染半滑舌鳎24 h后其肠道菌群相对丰度及多样性发生了明显变化。
2. 4 半滑舌鳎肠道组织相关功能基因的表达情况
由图5可看出,半滑舌鳎肠道中ALDOA基因的相对表达量在24hpi时显著高于CG(P<0.05,下同); PLB1、GGT1、HSPA1A和HINT1基因在12hpi时的相对表达量显著高于CG和24hpi,而TREH基因的相对表达量在12hpi时显著低于CG和24hpi,说明肠道组织中免疫功能相关基因(HSPA1A和HINT1)参与半滑舌鳎抗细菌感染的免疫应答过程,而代谢功能相关酶类基因(ALDOA、PLB1、GGT1和TREH)与肠道代谢功能紊乱和疾病发生密切相关。
3 讨论
目前,关于鱼类肠道菌群的研究已有较多报道,但主要集中在健康鱼类,而针对病原菌感染引起鱼类肠道菌群结构持续变化的研究相对较少(Han et al.,2010;Huang et al.,2016)。与健康鱼相比,患病鱼类肠道微生物多样性显著降低(Li et al.,2016a)。本研究采用16S rDNA高通量测序技术分析致病性海藻希瓦氏菌感染对半滑舌鳎肠道菌群结构的影响,结果表明,健康半滑舌鳎肠道微生物丰度及多样性均高于感染后,与张正等(2014)在患腹水病和皮肤溃烂病半滑舌鳎、刘志刚等(2018)在患链球菌病的尼罗罗非鱼(Oreochromis niloticus)的研究结果一致。由此推测,鱼类健康状况与肠道微生物多样性具有一定相关性。本研究还发现,半滑舌鳎肠道中变形菌门、拟杆菌门和厚壁菌门占绝对优势,与张正等(2014)的研究结果相似,表明这些菌门可能是半滑舌鳎肠道内的核心菌群。虽然感染海藻希瓦氏菌半滑舌鳎肠道中这些优势菌群的相对丰度与健康半滑舌鳎不同,但这些菌门均是丰度最高的类群。此外,本研究发现在海藻希瓦氏菌感染前后,半滑舌鳎肠道内均存在弧菌属、希瓦氏菌属和芽孢杆菌属,且这些菌属的相对丰度在感染后明显增加。弧菌是鱼类肠道中常见的微生物类群(Jensen et al.,2004;Hovda et al.,2007),且某些种能引起水产动物暴发恶性传染病,如鳗弧菌(V. anguillarum)和溶藻弧菌(杨少丽等,2005);希瓦氏菌和芽孢杆菌也是水产动物的主要条件性致病菌,可引起鱼类暴发疾病(Han et al.,2017b)。健康鱼类肠道内存在大量条件性致病菌,当机体肠道微生态被破坏时会迅速增殖,进而威胁鱼类健康,因此这些菌属应被给予高度关注。
除了肠道微生物组成与宿主健康状况密切相关外,肠道本身在鱼类的黏膜免疫系统中也发挥重要作用(Byadgi et al.,2014;Tafalla et al.,2016)。本研究结果表明,半滑舌鳎感染海藻希瓦氏菌后其肠道组织中的ALDOA、PLB1、HSPA1A、HINT1、GGT1和TREH基因表达水平均发生明显变化。HSPA1A基因编码Hsp72蛋白,该蛋白是热休克蛋白70(Heat shock protein 70,HSP70)家族的重要成员之一。热休克蛋白是一类保守的细胞蛋白,存在于从细菌到哺乳动物的所有生物体中(Mu et al.,2013;Liu et al.,2015),在应激反应及应激损伤中发挥重要作用(Ming et al.,2010;Mu et al.,2013)。至今,已有多种HSP70家族成员从患细菌性疾病的水产动物不同组织中被鉴定出来。哈维氏弧菌(V. harveyi)感染武昌鱼(Megalobrama amblycephala)6 h后其肝脏HSP70基因表达量显著升高并达峰值,随后开始降低(Ming et al.,2010);厚壳贻贝(Mytilus coruscus)血细胞中HSP70基因表达水平在鳗弧菌(V. anguillarum)感染后12~48 h显著升高,至感染72 h时降到正常水平(Liu et al.,2014);哈维氏弧菌感染后,花鲈(Lateolabrax maculatus)头肾、肠道和鳃组织中HSPA1A、HSC70-1、HSC70-2、HSPA4和HSPA14基因表达量呈先升高后下降的变化趋势(Han et al.,2017a)。在本研究中,半滑舌鳎感染海藻希瓦氏菌后其肠道组织中HSPA1A基因相对表达量在感染12 h时显著增高,但至感染24 h时又降至正常水平,与在武昌鱼(Ming et al.,2010)和花鲈(Han et al.,2017a)受到病原感染后SPA1A基因及HSP70家族其他成员的表达模式具有一致性。HINT1基因编码的蛋白分子是组氨酸三聚体蛋白超家族中分布最广的成员,其在低等和高等动物中的结构及功能高度保守,可通过参与Wnt通路、核因子κB、激活蛋白1信号通路而控制多种转录过程,调节细胞的分化、增殖和凋亡,具有免疫监视功能作用(Korsisaari et al.,2003)。感染海藻希瓦氏菌后,半滑舌鳎肠道组织HSPA1A和HINT1基因相对表达量均呈先升高后下降的规律,表明这两种蛋白(HSPA1A和HINT1)参与半滑舌鳎肠道抗细菌感染的免疫应答,但具体作用机理有待进一步探究。
ALDOA、PLB1、GGT1和TREH是参与动物机体生理过程的重要代谢酶类。其中,ALDOA是一种广泛存在于生物体中并参与糖酵解的重要酶,主要存在于肌肉及血液红细胞中(Zhang et al.,2017);PLB1是一种磷脂酶,作用于溶血磷脂Sn-1位酯键,广泛存在于动植物及微生物体内;PLB能水解磷脂生成相应的甘油酰磷脂和脂肪酸,最后生成各类小分子物质,如氨基酸和乙醇胺等被机体利用(Xu et al.,2009);GGT1在微生物、植物和动物中广泛存在,从谷氨酰胺化合物水解和转移氨酰基团到受体,全程参与谷胱甘肽的代谢,在转化白三烯C4过程中发挥重要作用(West et al.,2013);TREH是葡萄糖苷酶的一种,对海藻糖有特异作用,水解后生成2个分子的葡萄糖(Kamiya et al.,2004)。本研究結果表明,海藻希瓦氏菌感染半滑舌鳎12 h后,其肠道组织中的ALDOA、PLB1和GGT1基因表达升高,而TREH基因表达降低,故推测这些基因的差异表达还与半滑舌鳎肠道消化吸收代谢功能紊乱有关,即参与机体疾病的发生过程。
4 结论
半滑舌鳎感染海藻希瓦氏菌后其肠道菌群多样性降低、菌群结构发生变化,肠道组织中ALDOA、PLB1、GGT1、HSPA1A、HINT1和TREH等相关功能基因呈差异表达,说明海藻希瓦氏菌感染引起半滑舌鳎肠道微生态紊乱,且肠道组织中免疫功能相关基因和代谢功能相关酶类基因分别参与机体的免疫应答及疾病发生过程。
参考文献:
高桂生,张艳英,张召兴,吉志新,史秋梅,陈娟,高光平,赵欣欣,韩红升,宋青春. 2016. 半滑舌鳎皮肤溃疡病的病原分离鉴定与药物敏感性分析[J]. 中国兽医学报,36(4):595-599. [Gao G S,Zhang Y Y,Zhang Z X,Ji Z X,Shi Q M,Chen J,Gao G P,Zhao X X,Han H S,Song Q C. 2016. Isolation and identification of pathogen of skin ulcer from Cynoglossus semilaevis and anaiysis of antibio-tic sensitivity[J]. Chinese Journal of Veterinary Science,36(4):595-599.]
胡璇,孙敬锋,陈成勋,邢克智. 2014. 养殖半滑舌鳎腹水病的病原分离鉴定及药物敏感性分析[J]. 天津农学院学报,21(3):12-16. [Hu X,Sun J F,Chen C X,Xing K Z. 2014. Isolation and identification of pathogen of ascites disease from Cynoglossus semilaevis Günther and its antibiotic sensitivity analysis[J]. Journal of Tianjin Agricultural University,21(3):12-16.]
刘志刚,卢迈新,可小丽,王淼,张德锋. 2018. 尼罗罗非鱼肠道及养殖环境中菌群结构与链球菌病的相关性[J]. 水产学报,42(10):1635-1647. [Liu Z G,Lu M X,Ke X L,Wang M,Zhang D F. 2018. Correlation between microflora structure in intestinal tract and aquaculture environment of tilapia(Oreochromis niloticus) and streptococcicosis[J]. Journal of Fisheries of China,42(10):1635-1647.]
杨少丽,王印庚,董树刚. 2005. 海水养殖鱼类弧菌病的研究进展[J]. 渔业科学进展,26(4):75-83. [Yang S L,Wang Y G,Dong S G. 2005. Progress of research on vibriosis in marine cultured fish[J]. Marine Fisheries Research,26(4):75-83.]
张正,廖梅杰,李彬,王印庚,王岚,荣小军,陈贵平. 2014. 两种疾病发生对养殖半滑舌鳎肠道菌群结构的影响分析[J]. 水产学报,38(9):1565-1572. [Zhang Z,Liao M J,Li B,Wang Y G,Wang L,Rong X J,Chen G P. 2014. Study on cultured half-smooth tongue sole(Cynoglossus semilaevis Günther) intestinal microflora changes affec-ted by different disease occurrence[J]. Journal of Fisheries of China,38(9):1565-1572.]
周紅霞,姚俊杰,房文红,李新苍,赵姝,王元,吴俣学,周俊芳. 2017. 半滑舌鳎溃疡病原杀鲑气单胞菌的分离鉴定与药敏试验[J]. 海洋渔业,39(3):322-330. [Zhou H X,Yao J J,Fang W H,Li X C,Zhao S,Wang Y,Wu Y X,Zhou J F. 2017. Isolation and identification & antimicrobial susceptibility test of Aeromonas salmonicida associa-ted with the skin ulceration disease of Cynoglossus semilaevis Günther[J]. Marine Fisheries,39(3):322-330.]
Byadgi O,Puteri D,Lee J W,Chang T C,Lee Y H,Chu C Y,Cheng T C. 2014. The effect of TLR9 agonist CpG oligodeoxynucleotides on the intestinal immune response of cobia(Rachycentron canadum)[J]. Journal of Immunology Research. doi:10.1155/2014/273284.
Chen W Y,Ng Y H,Wu J H,Chen J W,Wang H C. 2017. Microbiome dynamics in a shrimp grow-out pond with possible outbreak of acute hepatopancreatic necrosis di-sease[J]. Scientific Reports,7(1):9395.
Cornejo-Granados F,Gallardo-Becerra L,Leonardo-Reza M,Ochoa-Romo J P,Ochoa-Leyva A. 2018. A meta-analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota[J]. PeerJ,6:e5382. doi:10.7717/peerj.5382.
Cornejo-Granados F,Lopez-Zavala A A,Gallardo-Becerra L,Mendoza-Vargas A,Sánchez F,Vichido R,Brieba L G,Viana M T,Sotelo-Mundo R R,Ochoa-Leyva A. 2017. Microbiome of Pacific white leg shrimp reveals differential bacterial community composition between wild,aquacultured and AHPND/EMS outbreak conditions[J]. Scientific Reports,7(1):11783. doi:10.1038/s41598-017- 11805-w.
Davey G C,Calduch-Giner J A,Houeix B,Talbot A,Sitjà-Bobadilla A,Prunet P,Pérez-Sánchez J,Cairns M T. 2011. Molecular profiling of the gilthead sea bream(Sparus aurata L.) response to chronic exposure to the myxosporean parasite Enteromyxum leei[J]. Molecular Immunology,48(15-16):2102-2112.
Desai A R,Links M G,Collins S A,Mansfield G S,Drew M D,van Kessel A G,Hill J E. 2012. Effects of plant-based diets on the distal gut microbiome of rainbow trout(Oncorhynchus mykiss)[J]. Aquaculture,350:134-142.
Ding Z F,Cao M J,Hu X S,Xu G H,Wang R L. 2017. Changes in the gut microbiome of the Chinese mitten crab(Eriocheir sinensis) in response to white spot syndrome virus(WSSV) infection[J]. Journal of Fish Disea-ses,40(11):1561-1571.
Duan Y,Zhang Y,Dong H,Wang Y,Zhang J. 2017. Effects of dietary poly-beta-hydroxybutyrate(PHB) on microbiota composition and the mTOR signaling pathway in the intestines of litopenaeus vannamei[J]. Journal of Microbiology,55(12):946-954.
Edgar R C. 2013. UPARSE:Highly accurate OTU sequences from microbial amplicon reads[J]. Nature Methods,10:996-998.
Giatsis C,Sipkema D,Smidt H,Heilig H,Benvenuti G,Verreth J,Verdegem M. 2015. The impact of rearing environment on the development of gut microbiota in tilapia larvae[J]. Scientific Reports,5:18206. doi:10.1038/srep18206.
Guo H,Xian J A,Wang A L. 2016. Analysis of digital gene expression profiling in hemocytes of white shrimp Litopenaeus vannamei under nitrite stress[J]. Fish & Shellfish Immunology,56:1-11.
Han S F,Liu Y C,Zhou Z G,He S X,Cao Y N,Shi P J,Yao B,Ringø E. 2010. Analysis of bacterial diversity in the intestine of grass carp(Ctenopharyngodon idellus) based on 16S rDNA gene sequences[J]. Aquaculture Research,42(1):47-56.
Han Y L,Hou C C,Du C,Zhu J Q. 2017a. Molecular cloning and expression analysis of five heat shock protein 70 (HSP70) family members in Lateolabrax maculatus with Vibrio harveyi infection[J]. Fish & Shellfish Immunology,60:299-310.
Han Z R,Lü A J,Shi H Y,Sun J F,Xing K Z,Hu X C,Sung Y Y. 2017b. Isolation,identification and characterization of Shewanella algae from reared tongue sole,Cynoglossus semilaevis Günther[J]. Aquaculture,468:356-362.
Han Z R,Sun J F,Lü A J,Wang A L. 2018. Biases from different DNA extraction methods in intestine microbiome research based on 16S rDNA sequencing:A case in the koi carp,Cyprinus carpio var. Koi[J]. Microbiology Open,8(1):e00626.
He W Q,Rahimnejad S,Wang L,Song K,Lu K,Zhang C X. 2017. Effects of organic acids and essential oils blend on growth,gut microbiota,immune response and disease resistance of Pacific white shrimp(Litopenaeus vannamei) against Vibrio parahaemolyticus[J]. Fish & Shellfish Immunology,70:164-173.
Hovda M B,Lunestad B T,Fontanillas R,Rosnes J T. 2007. Molecular characterisation of the intestinal microbiota of farmed Atlantic salmon(Salmo salar L.)[J]. Aquaculture,272(1-4):581-588.
Huang Z B,Li X Y,Wang L P,Shao Z Z. 2016. Changes in the intestinal bacterial community during the growth of white shrimp,Litopenaeus vannamei[J]. Aquaculture Research,47(6):1737-1746.
Jensen S,Øvreås L,Bergh Ø,Torsvik V. 2004. Phylogenetic analysis of bacterial communities associated with larvae of the Atlantic halibut propose succession from a uniform normal flora[J]. Systematic and Applied Microbio-logy,27(6):728-736.
Kamiya T,Hirata K,Matsumoto S,Arai C. 2004. Targeted disruption of the trehalase gene:Determination of the digestion and absorption of trehalose in trehalase-deficient mice[J]. Nutrition Research,24(2):185-196.
Korsisaari N,Rossi D J,Luukko K,Huebner K,Henkemeyer M,Makela T P. 2003. The histidine triad protein Hint is not required for marine development or Cdk7 function[J]. Molecular and Cellular Biology,23(11):3929-3935.
Lauzon H L,Gudmundsdottir S,Petursdottir S K,Reynisson E,Steinarsson A,Oddgeirsson M,Bjornsdottir R,Gudmundsdottir B K. 2010. Microbiota of Atlantic cod(Gadus morhua L.) rearing systems at pre-and post hatch stages and the effect of different treatments[J]. Journal of Applied Microbiology,109(5):1775-1789.
Li T T,Long M,Ji C,Shen Z X,Gatesoupe F J,Zhang X J,Zhang Q Q,Zhang L L,Zhao Y L,Liu X H,Li A H. 2016a. Alterations of the gut microbiome of largemouth bronze gudgeon(Coreius guichenoti) suffering from furunculosis[J]. Scientific Reports,6:30606. doi:10.1038/srep30606.
Li X M,Yan Q Y,Ringø E,Wu X B,He Y F,Yang D G. 2016b. The influence of weight and gender on intestinal bacterial community of wild large mouth bronze gudgeon (Coreius guichenoti,1874)[J]. BMC Microbiology,16(1):191.
Lin C S,Chang C J,Lu C C,Martel J,Ojcius D M,Ko Y F,Young J D,Lai H C. 2014. Impact of the gut microbiota,prebiotics,and probiotics on human health and disease[J]. Biomedical Journal,37(5):259-268.
Liu H H,He J Y,Chi C F,Shao J Z. 2014. Differential HSP70 expression in Mytilus coruscus under various stressors[J]. Gene,543(1):166-173.
Liu T,Pan L Q,Cai Y F,Miao J J. 2015. Molecular cloning and sequence analysis of heat shock proteins 70(HSP70) and 90(HSP90) and their expression analysis when exposed to benzo(a) pyrene in the clam Ruditapes philippinarum[J]. Gene,555(2):108-118.
Ming J,Xie J,Xu P,Liu W B,Ge X P,Liu B,He Y J,Cheng Y F,Zhou Q L,Pan L K. 2010. Molecular cloning and expression of two HSP70 genes in the Wuchang bream (Megalobrama amblycephala Yih)[J]. Fish & Shellfish Immunology,28(3):407-418.
Mu W J,Wen H S,Li J F,He F. 2013. Cloning and expre-ssion analysis of a HSP70 gene from Korean rockfish(Sebastes schlegeli)[J]. Fish & Shellfish Immunology,35(4):1111-1121.
Narrowe A B,Albuthi-Lantz M,Smith E P,Bower K J,Roane T M,Vajda A M,Miller C S. 2015. Perturbation and restoration of the fathead minnow gut microbiome after low-level triclosan exposure[J]. Microbiome,3:6.doi:10.1186/ s40168-015-0069-6.
Pérez T,Balcázar J L,Ruiz-Zarzuela I,Halaihel N,Vendrell D,de Blas I,Muzquiz J L. 2010. Host-microbiota interactions within the fish intestinal ecosystem[J]. Mucosal Immunology,3(4):355-360.
Rungrassamee W,Klanchui A,Maibunkaew S,Chaiyapechara S,Jiravanichpaisal P,Karoonuthaisiri N. 2014. Characte-rization of intestinal bacteria in wild and domesticated adult black tiger shrimp(Penaeus monodon)[J]. PLoS One,9(3):e91853.
Suo Y,Li E,Li T,Jia Y,Qin J G,Gu Z,Chen L. 2017. Response of gut health and microbiota to sulfide exposure in Pacific white shrimp Litopenaeus vannamei[J]. Fish & Shellfish Immunology,63:87-96.
Tafalla C,Leal E,Yamaguchi T,Fischer U. 2016. T cell immunity in the teleost digestive tract[J]. Developmental and Comparative Immunology,64:167-177.
Talwar C,Nagar S,Lal R,Negi R K. 2018. Fish gut microbio-me:Current approaches and future perspectives[J]. Indian Journal of Microbiology,58(4):397-414.
Wang J,Huang Y J,Xu K H,Zhang X Y,Sun H Y,Fan L F,Yan M T. 2019. White spot syndrome virus(WSSV) infection impacts intestinal microbiota composition and function in Litopenaeus vannamei[J]. Fish & Shellfish Immunology,84:130-137.
Wang Y Z,Sun J F,Lv A J,Zhang S L,Sung Y Y,Shi H Y,Hu X C,Chen S J,Xing K Z. 2018. Histochemical distribution of four types of enzymes and mucous cells in the gastrointestinal tract of reared half-smooth tongue sole Cynoglossus semilaevis[J]. Journal of Fish Biology,92(1):3-16.
West M B,Chen Y,Wickham S,Heroux A,Cahill K,Hanigan M H,Mooers B H M. 2013. Novel insights into eukaryo-tic γ-glutamyltranspeptidase 1 from the crystal structure of the glutamate-bound human enzyme[J]. The Journal of Biological Chemistry,289(16):11569.
Xia M J,Feng P,Mu C K,Ye Y F,Wang C L. 2018. Disruption of bacterial balance in the gut of Portunus trituberculatus induced by Vibrio alginolyticus infection[J]. Journal of Oceanology and Limnology,36(5):1891-1898.
Xiong J,Wang K,Wu J,Qiuqian L,Yang K,Qian Y,Zhang D. 2015. Changes in intestinal bacterial communities are closely associated with shrimp disease severity[J]. Applied Microbiology and Biotechnology,99(16):6911-6919.
Xu S Y,Zhao L S,Larsson A,Venge P. 2009. The identification of a phospholipase B precursor in human neutrophils[J]. The FEBS Journal,276(1):175-186.
Yang G,Xiu Y J,Chen Y D,Bai L,Sha Z X. 2017a. Identification and expression of complement component C8α,C8β and C8γ gene in half-smooth tongue sole (Cynoglossus semilaevis) and C8α recombinant protein antibacterial activity analysis[J]. Fish & Shellfish Immunology,72:658-669.
Yang H T,Zou S S,Zhai L J,Wang Y,Zhang F M,An L G,Yang G W. 2017b. Pathogen invasion changes the intestinal microbiota composition and induces innate immune responses in the zebrafish intestine[J]. Fish & Shellfish Immunology,71:35-42.
Ye L,Amberg J,Chapman D,Gaikowski M,Liu W T. 2014. Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish[J]. The ISME Journal,10(8):2076. doi:10.1038/ismej.2016.71.
Zhang F,Lin J D,Zuo X Y,Zhang Y X,Hong C Q,Zhang G J,Cui X J,Cui Y K. 2017. Elevated transcriptional levels of aldolase A(ALDOA) associates with cell cycle-related genes in patients with NSCLC and several solid tumors[J]. BioData Mining,10:6.doi:10.1186/s13040-016-0122-4.
(責任编辑 兰宗宝)
