一株高适应性Nitrosomonas eutropha CZ-4的脱氨特性
2019-08-28熊英,向斯,2,程凯*
熊 英,向 斯,2,程 凯*
一株高适应性Nitrosomonas eutropha CZ-4的脱氨特性
熊 英1,向 斯1,2,程 凯1*
(1.湖北工业大学资源与环境工程学院,河湖生态修复与藻类利用湖北省重点实验室,湖北 武汉 430068;2.武汉微盛科创环境生物科技有限公司,湖北 武汉 430068)
从垃圾渗滤液中分离得到了一株亚硝化单胞菌CZ-4,其16S rDNA序列与C91的相似性达99%.研究了pH值、温度、游离亚硝酸浓度、盐度等对其生长的影响,并测试了其在垃圾渗滤液、黑臭水和富营养化湖水中的脱氨效果.结果表明,该菌的最适生长pH值为7.3~8.7,最适生长温度为30.9℃,游离亚硝酸和盐度对该菌的半数抑制浓度分别约为0.11mg/L与2%.在最佳发酵条件下,该菌的最大氨氮去除速率为58mg/(L×h),最短倍增时间为8.2h;在不同类型的污水/地表水(初始氨氮浓度为0.66~603mg/L)中,该菌的最大氨氮去除速率为11.4mg/(L×h),最短倍增时间为10.9h,最低残余氨氮浓度为0.11mg/L.
自养氨氧化菌;垃圾渗滤液;亚硝化单胞菌;氨氮去除速率;倍增时间
氨氮是造成水体富营养化和黑臭的重要原因.生物硝化脱氨[1]是主流的脱氨方式,包括氨氧化和亚硝氮氧化两步[2],而氨氧化(将氨氮氧化为亚硝氮)正是硝化的限速步骤[3-4].污水处理系统中的氨氧化主要由自养氨氧化细菌(AOB)完成.AOB包括亚硝化单胞菌属()等5个属[3],而亚硝化单胞菌属在污水和富营养化地表水中分布最广、数量最多[5],对氨氧化的贡献最大[6-7].
亚硝化单胞菌以氨为唯一能源,以CO2为碳源,对环境条件极度敏感[3,8],易受温度、pH值、盐度、游离氨(FA)和游离亚硝酸(FNA)浓度等多种因素的影响[9-10].如不适的温度[11]和低pH值导致的高FNA浓度均会抑制氨氧化[12],而高盐度则会通过增大细胞渗透压影响氧和底物的传递,使氨氧化速率下降[13].目前,虽有适应上述单一因素的亚硝化单胞菌的报道,但单株亚硝化单胞菌对多种因素均具有较高适应性的案例尚未见报道.
不同的亚硝化单胞菌对FA的亲和力差异较大[14],如和等对FA的亲和力较高,适合在低FA环境中生长,而在高FA环境中则有较长的延迟期[15];而等对FA的亲和力较低,适合在高FA环境中生长,但在低FA条件下的竞争力较差[16-17].迄今为止,关于单株亚硝化单胞菌在不同FA浓度条件下均具有较高亲和力的研究也鲜见报道.
本文分离筛选得到一株AOB,通过研究pH值、温度、FNA浓度、盐度等对其生长的影响,发现该菌在不同FA浓度下均能有效生长(脱氨),且能够耐受较高浓度的FNA和盐度,并适应较宽的温度和pH值范围,对于深入研究其适应机理并开发其应用价值具有重要意义.
1 材料与方法
1.1 培养基
向液体基础培养基[18]中加入适量1mol/L的氢氧化钠溶液调节pH=7.8[19],固体培养基中的琼脂粉含量为1.8%.
1.2 分离纯化
将湖南郴州某垃圾渗滤液处理厂的活性污泥离心(4℃,6500r/min,15min)后,用灭菌生理盐水洗涤3次去除亚硝氮,按1%的体积比接种于基础培养基中,27℃,200r/min振荡培养,5d后用格里斯试剂检测亚硝氮的积累情况.将亚硝氮阳性样品涂布平板,27℃培养5d后挑取单菌落接种于基础培养基中,27℃,200r/min振荡培养5~7d,对亚硝氮阳性样品进行3轮平板划线纯化.纯化后的样品经结晶紫染色[20]后油镜镜检.
1.3 16S rDNA测序和系统发育树构建
采用通用引物27F和1492R[21]扩增16S rDNA片段,PCR反应体系:taq酶(15U/μL)0.1μL;缓冲液2μL;dNTP混合液(2.5mmol/L)1.6μL;模4μL;上下游引物各0.15μL;超纯水12μL.反应条件: 94℃预变性4min;94℃变性25s,58℃退火20s,72℃延伸30s,25个循环;72℃最终延伸5min.PCR产物经上海生工测序,再经Blastn比对,并通过MEGA5.0的邻接法构建系统发育树.
1.4 发酵条件的优化与验证
1.4.1 接种液的制备 将菌种保藏液离心(4℃, 6500r/min,15min)洗涤3次后,取1mL接种到100mL基础培养基中,27℃,200r/min振荡培养至亚硝氮积累速率达到约100mg/(L×d)时(需3~5d)作为接种液.
1.4.2 盐度和FNA浓度对氨氮去除效果的影响 (1)盐度的影响向100mL盐度(以NaCl计)分别为0.2%、0.5%、1%、2%、4%和6%的基础培养基中加入1mL接种液,27℃,200r/min振荡培养5d,间隔取样测定氨氮浓度.(2)FNA浓度的影响向100mL亚硝氮浓度分别为0,500,1000,2000和4000mg/L的基础培养基(初始pH值均为7.8,其FNA浓度分别为0,0.06,0.11,0.23和0.46mg/L)中加入1mL接种液,27 ℃,200r/min振荡培养5d,间隔取样测定氨氮浓度.
1.4.3 初始pH值和温度对氨氮去除率(NRR)的影响 (1)初始pH值的影响向100mL初始pH值分别为7.3、8.0和8.7的基础培养基中加入1mL接种液(设3平行),30℃,200r/min振荡培养3d后测定氨氮浓度.(2)温度的影响向100mL初始pH值为8.0的基础培养基中加入1mL接种液(设3平行),分别于26,30和34℃,200r/min振荡培养3d后测定氨氮浓度.(3)温度和初始pH值的联合作用采用响应面法,利用Design-Export软件设计2因素5水平3平行的试验分组,pH值范围7.5~8.5,温度范围27~33℃.向100mL基础培养基中加入1mL接种液,于200r/min振荡培养3d后,测定氨氮浓度,并绘制响应曲面.
1.4.4 补料发酵 向20L发酵罐中装入10L基础培养基,并接种500mL接种液.试验组(根据1.4.3的结果)发酵条件为:温度30.9℃,pH=8.2;对照组发酵条件为:温度27℃,pH=7.8.两组均采用1mol/L碳酸氢钠自动控制pH值.发酵期间,当氨氮浓度降至约300mg/L时,补充氯化铵溶液使其达到500mg/L.
1.5 对不同类型污水的脱氨测试
1.5.1 菌剂的制备方法 将补料发酵的产物离心(4℃,6500r/min,15min)洗涤3次去除亚硝酸盐作为菌剂.
1.5.2 垃圾渗滤液中的脱氨测试 向100mL垃圾渗滤液中接种1mL菌剂,对照组不接种,31℃,200r/ min振荡培养,48h后测氨氮与亚硝氮浓度.
1.5.3 在富营养化湖泊水中的脱氨测试 向100mL三种湖水(汤逊湖、南湖和野芷湖)中分别接种1mL菌剂,对照组不接种,31℃,200r/min振荡培养, 12h后测氨氮浓度.
1.5.4 在黑臭河水中的脱氨测试 向100mL黑臭水(巡司河水)中接种0.1mL菌剂(较低的接种剂量能够明显延长培养时间,从而有充足的时间进行多次测量以得到较准确的最短倍增时间),对照组不接种,31℃, 200r/min振荡培养,每隔12h测氨氮浓度.
上述实验组与对照组均设3平行.
1.6 测试与计算方法
分别采用纳氏试剂法和N-(1-奈基)-乙二胺光度法测定氨氮和亚硝氮[22];根据氨氮去除速率的变幅[19]计算倍增时间(h)[23];采用Anthonisen等[24]的方法计算FA浓度(mg/L)和FNA浓度(mg/L).
1.7 统计方法
采用Origin作图;采用SPSS进行正态性检验和方差分析(多重比较采用LSD法);采用Design- Export软件的二阶模型进行响应面试验的方差分析与回归分析.
2 结果与讨论
2.1 细菌的形态和系统发育
分离纯化得到一株AOB,该菌呈短杆状(0.66~ 0.86)×(1.52~1.75)mm(见图1),具有明显的运动性,符合的特征[3].且其16S rDNA序列(Genbank序列号为MH999419[25])与已知的亚硝化单胞菌的相似度为97%~99%,其系统发育树(图2)显示该菌与C91的遗传距离最近(相似度为99%),考虑到该菌分离于郴州垃圾渗滤液,故将其命名为CZ-4.
与同属的其它亚硝化单胞菌相比,在高氨氮环境中占比最高[12],常分布于污水处理厂等高氨环境中[3,26-27],如Schmidt等[25]从牛粪中分离到了N904.而CZ-4菌也来源于垃圾渗滤液(其氨氮浓度高达500~1000mg/L),与同样来源于污水处理厂的C91[6]的相似度高达99%.

图1 N.eutropha CZ-4的光学显微照片
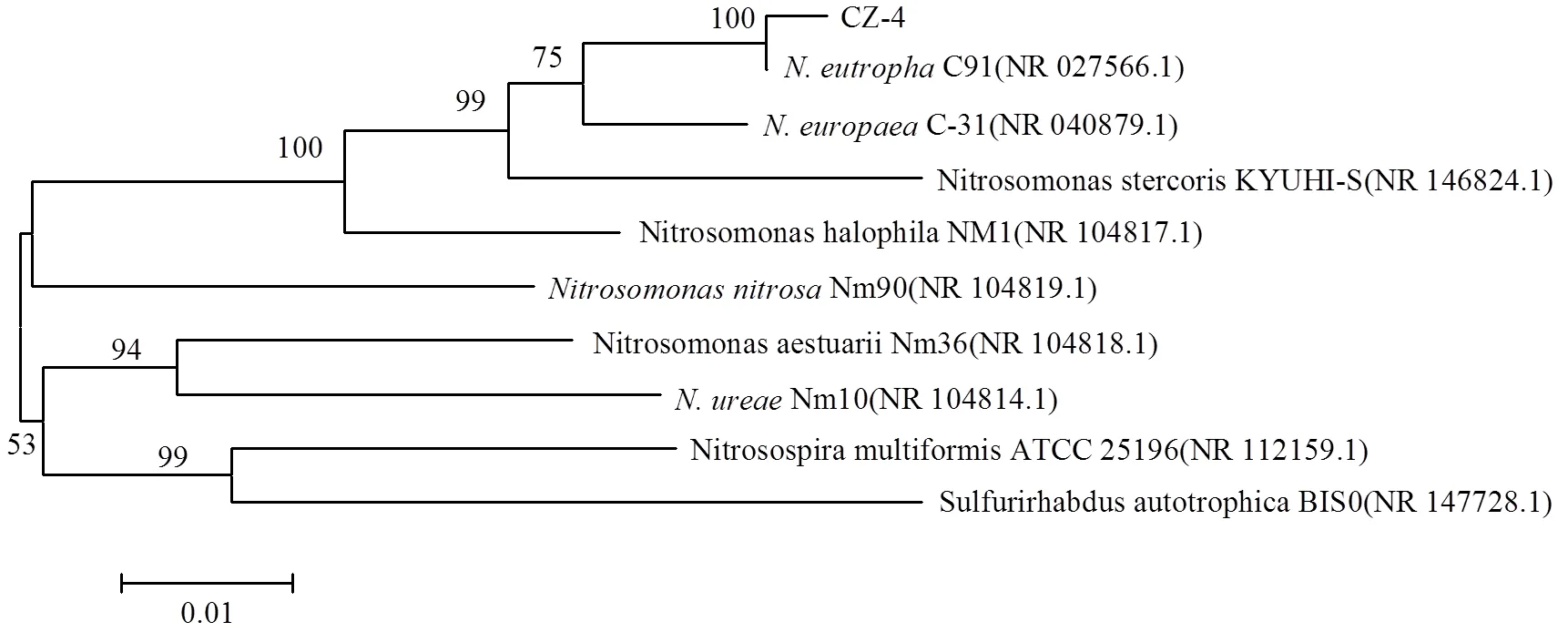
图2 N.eutropha CZ-4的系统发育树
2.2 环境因素对CZ-4菌的影响
2.2.1 盐度和FNA浓度对CZ-4菌的抑制效应 由图3a可见,盐度能够显著影响氨氮去除效果(<0.05):培养5d时,盐度2%条件下的NRR为52%,而盐度0.2%时的NRR为94%,说明CZ-4的半数抑制盐度约为2%,明显高于已知的亚硝化单胞菌(见表1).盐度是影响AOB生长的重要因素,高盐度使细胞渗透压增大,进而干扰底物和氧的传递并影响氨氧化速率[13],而微生物的耐盐能力则可能与蛋白质中的酸性氨基酸的含量有关[28-29].从应用上看,由于AOB的发酵过程中需要补碱(维持pH值稳定),盐度会随之增加,较高的耐盐能力将有利于该菌的补料发酵生产及在高盐废水中的应用.
FNA既是氨氧化的产物,也会对AOB产生毒性[24,27],因此FNA浓度也是AOB的重要影响因素[38].由图3b可见,FNA浓度能显著影响氨氮去除效果(<0.05):培养5d时,0.11mg/L FNA组的NRR为56%,而空白组的NRR则为94%,说明FNA对该菌的半数抑制浓度大于0.11mg/L;此外,0.23mg/L FNA组仍能够有效去除氨氮,说明该菌对FNA的最大耐受浓度应高于0.23mg/L,与Ms1[33]类似,高于19718[23,31-32]、spAL212[35-36]和C91[3,31]等多数亚硝化单胞菌,仅明显低于KYUHI-S[37].迄今为止,FNA对AOB影响机制的研究尚不充分[39],仅Stein等[31-32]发现FNA对3种AOB的影响机制各不相同,包括了特异性抑制氨单加氧酶(AMO),抑制AMO基因或一氧化氮还原酶基因的转录等.从应用的角度,较高的FNA耐受能力不但有利于该菌的补料发酵生产,而且有利于将该菌应用于短程硝化-反硝化等以亚硝氮为关键中间产物的脱氮工艺[40].

图3 盐度和FNA浓度对脱氨效果的影响

表1 不同亚硝化单胞菌对若干环境因子的适应性
注:a不低于最适pH值条件下的NRR的80%;b半数抑制盐度;c最大耐受盐度,n.d.为未检出.

图4 初始pH值和温度对NRR的影响
2.2.2 温度和pH值对CZ-4菌的影响 由图4可见,尽管30和34℃的NRR均显著高于26℃的NRR (<0.05),但该菌在26℃的NRR仍达到了最大NRR的58%.类似的,尽管初始pH=8.0的NRR显著高于pH=7.3的NRR(<0.05),但pH=8.7的NRR也能达到最大NRR的91%,说明适宜CZ-4菌生长的pH值范围至少为pH=7.3~8.7.
根据响应面试验的结果,温度和初始pH对NRR影响的回归方程为:
NRR=-2356.90+333.99 ×+68.65×+0.22××
-20.87×2-1.14×2(1)
式中:为温度,℃;为初始pH值.求解后的最佳培养条件为:温度30.9℃(<0.05),初始pH值为8.17 (<0.05),温度与初始pH值之间无交互作用(>0.05).图5是该回归方程的三维立体响应曲面,显示温度的响应面弯曲度比初始pH值更大,说明温度对氨氮去除率的影响较大.
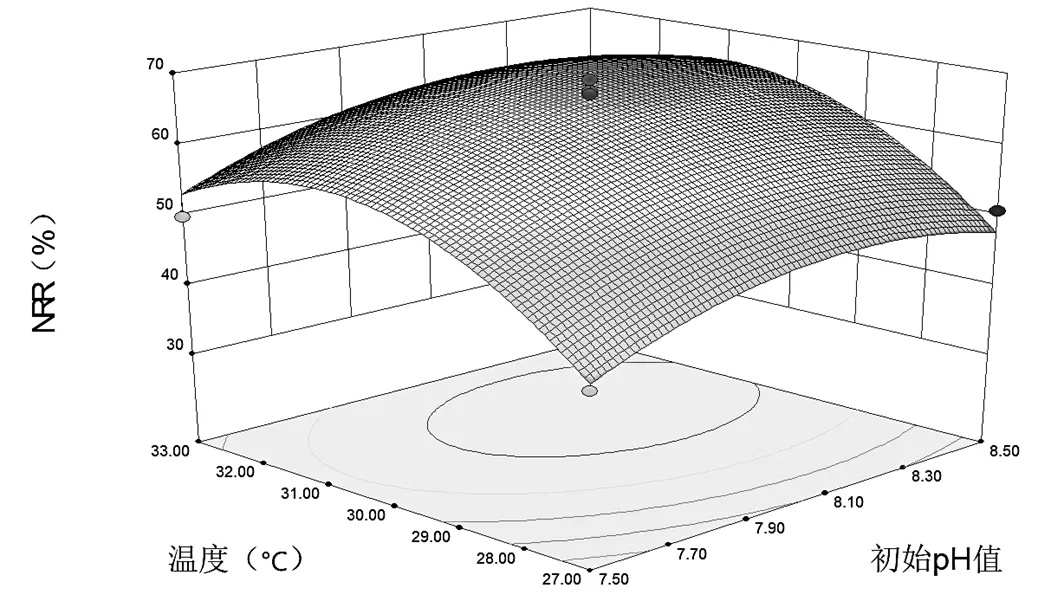
图5 温度和初始pH值的联合作用对NRR的影响
根据响应面试验的结果,优化了补料发酵的温度和pH值,结果表明(图6):尽管优化前后的平均氨氮去除速率(分别为21.85和18.65mg/(L×h))和最大氨氮去除速率(分别为48.45和57.59mg/(L×h))的变化均不大,但达到最大氨氮去除速率的时间从239.5h大幅度缩短至79.5h,而最短倍增时间也由48.5h减少至8.2h.说明优化后的温度和pH值条件能够明显加速CZ-4菌的生长,从而大幅缩短发酵周期.
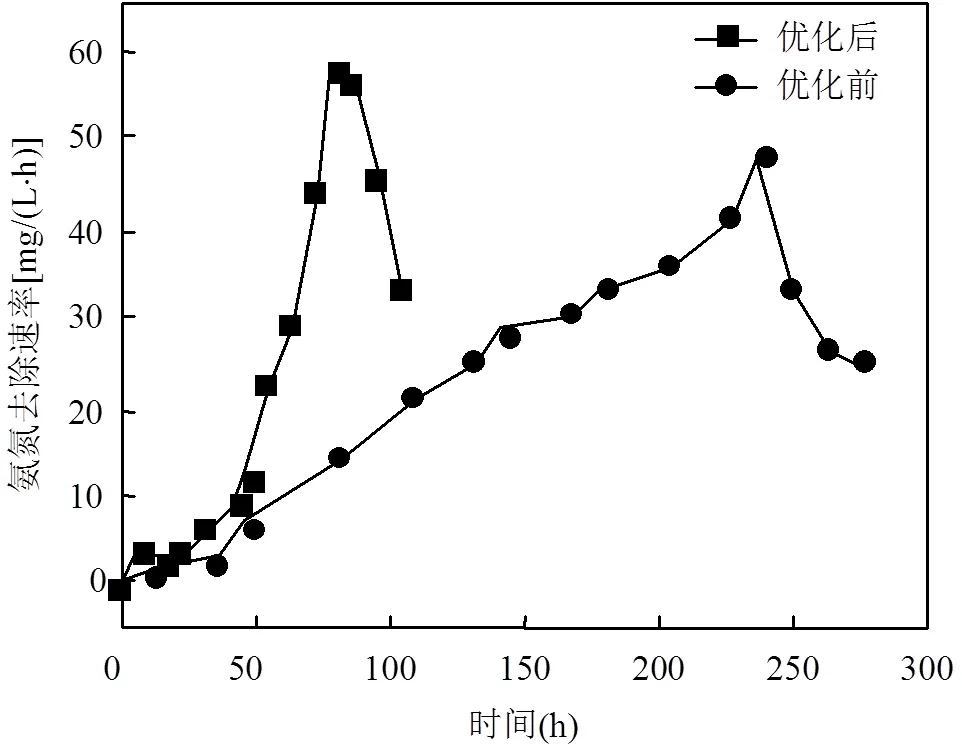
图6 补料发酵过程中氨氮去除速率的变化曲线
温度是影响AOB活性的重要因素[41],多数AOB的脱氨活性在10~30℃之间随升温而增加[42-43].但CZ-4菌的最适生长温度为30.9℃,明显高于其它亚硝化单胞菌(表1),将有利于降低该菌在发酵生产时的降温难度.此外,该菌在26~34℃时均保持较高的脱氨活性,也有利于在不同季节和不同温度的污水中应用该菌.
氨氮是AOB的能源与氮源,但只有FA才能透过细胞膜而被AOB所利用[2],故必须提供足够的FA才能支撑氨氧化.但是FA本身具有毒性[38],过高浓度的FA也可能直接抑制AOB的生长[44].FA的两面性使得不同类型的AOB具有不同的最适FA浓度(范围).而pH值则能够通过改变NH4+/NH3的电离平衡[12]而影响FA浓度,进而影响AOB的生长和代谢:多数AOB的最适pH值范围是7~8.5[27],且氨氧化速率随着pH值的下降而减小[45].表1比较了多株亚硝化单胞菌株的最适FA浓度范围,其中,多数菌的最适FA范围比较狭窄,如sp. AL212[35-36]的最适FA为0.5mg/L,而19718[30]的最适FA范围也仅为5.9~18.1mg/L.而CZ-4菌在pH=7.3~8.7的范围内均具有较高的氨氧化活性,对应的FA浓度范围为10.4~196.6mg/L,说明该菌对FA的亲和能力较强但又对高浓度的FA不甚敏感.目前,仅发现KYUHI-S[37]具有与CZ-4菌类似的较宽FA适应范围,但二者的16S rDNA序列的差异较大(图1,二者Blastn的相似度也仅为96%).从应用上看,较宽的最适FA范围不但有利于将CZ-4菌用于处理不同FA浓度的污水,而且有利于降低发酵生产时的pH值控制难度.
2.3 对不同类型污水的脱氨效果
菌剂对垃圾渗滤液的脱氨效果明显(图7):实验组的氨氮浓度显著低于对照组(<0.05),亚硝氮浓度则显著高于对照组(<0.05);实验组的氨氮平均去除速率达到11.4mg/(L×h),类似的,同期的亚硝氮的平均积累速率也达到了11.5mg/(L×h),说明脱氨的主要方式为氨氧化(将氨氮氧化为亚硝氮).
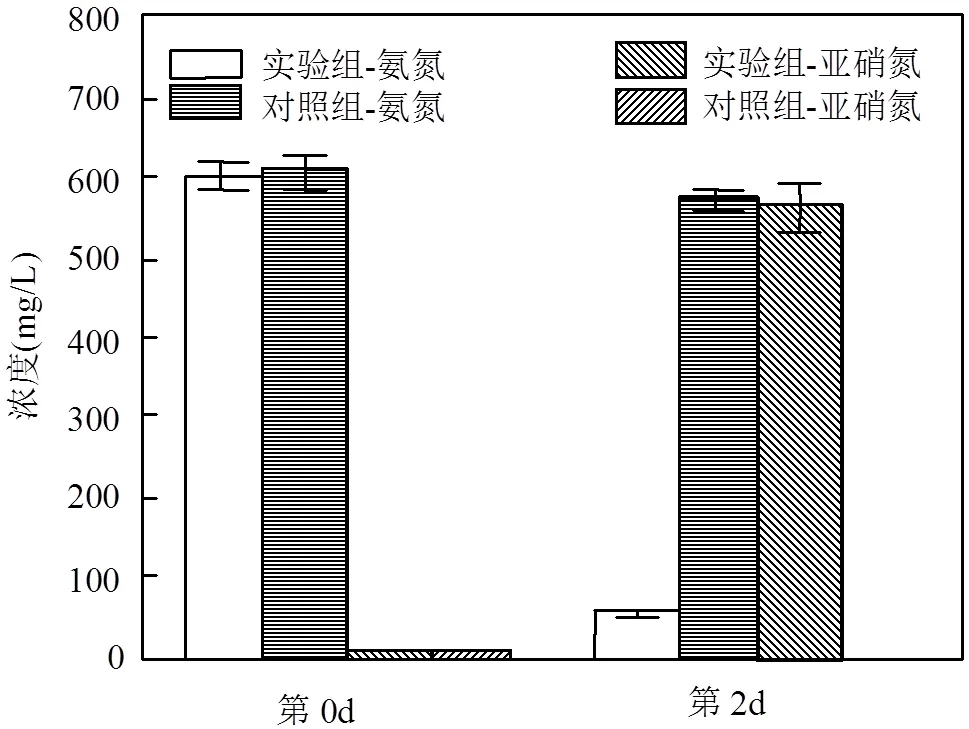
图7 垃圾渗滤液中的氨氧化效果
接种12h后,菌剂对3个不同来源的富营养化湖水也有显著的脱氨效果(图8a,<0.05):野芷湖水的氨氮浓度从地表水III类标准提高至地表水I类标准(残余氨氮浓度仅为0.11mg/L),汤逊湖水和南湖水的氨氮浓度也从地表水劣V类标准提高至地表水II类标准.类似的,在黑臭水中,按0.1%剂量投加的菌剂也能够有效脱氨(图8b,<0.05),特别是氨氮去除速率会持续增加:从12~24h时的0.09mg/(L×h),逐步增加至48~60h时的0.45mg/(L×h).

根据亚硝化单胞菌的底物亲和力和生长速率的不同,可分为r策略者和k策略者[5,46].其中,r策略者的底物亲和力低,生长快,如、.和.等[14];k策略者的底物亲和力高,但生长慢,如.和.等[15].鲜见单株亚硝化单胞菌在不同氨氮浓度培养基或污水中均具有较高脱氨活性的报道.由于缺少将纯种亚硝化单胞菌应用于污水的报道,表2比较了前人将亚硝化单胞菌接种至不同初始氨氮浓度的培养基中的平均氨氮去除速率,结果表明CZ-4菌不但在高氨氮培养基和高氨垃圾渗滤液中具有最高的氨氮去除速率,而且在中低浓度氨氮的地表水中也具有较高的氨氮去除速率,如该菌仅以0.1%的接种比处理初始氨氮浓度仅为15.88mg/L的黑臭河水时的氨氮去除速率,与.Ms1[33]和.KYUHI-S[37]等菌在按5%~10%比例接种于初始氨氮浓度为100~532mg/L的培养基中的速率相当(表2),甚至明显高于sp. THD-1[47]在氨氮浓度为58mg/L的培养基中的情况.而CZ-4菌对中低浓度氨氮具有较高亲和力的原因则可能与AMO有关:在基因组层面上,尽管多数AOB仅具有单拷贝的AMO基因,但少数AOB却具有多拷贝的AMO基因[15],特别是与CZ-4菌遗传关系最接近的C91也具有2个拷贝的AMO基因[48],而较多的拷贝数有利于增强AOB的低氨适应性[15,49];在表达调控的层面上,也有证据表明,AMO的数量也会随着FA浓度的降低而升高[50],从而增强AOB的低氨适应性.
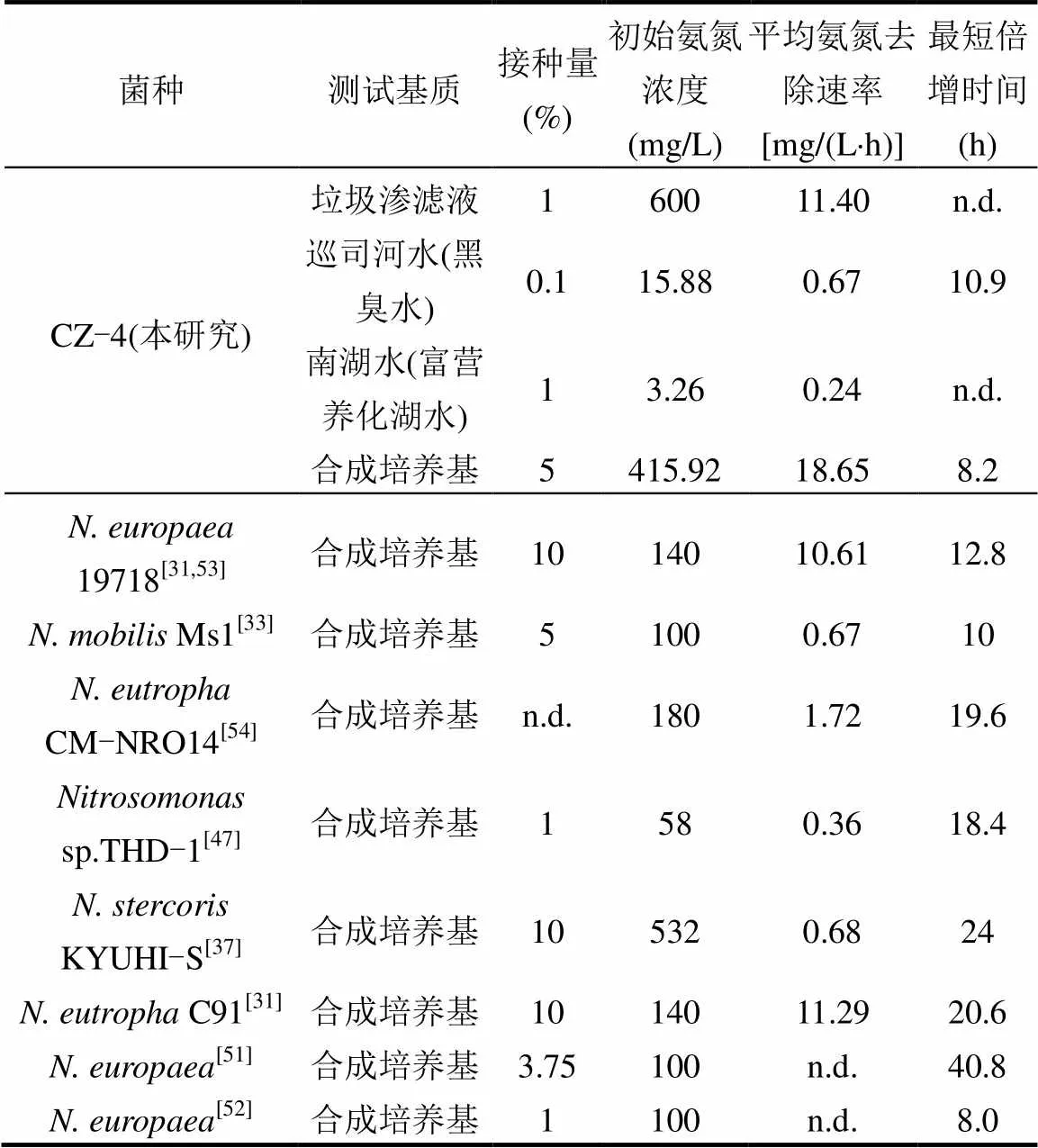
表2 不同亚硝化单胞菌脱氨效果的比较
最短倍增时间是指示微生物生长的重要指标,已知的亚硝化单胞菌的最短倍增时间均不低于8.0h[51-52],而CZ-4菌在培养基中的最短倍增时间低至8.2h,明显低于与CZ-4菌遗传关系最接近的C91[31]的最短倍增时间(表2).特别是,CZ-4菌在黑臭水中的最短倍增时间也仅为10.9h,甚至低于多数亚硝化单胞菌在合成培养基中的最短倍增时间(表2).综上所述,CZ-4菌既对氨氮具有较高的亲和力,又具有较快的生长速度,兼具r策略和k策略的优点,说明该菌具有重要的应用价值.
3 结论
3.1 从垃圾渗滤液中分离了一株.CZ-4,其16S rDNA序列与.C91的相似度为99%.
3.2 CZ-4菌的最适生长温度为30.9℃,最适生长pH值为7.3~8.7;其半数抑制盐度约为2%,半数抑制FNA浓度约为0.11mg/L.发酵条件优化后,最大氨氮去除速率为57.59mg/(L×h),最短倍增时间为8.2h.
3.3 CZ-4菌能够有效去除初始氨浓度为0.66~603mg/L的地表水或污水中的氨氮,对垃圾渗滤液的最大氨氮去除速率为11.4mg/(L×h),在黑臭水中的最短倍增时间为10.9h.
[1] Eighmy T T, Bishop P L. Distribution and role of bacterial nitrifying populations in nitrogen removal in aquatic treatment systems [J]. Water Research, 1989,23(8):947-955.
[2] Guo J, Peng Y, Wang S, et al. Pathways and organisms involved in ammonia oxidation and nitrous oxide emission [J]. Critical Reviews in Environmental Science and Technology, 2013,43(21):2213-2296.
[3] Koops H P, Purkhold U, Pommerening-Röser A, et al. The lithoautotrophic ammonia-oxidizing bacteria [J]. Prokaryotes, 2006, 5:778-811.
[4] Kowalchuk G A, Stephen J R. Ammonia-oxidizing bacteria: a model for molecular microbial ecology [J]. Annual Review of Microbiology, 2001,55:485-529.
[5] Koops H P, Andreas P R. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species [J]. FEMS Microbiology Ecology, 2001,37(1):1-9.
[6] Watson S W, Mandel M. Comparison of the morphology and deoxyribonucleic acid composition of 27strains of nitrifying bacteria1 [J]. Journal of Bacteriology, 1971,107(2):563-569.
[7] Koops H P, Harms H. Deoxyribonucleic acid homologies among 96strains of ammonia-oxidizing bacteria [J]. Archives of Microbiology, 1985,141(3):214-218.
[8] Papp B, Torok T, Sandor E,et al. High cell density cultivation of the chemolithoautotrophic bacterium[J]. Folia Microbiologica (Praha), 2016,61(3):191-198.
[9] Fiencke C, Spieck E, Bock E. Nitrifying bacteria [M]. Springer Netherlands, 2005:255-276.
[10] 张宇坤,王淑莹,董怡君,等.游离氨和游离亚硝酸对亚硝态氮氧化菌活性的影响[J]. 中国环境科学, 2014,34(5):1242-1247. Zhang Y K, Wang S Y, Dong Y J,et al. Effect of FA and FNA on activity of nitrite-oxidizing bacteria [J]. China Environmental Science, 2014,34(5):1242-1247.
[11] Groeneweg J, Sellner B, Tappe W. Ammonia oxidation in nitrosomonas at NH3concentrations near Km: Effects of pH and temperature [J]. Water Research, 1994,28(12):2561-2566.
[12] Claros J, Jimenez E, Aguado D,et al. Effect of pH and HNO2concentration on the activity of ammonia-oxidizing bacteria in a partial nitritation reactor [J]. Water Science & Technology, 2013, 67(11):2587-2594.
[13] 张宇坤,王淑莹,董怡君,等.NaCl盐度对氨氧化细菌活性的影响及动力学特性[J]. 中国环境科学, 2015,35(2):465-470. Zhang Y K, Wang S Y, Dong Y J, et al. Effect of NaCl salinity on activity of ammonia-oxidizing bacteria and kinetic characterization [J]. China Environmental Science, 2015,35(2):465-470.
[14] Suwa Y, Imamura Y, Suzuki T, et al. Ammonia-oxidizing bacteria with different sensitivities to (NH4)2SO4in activated sludges [J]. Water Research, 1994,28(7):1523-1532.
[15] Bollmann A, Laanbroek H J. Continuous culture enrichments of ammonia-oxidizing bacteria at low ammonium concentrations [J]. FEMS Microbiology Ecology, 2001,37(3):211-221.
[16] Verhagen F J, Laanbroek H J. Competition for ammonium between nitrifying and heterotrophic bacteria in dual energy-limited chemostats [J]. Applied and Environmental Microbiology, 1991,57(11):3255- 3263.
[17] Verhagen F J M, Hageman P E J, Woldendorp J W,et al. Competition for ammonium between nitrifying bacteria and plant roots in soil in pots: effects of grazing by flagellates and fertilization [J]. Soil Biology and Biochemistry, 1994,26(1):89-96.
[18] Oshiki M, Takagi R, Hatamoto M,et al. High-cell-density cultivation of Nitrosomonas europaea in a membrane bioreactor for performing protein purification and characterization studies [J]. The Journal of General and Applied Microbiology, 2017,62(6):330-333.
[19] Bollmann A, French E, Laanbroek H J. Isolation, cultivation, and characterization of ammonia-oxidizing bacteria and archaea adapted to low ammonium concentrations [J]. Methods in Enzymology, 2011,486:55-88.
[20] 范秀容,李广武,沈 萍.微生物学实验[M]. 2版.北京:高等教育出版社, 1989:51-53.Fan X R, Li G W, Shen P.Microbiology experiment [M]. 2nd Ed. Beijing: Higher Education Press, 1989:51-53.
[21] Weisburg W G, Barns S M, Pelletier D A, et al. 16S ribosomal DNA amplification for phylogenetic study. [J]. Journal of Bacteriology, 1991,173(2):697-703.
[22] 国家环境保护总局.水和废水监测分析方法 [M]. 4版.北京:中国环境科学出版社, 2002:271-281.State Environmental Protection Administration. Water and wastewater monitoring and analysis method [M]. 4th Ed. Beijing: China Environmental Science Press, 2002:271-281.
[23] Belser L W, Schmidt E L. Growth and oxidation kinetics of three genera of ammonia oxidizing nitrifiers [J]. FEMS Microbiology Letters, 2010,7(3):213-216.
[24] Anthonisen A C, Loehr R C, Prakasam T B et al. Inhibition of nitrification by ammonia and nitrous acid [J]. Water Pollution Control Federation, 1976,48(5):835-852.
[25] Xiong Y, Cheng K.strainCZ-4 16S ribosomal RNA gene, partial sequence, GenBank: MH999419.1 [EB/OL]. https://www.ncbi.nlm.nih.gov/nuccore/MH999419.1/,2019-4-10/2019-4-13.
[26] Schmidt I, Bock E. Anaerobic ammonia oxidation with nitrogen dioxide by[J]. Archives of Microbiology, 1997,167(2/3):106-111.
[27] Jiménez E, Giménez J B, Seco A, et al. Effect of pH, substrate and free nitrous acid concentrations on ammonium oxidation rate [J]. Bioresource Technology, 2012,124(11):478-484.
[28] Paul S, Bag S K, Das S, et al. Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes [J]. Genome biology, 2008,9(4):R70.
[29] Soppa J. From genomes to function: haloarchaea as model organisms [J]. Microbiology, 2006,152(3):585-590.
[30] Koops H P, Bottcher B, Moller,et al. Classification of eight new species of ammonia-oxidizing bacteria:sp.nov.,sp.nov.,sp.nov.,sp.nov.,sp.nov.,sp.nov.,sp.nov.andsp.nov.[J].Journal of General Microbiology, 1991,137:1689-1699.
[31] Cua L S, Stein L Y. Effects of nitrite on ammonia-oxidizing activity and gene regulation in three ammonia-oxidizing bacteria [J]. FEMS Microbiology Letters, 2011,319(2):169-175.
[32] Stein L Y, Arp D J. Loss of ammonia monooxygenase activity inupon exposure to nitrite [J]. Applied and Environmental Microbiology, 1998,64(10):4098-4102.
[33] Thandar S M, Ushiki N, Fujitani H, et al. Ecophysiology and comparative genomics ofMs1isolated from autotrophic nitrifying granules of wastewater treatment bioreactor [J]. Frontiers in Microbiology, 2016,7:1869.
[34] Itoh Y, Sakagami K, Uchino Y, et al. Isolation and characterization of a thermotolerant ammonia-oxidizing bacteriumsp. JPCCT2from a thermal power station [J]. Microbes and Environments, 2013,28(4):432-435.
[35] Limpiyakorn T, Kurisu F, Sakamoto Y, et al. Effects of ammonium and nitrite on communities and populations of ammonia-oxidizing bacteria in laboratory-scale continuous-flow reactors [J]. FEMS microbiology ecology, 2007,60(3):501-512.
[36] Yuichi S, Norton J M, Bollmann A, et al. Genome sequence ofsp.strain AL212, an ammonia-oxidizing bacterium sensitive to high levels of ammonia [J]. Journal of Bacteriology, 2011,193(18):5047-5048.
[37] Nakagawa T, Takahashi R.sp.nov., a chemoautotrophic ammonia-oxidizing bacterium tolerant of high ammonium isolated from composted cattle manure [J]. Microbes and Environments, 2015,30(3):221-227.
[38] Park S, Bae W. Modeling kinetics of ammonium oxidation and nitrite oxidation under simultaneous inhibition by free ammonia and free nitrous acid [J]. Process Biochemistry, 2009,44(6):631-640.
[39] Tan N C G, Kampschreur M J, Wanders W, et al. Physiological and phylogenetic study of an ammonium-oxidizing culture at high nitrite concentrations [J]. Systematic and Applied Microbiology, 2008,31(2): 114-125.
[40] Loosdrecht M C M. The SHARON-Anammox process for treatment of ammonium rich wastewater [J]. Water Science and Technology, 2001,44(1):153-160.
[41] Chen M, Chen Y, Dong S, et al. Mixed nitrifying bacteria culture under different temperature dropping strategies: Nitrification performance, activity, and community [J]. Chemosphere, 2018,195: 800-809.
[42] Lee S, Cho K, Lim J, et al. Acclimation and activity of ammonia- oxidizing bacteria with respect to variations in zinc concentration, temperature, and microbial population [J]. Bioresource Technology, 2011,102(5):4196-4203.
[43] Zeng J, Zhao D, Yu Z, et al. Temperature responses of ammonia- oxidizing prokaryotes in freshwater sediment microcosms [J]. PLOS ONE, 2014,9(6):1-8.
[44] 孙洪伟,于 雪,高宇学,等.游离氨对氨氧化菌(AOB)活性抑制动力学试验[J]. 环境科学, 2018,39(9):4294-4301. Sun H W, Yu X, Gao Y X, et al. Inhibitory kinetics of free ammonia (FA) on ammonia-oxidizing bacteria (AOB) [J]. Environmental Science, 2018,39(9):4294-4301.
[45] Fumasoli A, Morgenroth E, Udert K M. Modeling the low pH limit ofin high-strength nitrogen wastewaters [J]. Water Research, 2015,83:161-170.
[46] Serralta S, Joaquín, Gatti., et al. Enrichment of AOB and NOB population by applying a BABE Reactor in an activated sludge pilot plant [J]. Water Environment Research, 2015,87(4):369-377.
[47] 楚 江,王一农,章文军,等.亚硝化单胞菌(sp.)THD-1分离鉴定及高密度培养[J]. 生物学杂志, 2012,29(5):81-84. Chu J, Wang Y N, Zhang W J, et al. Isolating and high-density culturing ofsp.THD-1 [J]. Journal of Biology, 2012, 29(5):81-84.
[48] Stein L Y, Arp D J, Berube P M, et al. Whole-genome analysis of the ammonia-oxidizing bacterium,C91: Implications for niche adaptation [J]. Environmental Microbiology, 2008,9(12):2993-3007.
[49] Hommes N G. Mutagenesis and expression of amo, which codes for ammonia monooxygenase in[J]. Journal of Bacteriology, 1998,180(13):3353-3359.
[50] Bock E, Wagner M. Oxidation of inorganic nitrogen compounds as an energy source [J]. Prokaryotes, 2013,64:83-118.
[51] Keen G A, Prosser J I. Steady state and transient growth of autotrophic nitrifying bacteria [J]. Archives of Microbiology, 1987,147(1):73-79.
[52] Skinner F A, Walker N. Growth ofin batch and continuous culture [J]. Archives of Microbiology, 1961,38(4): 339-349.
[53] Cruvellier N, Poughon L, Creuly C,et al. Growth modelling ofATCC(R) 19718 andATCC(R) 25391: A new online indicator of the partial nitrification [J].Bioresource Technology, 2016,220:369-377.
[54] 杨浩锋,谢 柳,周俊利,等.一株氨氧化细菌的分离鉴定及其氨氧化特性[J]. 基因组学与应用生物学, 2013,32(4):453-458. Yang H F, Xie L, Zhou J L, et al. Isolation, identification and characteristics of an ammonia oxidizing bacterium [J]. Genomics and Applied Biology, 2013,32(4):453-458.
Nitrogen removal characteristics of a highly adaptableCZ-4.
XIONG Ying1, XIANG Si1,2, CHENG Kai1*
(1.Hubei Key Laboratory of Ecological Restoration for River-Lakes and Algal Utilization for College of Resources and Environmental Engineering, Hubei University of Technology, Wuhan 430068, China;2.Wuhan Micro-Glory Environmental Technology Innovation Co.Ltd, Wuhan 430068, China)., 2019,39(8):3365~3372
Astrain was isolated from the landfill leachate. It was named asCZ-4 for its 16s rDNA sequence was highly similar toC91at an identity of 99%. This work studied the influences of pH, temperature, free nitrous acid concentration and salinity on the growth ofCZ-4, as well as its NH3-N removal abilities in landfill leachate, black odor water and eutrophic lake water. The results showed that the optimum growth pH of the strain was 7.3~8.7, the optimum growth temperature was 30.9°C, and the IC50of free nitrous acid and salinity was about 0.11mg/L and 2% (in terms of NaCl), respectively. Under the optimal fermentation condition, the maximum NH3-N removal rate reached 58mg/(L×h), and the shortest doubling time was 8.2h. In different types of sewage/surface water (their initial ammonia concentration ranged from 0.66mg/L to 603mg/L), the NH3-N removal rate was up to 11.4mg/(L×h), the doubling time was as short as 10.9h, and the residual NH3-N concentration was down to 0.11mg/L.
autotrophic ammonia oxidizing bacteria;landfill leachate;;ammonia nitrogen removal rate;doubling time
X172,X703.1
A
1000-6923(2019)08-3365-08
熊 英(1994-),女,湖北孝感人,硕士研究生,主要研究方向为氨氧化微生物技术.发表论文1篇.
2019-01-04
国家科技重大专项(2017ZX07602002)
* 责任作者, 教授, chengkaicn@163.com
