基于随机森林特征选择算法的鼻咽肿瘤分割
2019-08-01李鲜王艳罗勇周激流
李鲜 王艳 罗勇 周激流


摘 要:针对医学图像中存在的灰度对比度低、器官组织边界模糊等问题,提出一种新的随机森林(RF)特征选择算法用于鼻咽肿瘤MR图像的分割。首先,充分提取图像的灰度、纹理、几何等特征信息用于构建一个初始的随机森林分类器;随后,结合随机森林特征重要性度量,将改进的特征选择方法应用于原始手工特征集;最终,以得到的最优特征子集构建新的随机森林分类器对测试图像进行分割。实验结果表明,该算法对鼻咽肿瘤的分割精度为:Dice系数79.197%,Acc准确率97.702%,Sen敏感度72.191%,Sp特异性99.502%。通过与基于传统随机森林和基于深度卷积神经网络(DCNN)的分割算法对比可知,所提特征选择算法能有效提取鼻咽肿瘤MR图像中的有用信息,并较大程度地提升小样本情况下鼻咽肿瘤的分割精度。
关键词:鼻咽肿瘤;随机森林;特征重要性;特征选择;最优特征子集
中图分类号:TP391.41
文献标志码:A
Abstract: Due to the low greylevel contrast and blurred boundaries of organs in medical images, a Random Forest (RF) feature selection algorithm was proposed to segment nasopharyngeal neoplasms MR images. Firstly, graylevel, texture and geometry information was extracted from nasopharyngeal neoplasms images to construct a random forest classifier. Then, feature importances were measured by the random forest, and the proposed feature selection method was applied to the original handcrafted feature set. Finally, the optimal feature subset obtained from the feature selection process was used to construct a new random forest classifier to make the final segmentation of the images. Experimental results show that the performances of the proposed algorithm are: dice coefficient 79.197%, accuracy 97.702%, sensitivity 72.191%, and specificity 99.502%. By comparing with the conventional random forest based and Deep Convolution Neural Network (DCNN) based segmentation algorithms, it is clearly that the proposed feature selection algorithm can effectively extract useful information from the nasopharyngeal neoplasms MR images and improve the segmentation accuracy of nasopharyngeal neoplasms under small sample circumstance.
英文关键词Key words: nasopharyngeal neoplasms; random forest; feature importance; feature selection; optimal feature subset
0 引言
医学图像分割是当前图像处理领域的热点问题之一,对医学图像进行精准的分割是后续治疗的重要保障; 然而,由于当前医学成像普遍存在灰度对比度低、器官组织边界模糊的问题,医学图像的分割精度始终无法得到有效的提升。
在诸多头颈部肿瘤中,鼻咽肿瘤是最常见的肿瘤之一,在全球尤其是中国的广东地区有着较高的发病率。与其他部位的肿瘤相比,鼻咽肿瘤结构复杂,周边血管、淋巴管、腺体较多,且不同患者之间肿瘤形状和大小有较大的差异,因此目前临床上通常依赖医生结合解剖学及肿瘤形态学知识对其进行手动分割,过程枯燥耗时,具有极大的主观性且可重复性差[1-4],因此,相关领域的研究者一直致力于开发一种自动/半自动的分割方法,实现鼻咽肿瘤的精准分割。
Zhou等[5-7]近十余年来在鼻咽肿瘤分割领域做了大量的工作,其在文献[5]中提出一种基于知识的模糊聚类方法,首先使用半监督模糊C均值算法对图像进行初始分割,随后再基于对称性、连通性及聚类中心这三种空间解剖信息得到最终的分割结果;文献[6]提出一种新的图像纹理测量方法对文献[5]所提出的算法进行了改进;文献[7]则通过构造一个二分类支持向量机(Support Vector Machine, SVM)模型对鼻咽肿瘤图像进行了分割; Lee等[8]基于图像掩模、贝叶斯概率统计、阈值平滑、种子生长等技术实现了鼻咽肿瘤的分割; Chanapai[9]首先对鼻咽肿瘤图像进行了分层定位,并根据各层位置将肿瘤分为三个部分,然后基于自组织映射(Self Organizing Map, SOM)技术构造这三个部分的表征图用于构建初始肿瘤区域,最后基于区域生长算法实现鼻咽肿瘤的最终分割; Huang等[10]提出一种混合算法,将Adaboost、SVM、贝叶斯(Bayes)分類器结合起来用于鼻咽肿瘤的分割; 洪容容等[11]提出一种基于区域生长的改进分割方法,该方法从基于区域生长的自动分割入手,利用概率矩阵完成初始种子的自动生成,再使用SUSAN(Small Univalue Segment Assimilating Nucleus)算子作为区域生长的终止准则,最终实现鼻咽肿瘤磁共振(Magnetic Resonance, MR)图像的分割; Huang等[12]首先使用距离正则化水平集演化方法勾画得到一个初始的肿瘤边界,再借助最大熵隐马尔可夫随机场得到最终的分割结果; 文献[13-15]则将目前主流的深度学习方法引入鼻咽肿瘤的分割领域,并取得了一定的成果。
随机森林(Random Forest,RF)算法最早由Breiman[16]提出,因其具有实现简单、训练速度快、抗过拟合能力强、可并行处理等优势,因此被广泛应用于数据处理、文本分类、语义分割等领域。
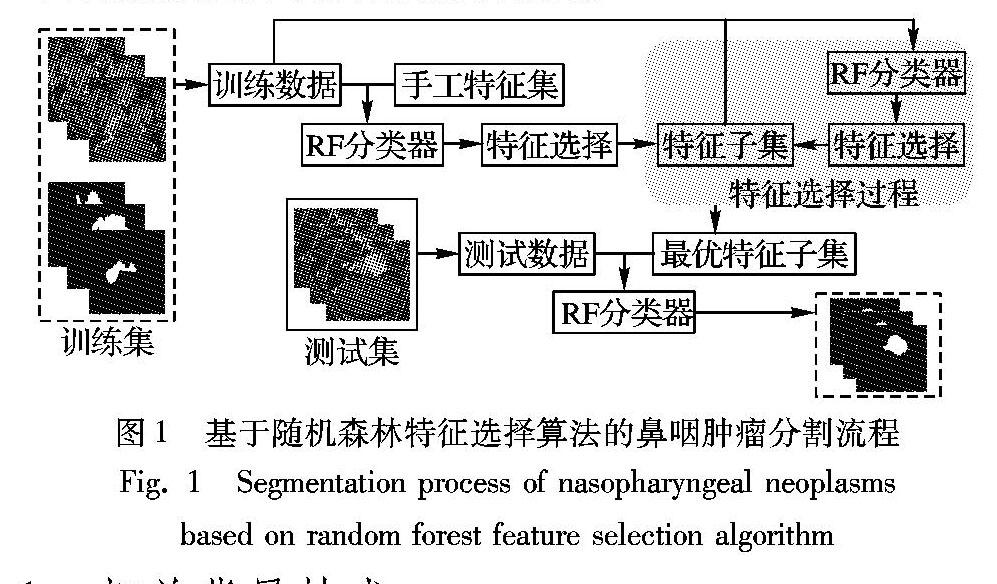
在此基础上,本文提出了一种随机森林特征选择算法用于鼻咽肿瘤MR图像的分割,算法的具体流程如图1所示。通过与基于普通RF、深度卷积神经网络(Deep Convolutional Neural Network, DCNN)的分割方法进行比较可知,本文算法可以有效提升鼻咽肿瘤的分割精度。
5 结语
本文借助随机森林特征重要性度量特性提出了一种新的鼻咽肿瘤MR图像分割方法,该方法可以实现对原始手工特征的选择优化,从而更好地实现对鼻咽肿瘤MR图像的分割。本文算法还有待改进,下一步拟借助深度学习方法的特征学习能力,将深度学习算法与RF结合起来,充分提取图像的中高级语义特征,进一步提升鼻咽肿瘤MR图像的分割精度。
参考文献 (References)
[1] 蒋君.多模态肿瘤图像联合分割方法研究[D].广州:南方医科大学, 2014: 40-45.(JIANG J. The research on tumor cosegmentation using multimodal images[D]. Guangzhou: Southern Medical University, 2014: 40-45.)
[2] CHU E A, WU J M, TUNKEL D E, et al. Nasopharyngeal carcinoma: the role of the EpsteinBarr virus [J]. Medscape Journal of Medicine, 2008, 10(7): 165.
[3] KLEIN G, KASHUBA E. Nasopharyngeal carcinoma[J]. Brenners Encyclopedia of Genetics, 2013, 13(1): 4-5.
[4] CHUA M L K, WEE J T S, HUI E P, et al. Nasopharyngeal carcinoma[J]. Lancet, 2015, 387(10022): 1012.
[5] ZHOU J, LIM T K, CHONG V. Tumor volume measurement for nasopharyngeal carcinoma using knowledgebased fuzzy clustering MRI segmentation[J]. Proceedings of SPIE, 2002, 4684: 1698-1708.
[6] ZHOU J, LIM T K, CHONG V, et al. A texture combined multispectral magnetic resonance imaging segmentation for nasopharyngeal carcinoma[J]. Optical Review, 2003, 10(5): 405-410.
[7] ZHOU J, CHAN K L, XU P, et al. Nasopharyngeal carcinoma lesion segmentation from MR images by support vector machine[C]// Proceedings of the 3rd IEEE International Symposium on Biomedical Imaging: Nano to Macro. Piscataway, NJ: IEEE Press, 2006: 1364-1367.
[8] LEE F K, YEUNG D K, KING A D, et al. Segmentation of NasoPharyngeal Carcinoma (NPC) lesions in MR images[J]. International Journal of Radiation Oncology Biology Physics, 2005, 61(2): 608-620.
[9] CHANAPAI W. Nasopharyngeal carcinoma segmentation using a region growing technique[J]. International Journal of Computer Assisted Radiology and Surgery, 2012, 7(3): 413-422.
[10] HUANG WC, LIU CL. A hybrid supervised learning nasal tumor discrimination system for DMRI[J]. Journal of the Chinese Institute of Engineers, 2012, 35(6): 723-733.
[11] 洪容容, 葉少珍. 基于改进的区域生长鼻咽癌MR医学图像分割[J]. 福州大学学报(自然科学版), 2014, 42(5): 683-687.(HONG R R, YE S Z. Segmentation of nasopharyngeal MR medical image based on improved region growing[J]. Journal of Fuzhou University (Natural Science Edition), 2014, 42(5): 683-687.)
[12] HUANG K W, ZHAO Z Y, GONG Q, et al. Nasopharyngeal carcinoma segmentation via HMRFEM with maximum entropy[C]// Proceedings of the 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Piscataway NJ: IEEE, 2015: 2968-2972.
[13] FENG A, CHEN Z H, WU X, et al. From convolutional to recurrent: case study in nasopharyngeal carcinoma segmentation[C]// Proceedings of the 2017 International Conference on the Frontiers and Advances in Data Science. Piscataway NJ: IEEE, 2017: 18-22.
[14] MEN K, CHEN X Y, ZHANG Y, et al. Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images[J]. Frontiers in Oncology, 2017, 7: 315.
[15] WANG Y, ZU C, HU G, et al. Automatic tumor segmentation with deep convolutional neural networks for radiotherapy applications[J]. Neural Processing Letters, 2018, 48(3): 1323-1334.
[16] BREIMAN L. Random forest[J]. Machine Learning, 2001, 45(1): 5-32.
[17] BREIMAN L. Bagging predictors[J]. Machine Learning, 1996, 24(2): 123-140.
[18] BREIMAN L. Manual on setting up, using, and understanding random forests V3.1[EB/OL]. [2012-05-05]. http://oz.berkeley.edu/users/breiman/Using_random_forests_V3.1.pdf.
[19] UWE H, RALF M, MICHAELl K B, et al. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data[J]. BMC Bioinformatics, 2009, 10(1): 1-16.
[20] ALTMANN A, TOLOSI L, SANDER O, et al. Permutation importance: a corrected feature importance measure[J]. Bioinformatics, 2010, 26(10): 1340-1347.
[21] CAROLIN S, ANNELAURE B, THOMAS K, et al. Conditional variable importance for random forests [J]. BMC Bioinformatics, 2008, 9(1): 307-307.
[22] 姚登舉.面向医学数据的随机森林特征选择及分类方法研究[D].哈尔滨:哈尔滨工程大学, 2016: 71-88. (YAO D J. Research on feature selection and classification method based on random forest for medical datasets[D]. Harbin: Harbin Engineering University, 2016: 71-88.)
