Screening, Identification and Characterization of Selenium-Enriched Bacillus pumilus D1-019
2019-07-26SUNYuXIAOYixinZHANGRanWANGChenghuaLIUXiaolingZHAOMouming
SUN Yu, XIAO Yixin, ZHANG Ran, WANG Chenghua*, LIU Xiaoling, ZHAO Mouming
(College of Light Industry and Food Engineering, Guangxi University, Nanning 530004, China)
Abstract: In this study, out of 119 bacterial isolates from duck foods, a novel selenium-enriched probiotic strain, designated as D1-019, was selected for its tolerance to selenite. This strain was obtained after four rounds of screening, which did not cause a red selenium phenomenon. Strain D1-019 was identified as Bacillus pumilus based on its colonial morphology,Gram staining, 16S rDNA analysis, and phylogenic tree analysis. Using an orthogonal experiment design, we obtained the optimum culture conditions for selenium enrichment as follows: pH 5, 30 ℃ for 48 h, and addition of 20 μg/mL selenite at 6 h of cultivation. Under these conditions, B. pumilus D1-019 eliminated selenite in the medium with a conversion rate of 100% and produced (1 327 ± 113) mg/kg of organic selenium, which was superior to the currently available seleniumenriched microbes. Our results indicated that B. pumilus D1-019 can be a promising candidate for selenium-enriched probiotics in the food industry.
Keywords: Bacillus pumilus; organic selenium; selenium-enriched; probiotics; selenite; 16S rDNA
Selenium-enriched probiotics combine the best qualities of probiotics and organic selenium supplements and have both probiotic potential and medical functions[1-2]. They can antagonize the colonization of pathogenic bacteria in the intestines of animals, suppress mutagenesis and cancer,and regulate the intestinal flora[3-5]. A moderate amount of selenium-enriched probiotic supplementation in the diet can significantly improve the production performance of young animals, as well as their immune function, antioxidant level,and gene expression level[6-7]. Bacillus has been used in food industry due to the excellent enzymatic activities of its protease, cellulose, and amylase, and the degradation of plant carbohydrates and stimulation of immune response. However,in contrast to the existing extensive studies on selenium-rich yeasts and Lactobacillus, there have been only a few reports on selenium-enriched Bacillus[3,8-9].
The currently employed methods to screen out seleniumenriched microbes can be classified into two categories:selenium-tolerance method and red selenium method. The former is based on the positive correlation between the inorganic selenium-resistance and selenium-biotransformation capacity of microbes, whereas the latter utilizes the negative correlation between the selenium-biotransformation capacity and the formation of red selenium[10]. These two methods are commonly used separately to achieve the development of the respective selenium-enriched microbes and red elemental selenium or selenium nanoparticles-producing microbes.However, hardly any selenium-enriched microbes have been screened by combining the two methods. Up to now, the best biotransformation efficiency for enriching selenium has been 94%, achieved by Saccharomyces cerevisiae at 20 μg/mL Na2SeO3. Additionally, a Bacillus subtilis strain isolated from naturally fermented Chinese vegetable pickles was found to transform 76% of the applied 45 μg/mL Na2SeO3[11]. To our best knowledge, no selenium-enriched probiotics with 100%transformation have been discovered to date.
In this study, we screened out a novel seleniumenriched Bacillus pumilus strain with 100% transformation rate using a synthetic method based on both its seleniumtolerance and red selenium formation. Moreover, we further optimized the culture conditions for enriching selenium by the implementation of an orthogonal experiment design and successfully characterized its selenium-enriching ability.
1 Materials and Methods
1.1 Strains, culture medium and chemical reagents
A collection of 119 unclassified bacterial strains, which were previously isolated from duck food samples (Guangxi Guilin Guiliu poultry Co. Ltd., China) and deposited in the Protein Engineering and Functional Food Laboratory of Guangxi University, were analyzed in this study. For convenience, the 119 strains were renamed sequentially as A1-A12, B1-B12, C1-C12, D1-D12, E1-E12, F1-F12, G1-G12, H1-H12, I1-I12, and J1-J11 in the screening process.All the bacterial strains were cultivated by using L-M17 medium, prepared by mixing 5 g of soybean peptone, 2.5 g of peptone, 2.5 g of casein peptone, 2.5 g of yeast extract powder, 5 g of beef powder, 0.5 g of sodium ascorbate, 19 g of sodium β-glycerophosphate, 0.25 g of magnesium sulfate,and 5 g of lactose in 1 L of water. The pH was adjusted to 7.0-7.4,and the medium was autoclaved at 121 ℃ for 15 min.
PrimeSTARTMHS DNA polymerase was acquired from China Dalian TaKaRa Bio Co. Ltd.; Plasmid DNA Purification Kit was acquired from America Omega Co.Ltd.; Na2SeO3was acquired from China Chengdu Jinshan Chemical Reagent Co. Ltd.. All of the chemicals and solvents used in this study were commercially available and of analytical grade.
1.2 Instruments and equipment
Tecan Infinite F200 PRO Microplate reader was produce by Tecan, Sweden Co. Ltd.; 7700e Inductively coupled plasma-mass spectrometry (ICP-MS) was produced by Aglient Co. Ltd.; Ultrapure deionized water was produced by France, Molsheim Co. Ltd.; T100TMThermal cyclers for PCR was produced by America Bio-Rad Co. Ltd..
1.3 Methods
1.3.1 Screening procedure for selenium-enriched bacteria
The screening procedure for selenium-enriched strains was performed in four steps. In the first step, all the 119 unclassified bacterial strains were screened out by their tolerance to Na2SeO3(50 μg/mL) using a densitometric method. Strains stored at -800 ℃ freezer were fetched out to spread over freshly prepared agar plates containing L-M17 medium. The streaked plates were incubated at 30 ℃ for 24 h.A single colony for each strain was picked to inoculate 5 mL of L-M17 medium, followed by culturing at 30 ℃ for 36 h.Then, 200 μL of L-M17 medium containing Na2SeO3at different concentrations was inoculated with 20 μL of each sample culture in 96-well deep-well plates and cultivated at 30 ℃ for 36 h. The Na2SeO3concentration used were 0,25, and 50 μg/mL, respectively. The strain concentration(OD600nm) in the well was measured by a microplate reader.The selenium tolerance of each strain was characterized by the ratio of OD600nmat a higher concentration of Na2SeO3to its lower concentration, and the strains with a ratio above 0.9 were chosen as candidates. In the second step, the selected candidates were screened again by elevating the Na2SeO3concentrations to 50, 200, and 500 μg/mL, respectively.The strains with a ratio above 1 were chosen as candidates for further screening. In the third step, the candidates were further evaluated by increasing the concentrations of Na2SeO3to 500, 865, and 1 730 μg/mL, correspondingly. Next, the strains were still-cultured in 5 mL of L-M17 medium at 30 ℃for 36 h. The top three strains with the best selenite tolerance were selected for the next screening stage. In the fourth step,the three candidates were rescreened by the red selenium phenomenon. By cultivating them at 30 ℃ for 36 h in 200 μL of L-M17 medium containing Na2SeO3concentrations ranging from 0 to 50 μg/mL, the strains which did not form red precipitation after treatment with 30 μg/mL Na2SeO3were chosen as the final strains for the next identification step.
1.3.2 Strain identification by colonial morphology, Gram staining and 16S rDNA analysis
The colonial morphology of the chosen strains was observed as described in the “Bergey’s Manual of Systematic Bacteriology”. The single colony of each strain was analyzed using the standard Gram staining method[12-13]. 16S rDNA genes were obtained by PCR using the isolated genomic DNAs as templates and a pair of universal primers, 27F (5’-AGA GTT TGA TCC TGG CTC AG-3’) and 1492R (5’-GGT TAC CTT GTT ACG ACT T-3’). The high-fidelity PrimeSTARTMHS DNA polymerase was employed following the manufacturer’s instructions. The amplified PCR products were then purified by agarose gel electrophoresis, followed by their direct sequencing at the Beijing Genomics Institute(Shenzhen, China). Next, the 16S rDNA sequences were identified in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and RDP database (http://rdp.cme.msu.edu/)by BLAST search and phylogenetic tree analysis via the MEGA software, edition 5.2.2[14].
1.3.3 Optimization of selenium-enrich conditions by orthogonal experiment design
To optimize the selenium-enriching conditions, the orthogonal experiment design was employed to study the main parameters, including temperature, pH, inorganic selenium (Na2SeO3) concentration, and selenium addition time[15]. An orthogonal design table L9(34) was used to arrange the four factors and three levels. A total number of nine combinations were tested, and the OD600nmand selenium conversion ratios were utilized to assess the effects of the combinations. The selenium conversion ratio was calculated by the ratio of the decrease of Na2SeO3concentration in culture medium to the initial Na2SeO3concentration. The selenium content was detected by ICP-MS according to the GB 5009.93-2017 Chinese standards for food safety:determination of selenium in foods. The standard curve for determining the selenium content by the ICP-MS is y=610 042.597 7x + 12.776 7 (R=0.997 0), where the x represented the Na2SeO3concentration (μg/mL), y was the count per second value; the detection limit and background equivalent concentration values were 1.438 × 10-5μg/mL and 2.094 × 10-5μg/mL, respectively.
1.3.4 Characterization of selenium-enriching bacteria
Ten milliliter selenium-enriched bacteria was inoculated into 100 mL of L-M17 medium in a 250 mL triangular flask(approximately OD600nm=0.03) and continuously cultivated at 30 ℃. After 6 h cultivation, Na2SeO3was added into the bacteria solution to a final concentration of 20 μg/mL,and the cultivation continued at 30 ℃ for 42 h. During the whole culture process, the cell concentrations (OD600nm) were measured every 2 h and used to plot growth curves. At the end of the culture period, the bacterial cells were collected by centrifugation at 12 000×g for 10 min. The precipitated cells were then washed twice with ultrapure deionized water and resuspended in the same ultrapure deionized water. Further,the cells were dried to a constant mass at 80 ℃, followed by determination of their dry mass. The selenium contents of inorganic and organic forms were determined by the ICP-MS method according to GB 5009.93-2017.
1.4 Statistical methods
The growth curves obtained from the experiments were fitted with Sigmoidal growth functions by GraphPad Prism software (GraphPad Software Inc., San Diego, Calif.), the obtained Hillslope values were used to compare their growth rates. Differences between two growth curves were evaluated by the unpaired, two-sided Student t-test, assuming the equal variation in the standard deviations as appropriate. P values of less than 0.05 were considered significant.
The selenium-enriched B. pumilus D1-019 strain screened out in this paper has been deposited in the China Center for Type Culture Collection (CCTCC) under the accession number of CCTCC 2018459. The nucleotide sequence of 16S rDNA of B. pumilus D1-019 has been deposited in the GenBank database under the accession No.MK300780.
2 Results and Analysis
2.1 Isolation of selenium-enriched microbes by synthetic screening
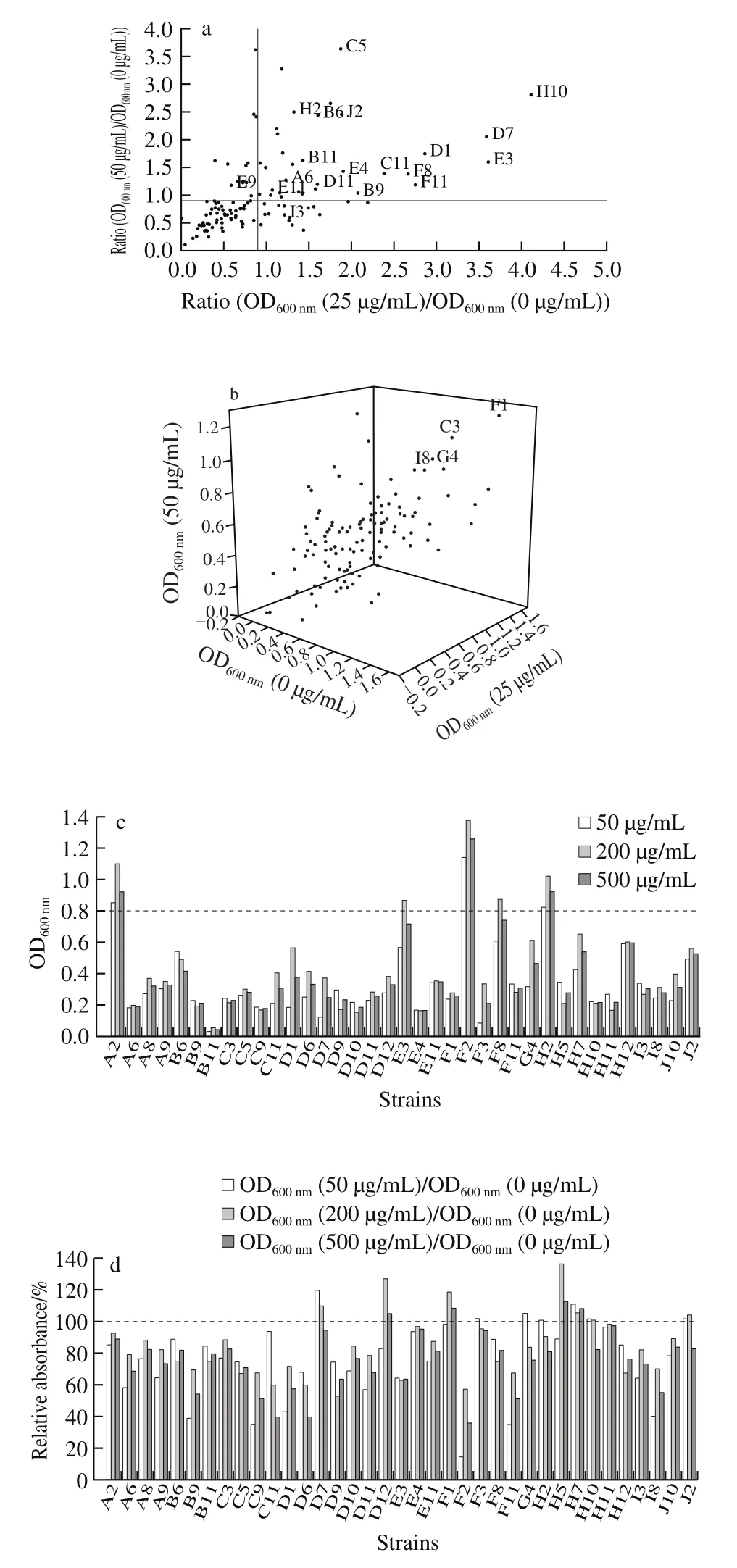
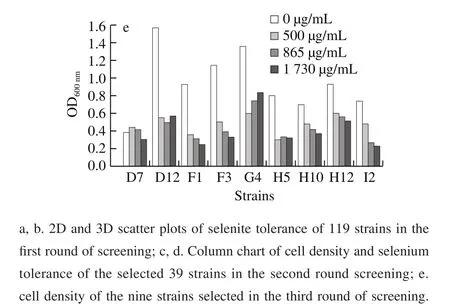
Fig. 1 Screening results of selenium-rich microorganisms
A collection of 119 unclassified microbial strains derived from duck food (unpublished data) were screened by a four-step synthetic screening process based on seleniumtolerance and red selenium phenomenon. As can be observed in Fig. 1a, in the first step, 33 strains showed ratios of OD600nm(25 μg/mL) to OD600nm(0 μg/mL) and OD600nm(50 μg/mL)to OD600nm(0 μg/mL) to be above 0.9. In addition, another four strains exhibited a faster-growing ratio at Na2SeO3concentrations of 25 and 50 μg/mL (Fig. 1b). The above 37 strains were chosen as candidtes for the next screening.In the second step, the growth of all selected 37 strains were inhibited to a certain extent, when the Na2SeO3concentration for screening were elevated to 200 and 500 μg/mL. Conversely, strains D12, F1, F3, and G4 had a moderately higher growth than that of the others at 500 μg/mL of selenite (Fig. 1c). Another 5 strains (D7, H5,H7, H10, and J2) exhibited better tolerance under Na2SeO3concentrations ranging from 50 μg/mL to 500 μg/mL(Fig. 1d). In the third step, the Na2SeO3concentration was further increased to 500, 864.7 μg/mL, and 1 729.40 μg/mL.As can be seen in Fig. 1e, the addition of selenite above 500 μg/mL considerably suppressed the growth of all 9 strains, except D7, whereas D12, G4, and H12 grew better than the other 5 strains at each concentration of Na2SeO3.
In the fourth step, all three strains grew well without red sediment formation at 10 μg/mL Na2SeO3. The increase in the Na2SeO3concentrations from 10 to 50 μg/mL Na2SeO3,D12 initially formed a large amount of red sediment at 20 μg/mL, followed by the H12 at 30 μg/mL Na2SeO3, while G4 brought only moderate red selenium at concentrations of up to 50 μg/mL Na2SeO3. Finally, G4, which was originally denoted as D1-019 when isolated from duck food, was the chosen one for the subsequent strain identification.
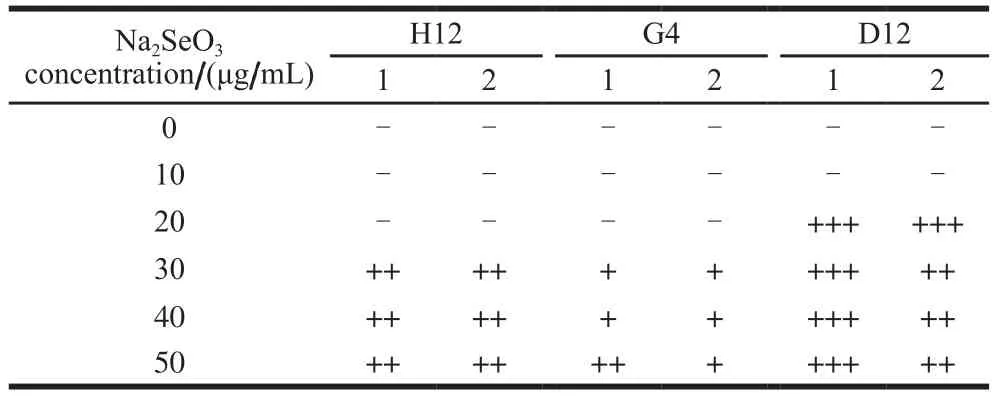
Table 1 Red selenium sedimentation phenomena caused by strains H12, G4, and D12 at different concentrations of selenite
2.2 Strain identification by colonial morphology, Gram staining, and 16S rDNA analysis
As can be seen in Fig. 2a, G4 had a yellowish, opaque,and flat colony with an irregular shape and edge. Its surface was wet, smooth, and sticky, and the rod-shaped cells were purple-dyed by Gram staining, indicating a Gram-positive bacterium (Fig. 2b). The 16S rDNA with a length of 1 454 bp were successfully amplified by PCR and sequenced (Fig.3); the sequence has been deposited in GenBank under the accession No. MK300780. BLAST search in GenBank database revealed that 16S rDNA G4 had the highest 100%homology with that of B. pumilus G1-3 (GenBank Accession No. JX120613.1) and B. pumilus GR24 (GenBank Accession No. KC771039.1)[16]. Similar results were obtained when the 16S rDNA sequence of G4 were analyzed in the RDP database. G4 matched best with B. pumilus B147 (RDP Accession No. S000635306)[17]. Our phylogenic tree analysis also revealed that G4 belonged to B. pumilus (Fig. 4).Based on the above analysis results, we identified G4 strain as B. pumilus D1-019. Generally, B. pumilus has wide applications in the production of food additives, such as enzymes and single cell proteins, and the degradation of toxic food substances, such as bisphenol A and aflatoxin M1[18-22].This bacterial species has also been approved for use as a feed additive and a directly-fed probiotic microorganism in many counties, including China and the USA. Therefore,the conditions for culturing B. pumilus D1-019 were further optimized for selenium-enriching, and its selenium-enriching performance was determined in our study.
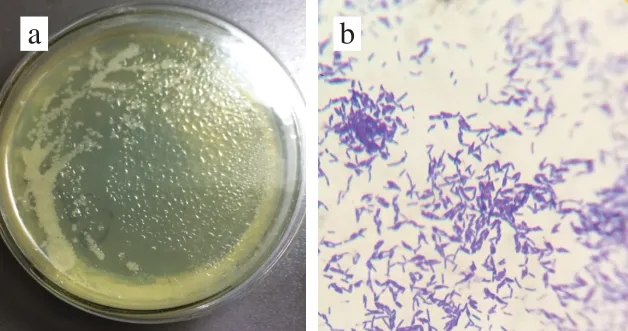
Fig. 2 Colonial morphology (a) and Gram staining (b)
Fig. 3 Agarose gel electrophoresis
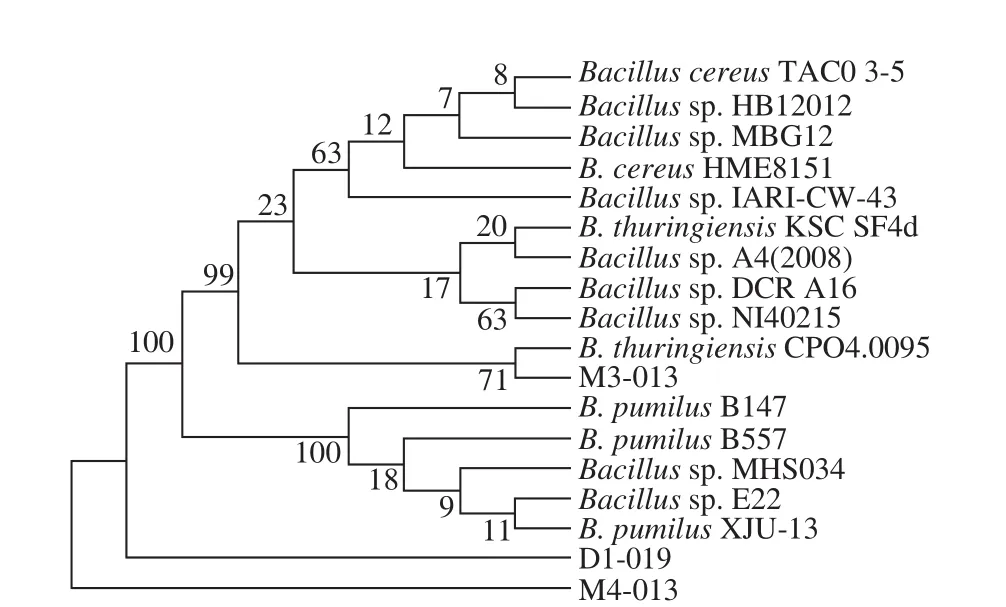
Fig. 4 Phylogenetic tree of strain D1-019
2.3 Optimization of the selenium-enriching conditions using the orthogonal experimental design
According to L9(34) orthogonal experimental design, a total number of 9 experiments were conducted to test the four factors, including culture temperature, initial pH, Na2SeO3concentration, and selenium addition time. The cell yield and selenium conversion results are presented in Tables 2 and 3.The average cell yield (OD600nm) was 1.30, and the maximum range of OD600nmof initial pH reached 0.45, whereas those of the culture temperature, Na2SeO3concentration, and time to add selenium were 0.25, 0.11, and 0.10, respectively (Table 2).Our results showed that the initial pH had the strongest impact on the growth of B. pumilus D1-019, followed in a descending order by the culture temperature, Na2SeO3concentration, and the selenium addition time. The optimal combination to obtain the highest selenium-enriched cell yield was the following: a temperature of 37 ℃, initial pH 6,a Na2SeO3concentration of 15 μg/mL, and addition of selenium at the beginning of the cultivation.

Table 2 L9 (34) Orthogonal experiment design with biomass yield(OD600 nm) of B. pumilus D1-019
Table 3 presents the orthogonal array of selenium conversion radios of B. pumilus D1-019. As can be seen,the selenium conversion ratio reached up to 83% in all experiments, except for groups 8 and 9. Based on the change in the mean value of each factor, the conversion ratio decreased sharply from 87.27% to 62.92% with the increase in the culture temperature from 30 ℃ to 42 ℃, whereas its increased markedly from 67.28% to 85.92% with the rise in the concentration of Na2SeO3from 10 to 20 μg/mL.However, the conversion ratio initially declined and then rose along with the increase in the initial pH of the culture medium from 5 to 7. Conversely, the conversion ratio was increased at first and then decreased with the prolongation of the time of Na2SeO3addition from 0 to 6 h and from 6 to 12 h. The culture temperature had the strongest impact on the conversion ratio with a maximum range of 24.35%,and followed by the initial pH, selenium addition time, and finally the Na2SeO3concentration, whose maximum values were 22.33%, 19.65%, and 18.64%, respectively. Therefore,we identified the optimum combination for obtaining the highest conversion ratio: a temperature of 30 ℃, initial pH 5,Na2SeO3concentration of 20 μg/mL, and selenium addition at 6 h of cultivation.

Table 3 L9 (34) Orthogonal experiment design with conversion rate of selenite
2.4 Confirmation of the optimal selenium-enriching conditions and selenium-enriched performance
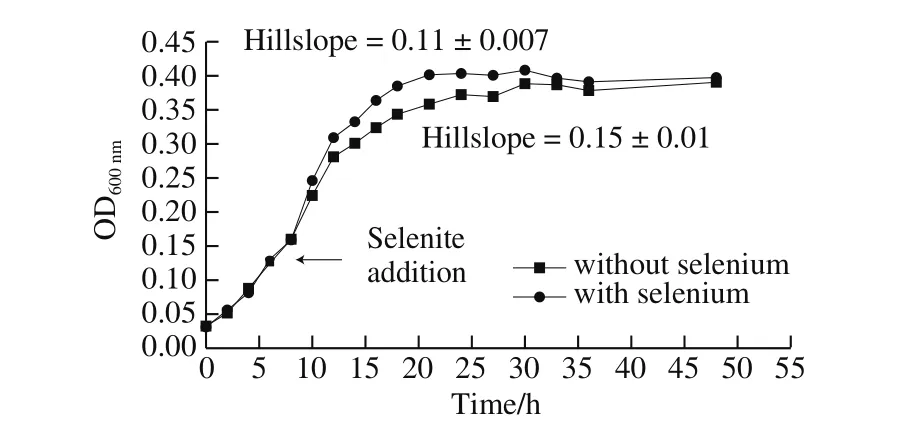
Fig. 5 Growth curves of B. pumilus D1-019 cultivated with or without 20 μg/mL of selenite
Under the optimal selenium-enriching conditions for conversion of selenite specified above, B. pumilus D1-019 showed typical sigmoidal curve similar to that without the addition of selenite, reaching OD600nmof 0.4, which was lower than those obtained in all aforementioned experiments(Fig. 5). When modeled using Sigmoidal growth functions,the growth curves with and without the addition of selenium showed Hillslope values of 0.11 ± 0.007 and 0.15 ± 0.01,respectively, which correspond well with the slight reduction of growth ratio and the prolonged time to the stationary stage after the selenite addition. The evaluation by the unpaired t-test with equal standard deviation the two-tailed established a P-value of 0.75, indicating that the differences between the two growth curves with and without addition of selenium were insignificant (P < 0.05). ICP-MS analysis revealed that the cultivated cells contained (1 327 ± 113) mg/kg organic selenium, (265 ± 30) mg/kg inorganic selenium,indicating that organic selenium accounted for 83% of the total selenium. However, there was no detectable inorganic selenium in the fermentation broth at the detection limit of 1.44 × 10-5μg/mL, indicating that the selenium conversion ratio reached 100%, which was higher than the above orthogonal experimental results (Table 3).
3 Discussion
Selenium-enriched probiotics can be used as both organic selenium supplements and probiotics and are attracting increasingly research attention[3-4,23]. However,hardly any such selenium-enriched probiotics have been discovered, except model strains of yeast and Lactobacillus.In the present study, we isolated and characterized a novel selenium-enriched probiotic, B. pumilus D1-019, from a collection of 119 strains originated from duck food.B. pumilus D1-019 survived in a Na2SeO3concentration of 1 730 μg/mL of Na2SeO3and eliminated all the Na2SeO3(20 μg/mL)from the culture medium at a conversion ratio of 100%, which equals the theoretical maximum ontain (1 327 ± 113) mg/kg of organic selenium (approximately 83% of their total selenium content), which ranks B. pumilus D1-019 as the best selenium-enriched microbe that satisfies the requirements to be an efficient organic selenium source for either food or feed supplements[15,24-25]. In addition, this powerful enriching capability may be of critical significance to the disposal of seleniferous soil and water[26-27].
Selenium has been exploited in its inorganic forms, such as Se2-, SeO32-, and SeO42-anions, as well as nanoparticles,elemental selenium, as well as organic selenium forms, such as selenoprotein, selenocysteine, and selenomethionine[28-29].Evidence exists that red selenium nanoparticles are safer than traditional inorganic selenium and organic selenium;biogenic selenium nanoparticles are more biocompatible and less toxic with inorganic selenium[9,30]. However, the toxicity of selenium nanoparticles varies largely among different species and the unclear mechanism underlying the transformation from selenite to elemental selenium still limits its commercial development[28,31]. Thus, the commercially available selenium sources are still confined to the traditional serious inorganic selenite and organic selenium yeast. In this study, we proposed a synthetic screening method by minimizing the unmanageable red selenium and maximizing the tolerance to inorganic selenite, and successfully screened out the probiotic B. pumilus D1-019 from a collection of 119 strains. Compared to the results of a previous investigation,in which a conversion ratio of 91.36% was reported for the conversion from inorganic selenium into elemental form by photosynthetic bacteria, the identified here B. pumilus D1-019 transformed 100% of the inorganic selenite present in the medium into other forms, without obvious formation of red elemental selenium[9]. The synthetic screening method for the novel selenium-enriched prebiotic B. pumilus D1-019 employed in the present examination may have important implications to the development of other novel seleniumenriched probiotics.
However, an obvious inconsistency is available between the optimal conditions for the selenium-tolerant growth and the selenium-enrichment of B. pumilus D1-019, and the cell yield under the latter conditions was lower than that under the former (Tables 2 and 3). These differences may be attributed to the different metabolism processes of selenium absorption, transformation, and utilization underlying the physiological growth states, as established in seleniumenriched Bifidobacterium[24]. To simultaneously achieve a better growth and selenium enrichment, it is worth trying to conduct two-stage experiments, satisfying the optimal culture conditions at each stage. Generally, usually no correlation exists between the total selenium and its inorganic forms in foods and those in living organisms; the distribution and forms of selenium in B. pumilus D1-019 are also still unclear[29]. Therefore, the metabolism and forms of selenium in B. pumilus D1-019 need further investigation in the future.
4 Conclusions
In conclusion, a novel selenium-enriched bacterium was isolated by a synthetic consideration of seleniumtolerance and red selenium phenomenon, which identified as B. pumilus D1-019 by colonial morphology, Gram staining,and 16S rDNA analysis. Furthermore, the conditions for growth and selenium enrichment of B. pumilus D1-019 were optimized by orthogonal experiments. Under the optimum conditions obtained by the application of an orthogonal experiment design, B. pumilus D1-019 converted all the Na2SeO3in the medium (20 μg/mL) at a conversion ratio of 100% and produced (1 327 ± 113) mg/kg of organic selenium(approximately 83% of the total selenium content). The origin of duck food, high selenium tolerance, and the conversion ratio of 100% of selenium enrichment suggest that B. pumilus D1-019 is a selenium-enriching probiotic that might have extensive applications in food industry.
