A Novel Reversible Data Hiding Scheme Based on Lesion Extraction and with Contrast Enhancement for Medical Images
2019-07-18XingxingXiaoYangYangRuiLiandWeimingZhang
Xingxing Xiao, Yang Yang, , Rui Li and Weiming Zhang
Abstract: The medical industry develops rapidly as science and technology advance.People benefit from medical resource sharing, but suffer from privacy leaks at the same time.In order to protect patients’ privacy and improve quality of medical images, a novel reversible data hiding (RDH) scheme based on lesion extraction and with contrast enhancement is proposed.Furthermore, the proposed scheme can enhance the contrast of medial image's lesion area directly and embed high-capacity privacy data reversibly.Different from previous segmentation methods, this scheme first adopts distance regularized level set evolution (DRLSE) to extract lesion and targets at the lesion area accurately for medical images.Secondly, the data is embedded into the lesion area by improved histogram shifting method to enhance the contrast of medial image’s lesion area.Lastly, the rest of data is embedded into the non-lesion area by the high-capacity embedding method to achieve the higher payload.At the receiving end, data can be extracted completely and images can be recovered losslessly by the third party with right.Experimental results have shown that the method of lesion extraction has an advantage over the existing segmentation methods in medical images.The image quality is improved well and the performance of contrast enhancement in the lesion area is better than other RDH methods with contrast enhancement.
Keywords: Reversible data hiding, contrast enhancement, lesion extraction, privacy protection, medical images.
1 Introduction
Cloud and big data have been derived from the explosion of data and information.The researches in such areas have promoted the rapid development of information storage, information mining, network application and computing technology.However, cloud and big data are easy to cause privacy leaks as personal information mounts and accumulates in the network.
Modern big data increasingly appears in a variety of forms.And image is a commonly used medium of information storage and transmission.In particular, the medical image is an important carrier of information about patients' physical condition.Besides, the medical image is labeled with private information, such as name, age, sex and so on, to distinguish its ownership.As a result, there are privacy leaks of patients in cloud and big data.A secure scheme will be desired to protect patients' privacy in the sharing of medical images.
Reversible data hiding (RDH) is a data hiding technique that can recover the original image without any distortion after the embedded data is extracted from the marked image and the hidden data is so unperceived that it is protected well.There are already many classic algorithms, such as difference expansion (DE) [Tian (2003)], histogram shifting (HS) [Ni, Shi, Ansari et al.(2006); Du, Yin and Zhang (2018)], and prediction error expansion (PEE) [Sachnev, Kim, Nam et al.(2009); Li, Yang and Zeng (2011)].DE method expands the difference value between two pixels to obtain redundant space to embed data.HS method embeds data by shifting the peak bin or lowest bin of the gray histogram.And PEE method expands a prediction error which is calculated according to the prediction rule to obtain redundant space to embed data.
This important technique is widely used in medical images, where no distortion of the original image is allowed.Different from natural images, medical images are often demanded that the contrast of images can be enhanced for helping doctors to diagnose [Wu, Dugelay and Shi (2015); Wu, Tang, Huang et al.(2018); Gao and Shi (2015); Yang, Zhang, Liang et al.(2016); Gao, Wan, Yao et al.(2017); Yang, Zhang, Liang et al.(2018)].And data to be embedded is not limited to authentication information, it may be information about the patient’s privacy, such as personal information, diagnostic records and results.These information is embedded into medical images to be protected.Only the third party with right, such as doctors and patient himself, can extract data and recover image completely.There are the state-of-the-art RDH methods with contrast enhancement as follows.Wu et al.[Wu, Dugelay and Shi (2015)] expanded the peakpairs of gray histograms to embed data to enhance image’s contrast, and they improved image preprocessing to achieve the higher payload later [Wu, Tang, Huang et al.(2018)].On the basis of HS, Gao et al.[Gao and Shi (2015)] added wavelet domain to embed more data.Yang et al.[Yang, Zhang, Liang et al.(2016)] prioritized to embed data into the texture region by prediction errors histogram.For medical images, there are existing RDH methods based on region of interest (ROI) segmentation, such as adaptive threshold detector (ATD) and Otsu.Gao et al.[Gao, Wan, Yao et al.(2017)] adopted HS to embed data into ROI which was segmented by Otsu.Yang et al.[Yang, Zhang, Liang et al.(2018)] adopted HS to embed data respectively into region of interest and region of noninterest (NROI) which were segmented by ATD.
Through analyzing the existing RDH methods with contrast enhancement, we find that the contrast enhancement of these methods [Wu, Dugelay and Shi (2015); Wu, Tang, Huang et al.(2018); Gao and Shi (2015); Yang, Zhang, Liang et al.(2016)] are not obvious at the low embedding rate and the segmentation ways [Gao, Wan, Yao et al.(2017); Yang, Zhang, Liang et al.(2018)] which are based on threshold are not intuitive for doctors to diagnose.Therefore, the more goal-oriented segmentation scheme and RDH method with obvious contrast enhancement are required for medical images.Thus, we propose a novel RDH scheme based on lesion extraction and with contrast enhancement for medical images in this paper.The main contributions of the proposed scheme include two aspects:
(1) Propose a novel way which can extract lesion by distance regularized level set evolution.Experimental results have shown that it is goal-oriented and intuitive for medical images.And it has an advantage over the state-of-the-art segmentation methods for doctors to diagnose.
(2) Propose two different embedding methods in the lesion area and the non-lesion area respectively.To improve quality of medical images, data first is embedded into empty bins of the gray histogram in the lesion area after stretched.To achieve the higher payload, the rest of data is embedded into the non-lesion area.Experimental results have shown that the contrast of lesion area is enhanced obviously and the performance of contrast enhancement in the lesion area is better than others which also are RDH methods with contrast enhancement.
The rest of this paper is organized as follows.Section 2 describes the details of the proposed scheme.Experimental results and analysis are shown in Section 3.Finally, conclusion is presented in Section 4.
2 Proposed method
To protect patients' privacy and improve quality of medical images, this paper proposed a novel reversible data hiding scheme based on lesion extraction and with contrast enhancement.Aiming at the lesion area, the proposed scheme adopts distance regularized level set evolution to extract lesion for helping doctors to diagnose.Privacy data related to the patient is embedded into the lesion area and the non-lesion area respectively to be recorded and protected.In addition, the image quality is improved well.Data can be extracted completely and image can be recovered losslessly by the third party with right.Fig.1 illustrates the diagram of the proposed scheme.

Figure 1: The diagram of the proposed scheme
2.1 Lesion extraction
At present, there are existing RDH methods which based on ROI segmentation, such as ATD and Otsu, for medical images.Those methods based on threshold segmentation are used to segment an image into foreground regarded as ROI and background regarded as NROI.In fact, doctors first distinguish between the normal and abnormal area of medical images after observing.Then, they analyze the lesion characteristics of the abnormal area intensively and make a clinical diagnosis eventually.So, the lesion area is an important basis for doctors to make a diagnosis.The proposed scheme adopts distance regularized level set evolution (DRLSE) [Li, Xu, Gui et al.(2010)] which is suitable for various types of medical images, such as computed tomography (CT), magnetic resonance imaging (MRI), ultrasound imaging (USI) and so on, to extract lesion.Due to space limitations, we briefly describe the process of lesion extraction as follows:
(1) Lesion initialization: The approximate position of lesion is marked in red rectangle manually as shown in Figs.2(b), 2(e) and 2(h).
(2) Lesion extraction: The accurate lesion area is adaptively circled by the method of distance regularized level set evolution as shown in Figs.2(c), 2(f) and 2(i).The area drawn by the red line is the lesion area which it is denoted as L.
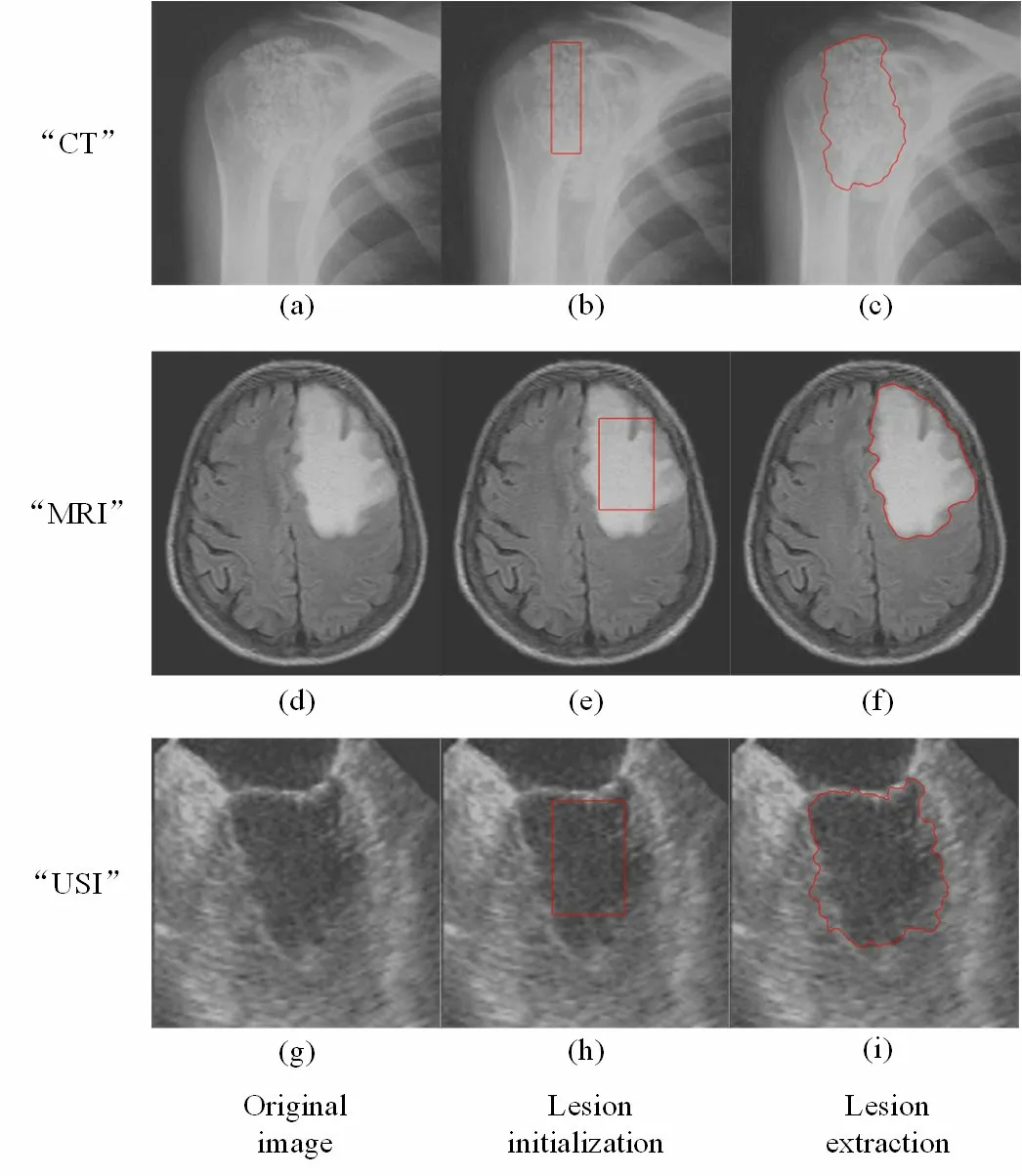
Figure 2: The process of lesion extraction
Let I be a medical image anI (x, y) be a pixel located at the coordinates (x, y) on I .
A binary image Ib(x, y) is generated by

where L is the region of lesion area.“1” inbI denotes the lesion area.“0” inbI denotes the non-lesion area.We can extract lesion from I in accordance withbI .
2.2 Data embedding
To protect patients' privacy, privacy data is embedded into the corresponding medical image.Data first is embedded into the lesion area to enhance its contrast.The rest of data is embedded into the non-lesion area to achieve the higher payload.To achieve reversibility, the auxiliary information needs to be embedded into the image.This section details the process of data embedding.
2.2.1 Embedding in the lesion area
Medical images’ lesion area is the important basis for doctors to make a diagnosis.We aim at enhancing the contrast of lesion area for improving the quality of medical images and achieving RDH meanwhile.To enhance contrast, the lesion area is stretched firstly.As a result, there are empty bins in the gray histogram.Data is sequentially embedded into the empty bins of the stretched histogram.And the contrast of lesion area is enhanced further, which is similar to the effect of histogram equalization.The embedding steps in the lesion area are detailed as follows:
(1) The value of pixel in the lesion area is stretched firstly.The original valueoI will be
stretched to Il, when the value is stretched from [Imin,Imax] into [Lmin,Lmax] as follow:

wherelI is the pixel in the lesion area after stretched.In general, Lmin=0 and Lmax=255.
(2) To avoid overflow and underflow problem, data is embedded by shifting histogram from left to right if the pixel Ilis in [0,126], or data is embedded by shifting histogram from right to left if the pixel lI is in [129,255].Calculate the gray histogram of lesion area.The modified pixel I′lis modified by
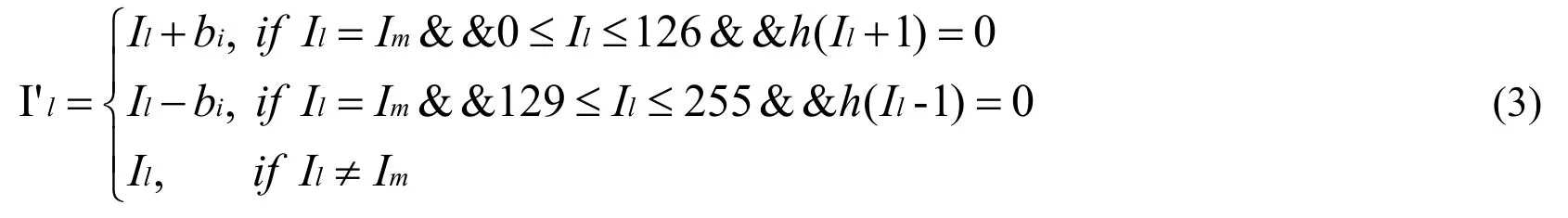
In which Ilis the unmodified pixel in lesion area.Imis the pixel value of peak bin in gray histogram.bi∈{0 ,1} is the secret data to be embedded.h( Il) is the frequency of pixel Ilin the gray histogram.
(3) Repeat Step (2) until there is no empty bin to be embedded or all data is embedded into the lesion area.The pixel value of peak bin in each round is embedded as the part of privacy data into the next round.
2.2.2 Embedding in the non-lesion area
To achieve the higher embedding rate, the rest data is embedded into the non-lesion area when there is no empty bin to embed data in the lesion area.We adopt a high-capacity RDH method [Li, Yang and Zeng (2011)] that can achieve the high embedding rate.The embedding steps in the non-lesion area are detailed as follows:
(1) Select one of four prediction modes in Weng et al.[Weng, Pan and Zhou (2017)] and calculate the prediction errors eji, by

In which the prediction value pji, is predicted by its neighbors simply using the interpolation technique in Luo et al.[Luo, Chen, Chen et al.(2010)].
(2) Calculate the value of smoothness σ which measures whether the pixel Iji,is smooth or complex by standard deviation of jiI ,and its eight surrounding neighbors.
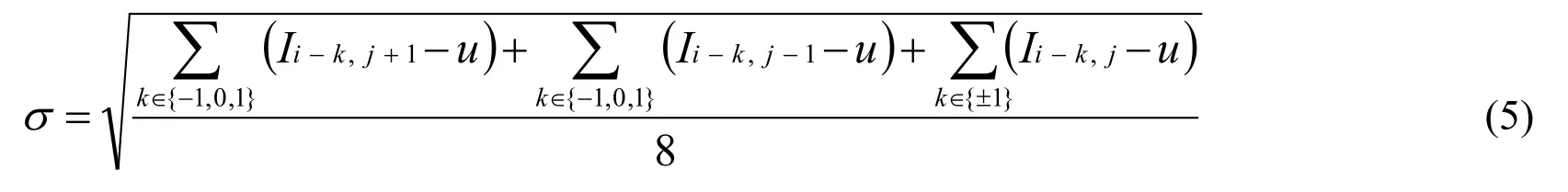
In which u is the mean of Iji, and its eight surrounding neighbors.
(3) The expanded prediction erroris calculate by Eq.(6) if Tv<σ.

In which bi∈{0,1,2,3}, Tvand Tpare thresholds detailed in Li et al.[Li, Yang and Zeng (2011)].
e′i,jis calculate by Eq.(7) if σ≥Tv.

In which bi∈{0 ,1}.

Steps (1)-(4) are the process of a layer embedding.If a layer embedding does not meet the required embedding rate, repeat Steps (1)-(4) until all data is embedded into the image.
2.2.3 Embedding of auxiliary information
In order to extract data completely and recover image losslessly, the auxiliary information, such as the edge of lesion area, the peak bin in the last round Imin the lesion area, the threshold Tvand Tp, the number of embedding layer, the end of symbol and the compressed location map in the non-lesion area, needs to be embedded into image.The four sides of medical image do not contain critical information, so they are used to embed all auxiliary information.As shown in Fig.3, the size of the image is X ∗ Y , h rows and h columns of the image’s sides without first M pixels is the place to embed auxiliary information by replacing least significant bit (LSB).It is worth noting that h rows and h columns of the image’s sides are segmented first and they do not participate in the embedding process of lesion or non-lesion area.h(2 bits), the number of LSB plane (2 bits) and the end of symbol(log2X +log2Ybits) are put in the first M (M=2+2+log2X +log2Y) pixels’ LSB.Original LSBs in h rows and h columns of image’s four sides are embedded into the non-lesion area to vacate room which is used to embed the auxiliary after all data is embedded into image.

Figure 3: The four sides of image
2.3 Data extraction and image recovery
The marked images are images with contrast enhancement and contain data which is not perceived.Only the third party with right, such as doctors and patient himself, can extract data and recover image.If doctors want to learn about information related patients or patient himself wants to know diagnostic records and results, data can be extracted from the marked image and the medical image can be recovered without distortion.This section details the process of data extraction and image recovery.
(1) Extract first M pixels’ LSB in four sides of image to obtain h, the number of LSB plane and the end of symbol.
(2) Extract all auxiliary information from four sides of image except for the first M pixels, such as the edge of lesion area, the peak bin in the last round mI in the lesion area, the threshold Tvand Tp, the number of embedding layer, the end of symbol and the compressed location map in the non-lesion area.And decompress the location map of non-lesion area.
(3) Extract the lesion area and the rest of image is the non-lesion area according to the edge of lesion area.
(4) In the non-lesion area, if Tv<σ, data biand the prediction error eji,are calculated by Eq.(9) and Eq.(10).

Where bi∈{0,1,2,3}.
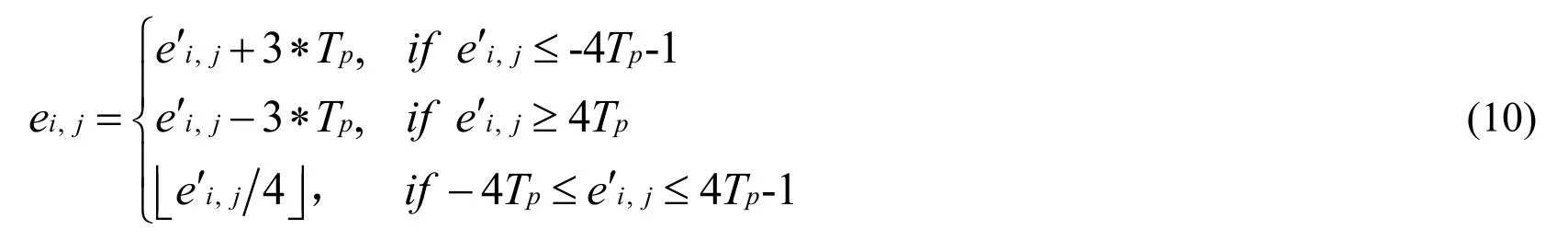
If σ≥Tv, data biand the prediction error ei,jare calculated by Eq.(11) and Eq.(12).

where bi∈{0 ,1}.
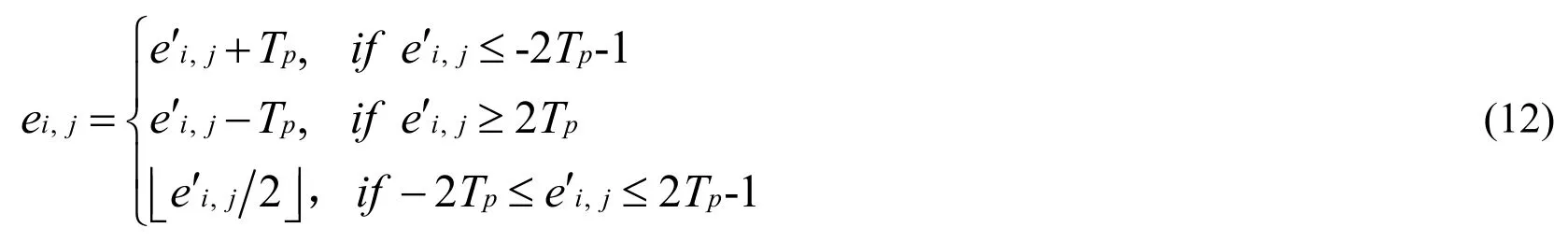
The original pixel Ii,jin the non-lesion area is recovered by

Repeat the process of extraction and recovery in the non-lesion area until all data is extracted.
(5) Obtain LSBs of image in four sides from data which just has been extracted from the non-lesion area to recover four sides of image.
(6) In the lesion area, data extraction and image recovery are as follows:

(7) The pixel Ilin the lesion area is recovered to Iowhich is the pixel before stretched by

3 Experimental results
We do a lot of experiments on medical images.However, due to the limitation of the space, we only randomly choose three medical images (in Fig.4) from three different kinds which are often made to examine body in hospital to show the experiment results: CT mainly examines bone, joint, organ in thorax etc through X-ray, MRI mainly examines brain, soft tissue of the whole body etc through electromagnetic wave, USI mainly examines abdomen, blood vessel etc through ultrasound.To illustrate the characteristic of the proposed scheme, we do two series experiments: discuss the experiment results in the lesion area, and compare the performance of contrast enhancement in the lesion area by the proposed method with others which are also RDH methods with contrast enhancement at different embedding rate.In this section, the experimental results and analysis are introduced in detail.

Figure 4: Three original medical images
3.1 Experiment results in the lesion area
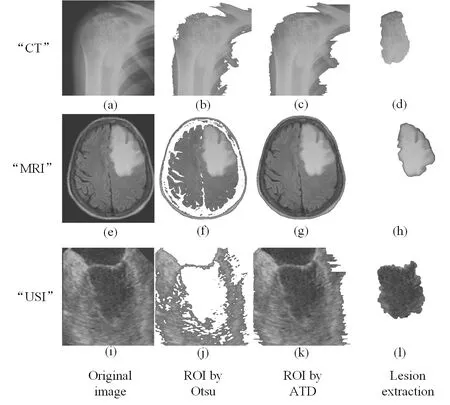
Figure 5: The comparison of lesion extraction with ROI methods
We discuss the experiment results of lesion area in this section.We first compare lesion extraction in the proposed scheme with ROI methods which are the state-of-the-art segmentation methods.ROI methods based on threshold segmentation divide images into foreground and background.Foreground is regarded as ROI which is interesting for doctors and background is regarded as NROI which do not contain the key information.As shown in Figs.5(b), 5(f) and 5(j) are ROI by Otsu [Gao, Wan, Yao et al.(2017)].In particular, Fig.5(j) ignores the important place where is the lesion area by Otsu.Figs.5(c), 5(g) and 5(k) are ROI by ATD [Yang, Zhang, Liang et al.(2018)].Fig.5(d), 5(h) and 5(l) are the lesion area by the way of lesion extraction.We can conclude that ROI is just the place where is sweeping, and it is not intuitive to observe.The lesion area is the doctor's focus and used as the basis for clinical diagnosis.Lesion extraction is performed on the lesion area, and the effect of segmentation is clear and intuitive to observe.So, the proposed scheme based lesion extraction has an advantage over other segmentation methods.From the subjective visual and objective data, we discuss the performance of contrast enhancement and the maximum embedding bits in the lesion area.As shown in Figs.6, 7 and 8, the contrast of lesion area in three different types of medical images is enhanced obviously in 0.1 bpp, 0.6 bpp, 0.8 bpp, and 1 bpp respectively.Since data is embedded into empty bins in the lesion area, there is the maximum embedding bits.As shown in Tab.1, there are 106859 bits, 131466 bits, 309087 bits in the lesion area of three medical images respectively, the corresponding value of no-reference contrast distortion image quality assessment (NR-CDIQA) is 2.6808, 2.5863, and 2.8334 respectively, and the corresponding value of no-reference improved contrast distortion image quality assessment (NR-ICDIQA) is 2.8265, 2.9753, and 2.8602 respectively.Here, NR-CDIQA [Fang, Ma, Wang et al.(2014)] and NR-ICDIQA [Wu, Zhu, Yang et al.(2018)] are noreference image quality assessment (IQA) method based on the principle of natural scene statistics (NSS) only for contrast enhancement.NR-CDIQA and NR-ICDIQA methods can effectively assess the quality of contrast-enhancement images.The higher the scores of NR-CDIQA and NR-ICDIQA are, the better the quality of images is.
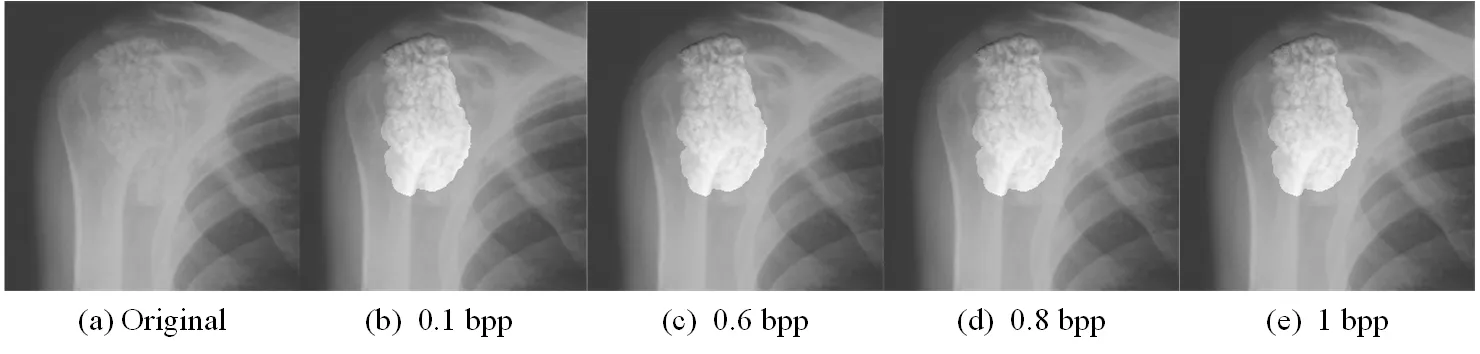
Figure 6: The original image and the marked images of “CT” in 0.1 bpp, 0.6 bpp, 0.8 bpp, 1 bpp
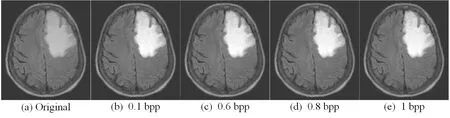
Figure 7: The original image and the marked images of “MRI” in 0.1 bpp, 0.6 bpp, 0.8 bpp, 1 bpp

Figure 8: The original image and the marked images of “USI” in 0.1 bpp, 0.6 bpp, 0.8 bpp, 1 bpp
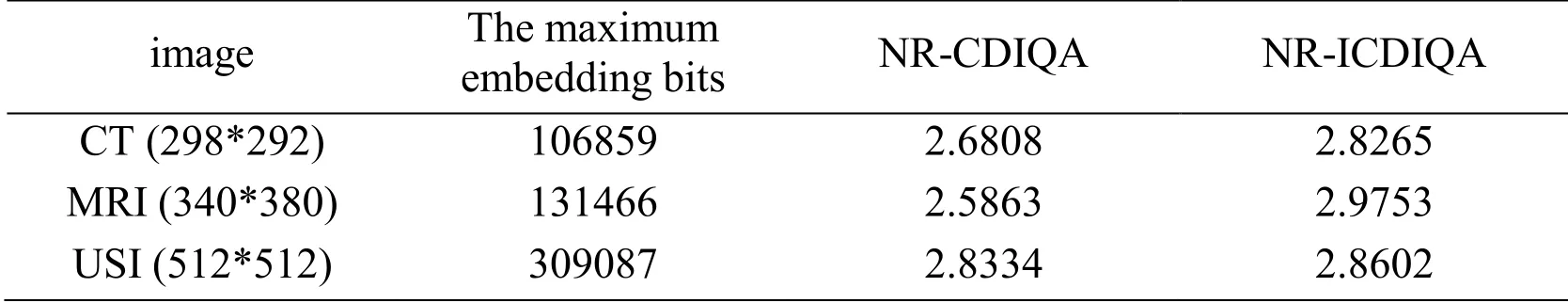
Table 1: The maximum embedding bits and the value of NR-CDIQA and NR-ICDIQA in the lesion area
3.2 Comparison of the proposed method with other RDH methods with contrast enhancement
We discuss the performance of contrast enhancement in the lesion area by the proposed method when compared with Yang et al.[Yang, Zhang, Liang et al.(2016)] and Wu et al.’s [Wu, Tang, Huang et al.(2018)] method which are also contrast-enhancement RDH methods at different embedding rates, as shown in Tabs.2, 3 and 4.We also calculate peak signal to noise ratio (PSNR) and structural similarity index measurement (SSIM).It is worth noting that PSNR and SSIM are traditional image quality assessment.PSNR is based on the error between the marked image and the original image.It is an image quality assessment for error sensitive image.But it does take into account the human visual characteristics.SSIM is a fullreference image quality assessment which reflects the structural characteristics of the image, but it ignores the underlying visual characteristics of the human visual system.Therefore, PSNR and SSIM do not evaluate the image with contrast enhancement well.We use NRCDIQA and NR-ICDIQA methods to assess the quality of marked images.The value of NRCDIQA and NR-ICDIQA by the proposed method are higher than Yang and Wu’s method as shown in Tabs.2, 3 and 4.It shows the contrast of image quality in the lesion area is enhanced obviously by the proposed method and is better than other contrast-enhancement RDH methods at different embedding rates.Next, we analyze the reasons why the proposed method is better than Yang and Wu’s method.Yang’s method prioritizes to embed data into the texture region by prediction errors histogram.Lesion is not necessarily the texture region.Yang’s method does not effectively enhance the contrast of lesion area.Wu’s method embeds data into images by histogram shifting.It first chooses the highest two bins, and keeps the bins between the two peaks unchanged and shifts the other bins outwards.Its contrast is obviously enhanced at the low embedding rate.The proposed method adopts DRLSE to extract lesion.It is goal-oriented for medical images.In the lesion area, the pixels are stretched to enhance contrast.Data is firstly embedded into empty bins of stretched histogram to enhance contrast further.The contrast of lesion area is obviously enhanced at the low embedding rate.In addition, it can avoid the overflow and underflow problem.In the non-lesion area, the rest of data is embedded to achieve the higher payload.It can achieve good image quality at high embedding rate.Through a lot of experiments, the quality of marked images in the lesion area is obviously better than other RDH methods with contrast enhancement.
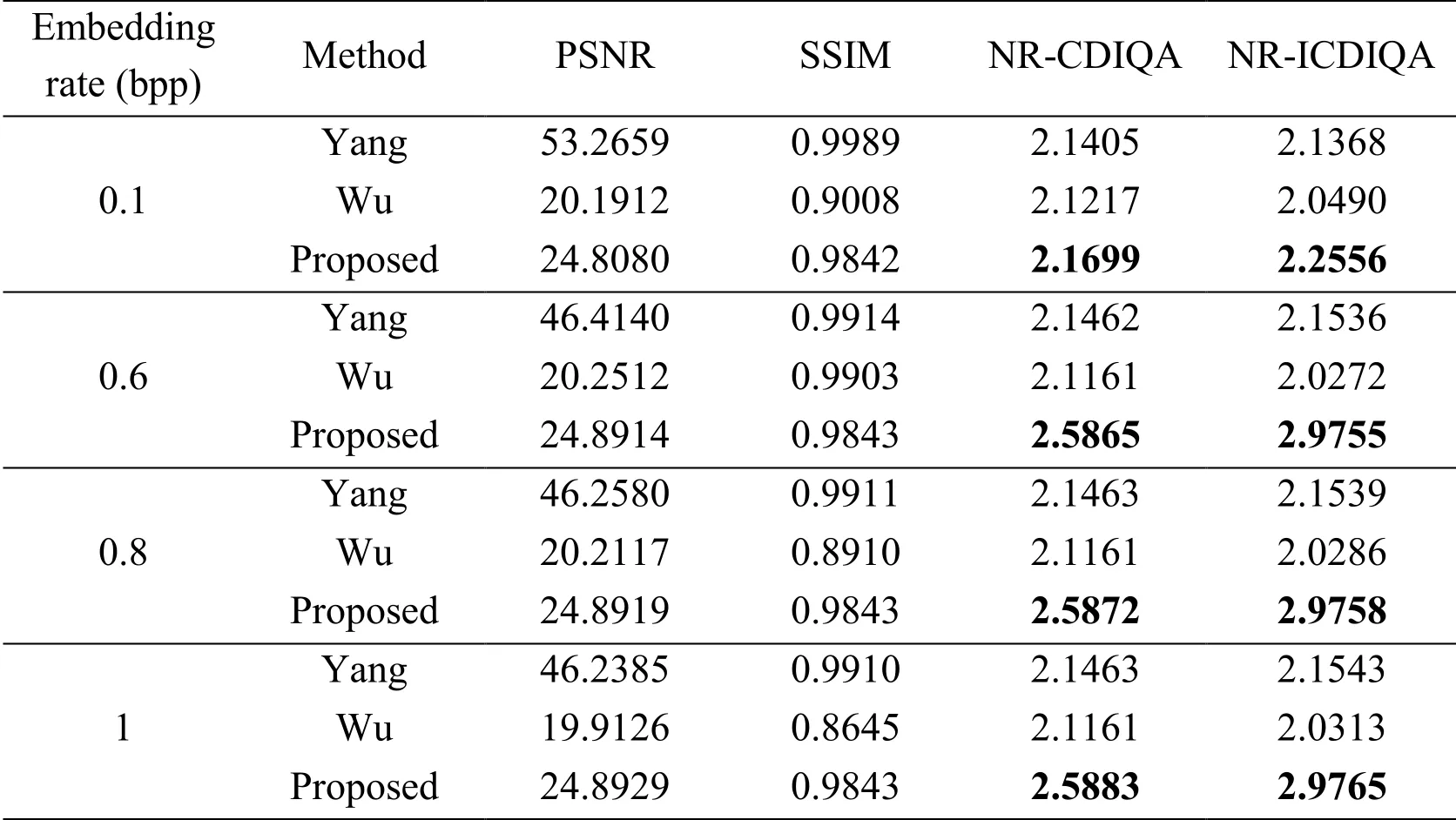
Table 3: Comparison of the proposed method with Yang and Wu’s method in “MRI”
4 Conclusion
Aiming at protecting patients' privacy and improving quality of medical images, this paper proposes a novel reversible data hiding scheme based on lesion extraction and with contrast enhancement.The proposed scheme first adopts DRLSE to extract lesion.In the lesion area, data is embedded preferentially into empty bins of stretched histogram to improve image quality.In addition, it can avoid the overflow and underflow problem.In the non-lesion area, the rest of data is embedded to achieve the higher payload.What is more, data can be extracted completely and image can be recovered losslessly by the third party with right.Through a lot of experiments, the proposed scheme based on lesion extraction has an advantage over other segmentation methods.Experimental results have shown that the quality of marked image is improved obviously and the performance of contrast enhancement in the lesion area is better than other contrast-enhancement RDH methods.In conclusion, the proposed scheme can protect patients' privacy and improve the quality of medical images effectively.In the future, the method of adaptive lesion extraction, such as machine learning method, and reversible data hiding for improving the quality of medical images will be researched further.
Acknowledgement:This work was supported in part by the Natural Science Foundation of China under Grants 61502007 and 61572452, the Natural Science Research Project of Anhui province under Grant 1608085MF125, the NO.58 China Post-doctoral Science Foundation under Grant 2015M582015, the Backbone Teacher Training Program of Anhui University and the Doctoral Scientific Research Foundation of Anhui University under Grant J01001319.
References
Du, Y.; Yin, Z.X.; Zhang, X.P.(2018): Improved lossless data hiding for JPEG images based on histogram modification.Computers, Materials & Continua, vol.55, no.3, pp.495-507.
Fang, Y.M.; Ma, K.D.; Wang, Z.; Lin, W.S.; Fang, Z.J.et al.(2014): No-reference quality assessment of contrast-distorted images based on natural scene statistics.IEEE Signal Processing Letters, vol.22, no.7, pp.838-842.
Gao, G.Y.; Shi, Y.Q.(2015): Reversible data hiding using controlled contrast enhancement and integer wavelet transform.IEEE Signal Processing Letters, vol.22, no.11, pp.2078-2082.
Gao, G.Y.; Wan, X.D.; Yao, S.M.; Cui, Z.M.; Zhou C.X.et al.(2017): Reversible data hiding with contrast enhancement and tamper localization for medical images.Information Sciences, vol.385, pp.250-265.
Li, C.M.; Xu, C.Y.; Gui, C.F.; Fox, M.D.(2010): Distance regularized level set evolution and its application to image segmentation.IEEE Transactions on Image Processing, vol.19, no.12, pp.3243-3254.
Li, X.L.; Yang, B.; Zeng, T.L.(2011): Efficient reversible watermarking based on adaptive prediction-error expansion and pixel selection.IEEE Transactions on Image Processing, vol.20, no.12, pp.3524-3533.
Luo, L.X.; Chen, Z.Y.; Chen, M.; Zeng, X.; Xiong, Z.(2010): Reversible image watermarking using interpolation technique.IEEE Transactions on Information Forensics and Security, vol.5, no.1, pp.187-193.
Ni, Z.C.; Shi, Y.Q.; Ansari, N.; Su, W.(2006): Reversible data hiding.IEEE Transactions on Circuits and Systems for Video Technology, vol.16, no.3, pp.354-362.
Sachnev, V.; Kim, H.J.; Nam, J.; Suresh, S.; Shi, Y.Q.(2009): Reversible watermarking algorithm using sorting and prediction.IEEE Transactions on Circuits and Systems for Video Technology, vol.19, no.7, pp.989-999.
Tian, J.(2003): Reversible data embedding using a difference expansion.IEEE Transactions on Circuits and Systems for Video Technology, vol.13, no.8, pp.890-896.
Weng, S.W.; Pan, J.S.; Zhou, L.Z.(2017): Reversible data hiding based on the local smoothness estimator and optional embedding strategy in four prediction modes.Multimedia Tools and Applications, vol.76, no.11, pp.1-23.
Wu, H.T.; Dugelay, J.L.; Shi, Y.Q.(2015): Reversible image data hiding with contrast enhancement.IEEE Signal Processing Letters, vol.22, no.1, pp.81-85.
Wu, H.T.; Tang, S.H.; Huang, J.W.; Shi, Y.Q.(2018): A novel reversible data hiding method with image contrast enhancement.Signal Processing: Image Communication, vol.62, pp.64-73.
Wu, Y.J.; Zhu, Y.H.; Yang, Y.; Zhang, W.M.; Yu, N.H.(2018): A no-reference quality assessment for contrast-distorted image based on improved learning method.Multimedia Tools and Applications, pp.1-20.
Yang, Y.; Zhang, W.M.; Liang, D.; Yu, N.H.(2016): Reversible data hiding in medical images with enhanced contrast in texture area.Digital Signal Processing, vol.52, pp.13-24.
Yang, Y.; Zhang, W.M.; Liang, D.; Yu, N.H.(2018): A ROI-based high capacity reversible data hiding scheme with contrast enhancement for medical images.Multimedia Tools and Applications, vol.77, no.14, pp.18043-18065.
杂志排行
Computers Materials&Continua的其它文章
- A DPN (Delegated Proof of Node) Mechanism for Secure Data Transmission in IoT Services
- A Hybrid Model for Anomalies Detection in AMI System Combining K-means Clustering and Deep Neural Network
- Topological Characterization of Book Graph and Stacked Book Graph
- Efficient Analysis of Vertical Projection Histogram to Segment Arabic Handwritten Characters
- An Auto-Calibration Approach to Robust and Secure Usage of Accelerometers for Human Motion Analysis in FES Therapies
- Balanced Deep Supervised Hashing
