Isolation and identification of novel casein-derived bioactive peptides and potential functions in fermented casein with Lactobacillus helveticus
2019-05-26MengzhuFnTingtingGuoWnruLiJingChenFushuoLiChoWngYiShiDviXiLiShohuiZhng
Mengzhu Fn,Tingting Guo,Wnru Li,Jing Chen,Fushuo Li,Cho Wng,Yi Shi,Dvi Xi-n Li,Shohui Zhng,b,∗
a School of Agriculture and Biology,Shanghai Jiao Tong University,China
b Zhejiang Go Peptides Life Science and Healthcare Technology Co.,Ltd.,China
c School of Systemic Biology and Medicine,Shanghai Jiao Tong University,China
d Zhejiang Panda Dairy Group Co.Ltd.,China
Keywords:
ABSTRACT
1. Introduction
Protein is the material basis of life, and all life events need to have protein participation. Bioactive peptides have been defined as“specific protein fragments that have a positive impact on body functions or conditions and may ultimately influence health” [1].A large number of bioactive peptides from animals or plants have been identified, which belong to current diet. For example, there were many bioactive peptides derived from food protein sources having been reported, such as from fermented milk [2], cheddar cheeses[3],hen egg white[4],tuna[5],soybean[6],rice[7],peanut[8], flaxseed [9], and pumpkin [10]. But milk proteins are still the major source of bioactive peptides and these peptides exhibit a wide range of biological activities [11]. Most of these bioactive peptides are derived from the degradation of casein. A number of studies showed that casein act as an excellent substrate to produce a variety of bioactive peptides with angiotensin I converting enzyme(ACE)-inhibitory,immunological,antioxidant,antithrombotic or opioid effects[12,13].In a recent study,medium enriched with casein as a sole nitrogen source (comprised of 3% casein, 5%lactose and 1.17%yeast carbon base)was fermented for 42 h with each of two lactic acid bacteria isolated from the commercial fermented milk Soful (Yakult®, Mexico) to produce ACE-inhibitory peptides [14]. The use of casein or lactoferrin as a sole nitrogen source to dentify the ACE-inhibitory peptides released by yeasts has been recently described[15].Compared to synthesed peptides,bioactive peptides produced by microbial fermentation are considered safer and healthier, without side effects, as they are derived from edible food proteins.
The approach to the production of these bioactive peptides involves the choice of microbial species.At present,the main strains are from Lactobacilli,Lactococcus,Leuconostoc and acetic acid bacteria, as well as different types of yeasts, and even kefir [16–18].Many studies have focused on the potential of LAB to release bioactive peptides with milk proteins[19].Compared to other lactic acid bacteria, Lactobacillus helveticus has a strong proteolytic activity with the most efficient proteolytic system, so it has been widely used in the food industry, especially in the dairy industry [20,21].The bioactive peptides VPP and IPP from β-casein were isolated and identified from the fermented milk with Lactobacillus helveticus CP790,and the significant ACE-inhibitory effect in vitro was tested[22,23]. To achieve maximum production of bioactive peptides, it is necessary to find the optimal microbial species and fermentation conditions.
Many studies using lactic acid bacteria also have demonstrated their ability to produce different bioactive peptides during the fermentation process by their surface specific proteolytic systems,especially cell envelope proteinases(CEPs)[24].The product is far more complex than imagined after different microbial hydrolysis.236 peptides were uniquely detected in milk fermented with kefir,but not in raw milk,which indicated that the fermentation increase the proteolytic activity and change the complexity of composition of the peptide fraction[25].H.E.Hatmi et al.[26]revealed the presence of 347 peptides in camel milk fermented with the proteolytic strain LMD-9 of S. Thermophilus and investigated their free radical scavenging power. Most of the peptides in their studies were derived from the four individual CNs, especially from β-CN, the major component.However,less is known about the composition and biological activity of total peptides, the relationship of extracellular and intracellular peptides and the connection between casein-derived peptides and bacterial protein-derived peptides.
In our previous work, we studied casein fermented by Lactobacillus helveticus CICC6024 at optimal process conditions,followed by the isolation and identification of novel casein-derived bioactive peptides. In addition, the comprehensive analysis of the total casein-derived peptide profile in both extracell and intracell is carried out, including composition of the peptide fractions revealing information about the mechanisms of peptide formation.Finally, the potential bioactivities of novel casein-derived bioactive peptides and quantification of the target peptide sequence are discussed and implemented.
2. Materials and methods
2.1. Materials
Casein, lactose and Man, Rogosa and Sharpe (MRS) medium were purchased from Sigma-Aldrich (St. Louis, MO, USA). Amicon Ultra-15 regenerated cellulose filters with a molecular weight(MW)cut-off of 3 kDa were obtained from Millipore(Carrigtwohill,Ireland). Purified target peptide DELQDKIHPF (above 95% purity)were synthesized by Shanghai Top-peptide Co., Ltd (Shanghai,China). All other chemicals were of the highest grade available commercially.
2.2. Microorganism and culture conditions
The Lactobacillus helveticus CICC6024 was placed in a 37◦incubator for 5 min and activated at 37◦in MRS medium for 2–3 times.The medium of casein concentration 8%,lactose concentration 6%and NaCl 0.5%were prepared respectively,65◦water bath with 1 mol/L NaOH solution to adjust the pH to 6.5 to make casein dissolved,115◦high temperature sterilization for 15 min after 1 h water bath. Pre-cultures were prepared by inoculating the activated Lactobacillus helveticus CICC6024 with 6% of inoculum level in sterile casein mediun and incubated at 37◦for 7 h to obtain a fermentation broth.
2.3. Preparation of peptide fractions from fermentation broth and Lactobacillus helveticus cells
After centrifugation at 4000 rpm/min for 15 min and 20◦,the supernatant that is extracellular peptides extract was collected. At the same time, cells from fermented casein sample were collected and washed three times with 50 mmol/L Tris−HCl(pH7.1) by centrifugation at 400045005000rpm/min (20◦,15 min) respectively. The washed cells were suspended in cell lysis buffer (Beyotime®Biotechnology, China) and 100 mM PMSF(Beyotime®Biotechnology, China), 0◦incubating for 30 min and further rushed by ultrasonic treatment (200 W) at 3 s intervals for 10 min in an ice bath. The crude homogenate was centrifuged at 9000 rpm/min for 20 min at 4◦. The supernatant was used as a intracellular peptides extract. The supernatants of extracellular and intracellular extracts obtained were subjected to ultrafiltration centrifugation using centrifugal filter units with a 3 kDa cutoff(Carrigtwohill,Ireland)at 4800 r/min for 30 min and 4◦for the removal of macromolecule casein.
Before solid-phase extraction(SPE),the ultrafiltrate was diluted 10 times with ddH2O.Solid phase extraction conditions were modificated slightly according to C.ClarianaS[27]by means of C18-SPE(100 mg C18-E, 1 mL,Waters, USA). SPE columns were first conditioned with 2 mL of methanol and 2 mL of water. After loading 2 mL of ultrafiltrate samples,the retained compounds were eluted with 400 μL of eluent (80:20 v/v methanol/water and 0.1% v/v formic acid). During all the above steps, control the flow rate of approximately 1 mL/min. The eluent was evaporated with nitrogen blowing instrument at room temperature to about 1–2 mL for concentration, and then it was loaded into EP tubes and vacuum frozen for 12 h and dried to a powder and stored at-80◦for spare.
2.4. Gastrointestinal digestion in vitro
The samples were lyophilized into powders followed by gastrointestinal digestion simulation whose main steps referred to R.JÁG et al.[28]and were modified according to M.M.Contreras et al.[29].Specific Methods:1.5 mg of sample powders was dissolved in 1.5 mL of ddH2O. Pepsin solution was added into sample solution(enzyme: substrate=1:50, w/w), and the pH was adjusted to 2.0,water bathing at 37◦for 90 min.Subsequently,the pH was adjusted to 7.5 and trypsin solution was added (enzyme: substrate=1: 25,w/w), water bathing at 37◦for 150 min. Finally, the enzyme was inactivate in 95◦hot water bathing for 5 min to make enzyme reaction terminated. The solution was freezing-dried into powders,stored at-80◦for subsequent analysis.
When it was past midnight, and the robbers saw from afar that the light was no longer burning in their house, and all appeared quiet, the captain said, We ought not to have let ourselves be frightened out of our wits; and ordered one of them to go and examine the house.18
2.5. Total peptides quantification
The total peptides concentration was measured by Lowry [30]assay with peptone [31] as a standard (Lowry Protein Assay Kit,Shanghai Sangon Biotech).
2.6. Identification of peptides by Nano LC–MS/MS
The peptide samples was analyzed using an LC system (Nano Pump, Ultimate 3000, Dionex, Thermofisher) coupled with an ESI-Q-TOFmass spectrometer (maXis, Impact, Bruker Daltonik,Germany).To desalt and concentrate samples,each peptide sample was re-dissolved in UP H2O with 0.1% formic acid and shaken for 5–10 min, followed by high-speed centrifugation at 12,000 r/min for 10 min. The Millipore C18 ZipTip was equilibrated with 0.1%FA and ACN, followed by loading peptide samples. Then the samples were washed with 0.1% FA and eluted with 20% ACN, 0.1%FA. The eluted sample was dried and further dissolved with 2%ACN, 0.1% FA and analyzed by NanoLC-MS with C18 capillary column (75 μm×15 cm, 3 μm, Dionex, Thermofisher). The elution system consisted of A: 0.1% (v/v) formic acid (TFA) in water and B: 0.1% (v/v) formic acid in acetonitrile. 1 μL sample was injected and eluted at a flow rate of 0.25 μL/min with a linear gradient as follows:0–10 min, 2% B; 10–12 min, 4%–8% B; 12–110min,8%–20% B; 110–125 min, 20%–30% B; 125–125.1min, 30%–80% B;125.1–135 min, 80% B; 135–135.1min, 80%–2% B; 135.1–150 min,2%B.The LC setup was coupled online to a Q-TOF using a nano-ESI source (Bruker Daltonik, Germany) in data dependent acquisition(DDA)mode(m/z 100–1500).The Source Capillary was set at 2000 v, the flow and temperature of dry gas was 2.0 L/min and 150◦C respectively.The mass spectrometerwas set as one fullMSscan followed by tenMS/MS scans on the tenmost intense ions from the MS spectrum. Peptide charges of +1, +2, and +3 were selected in each scan.The results from the product ion scans were subjected to a MASCOT(version 2.4,Matrix Science)search against database for peptide identification.
2.7. Database search and peptide profile analysis
Mascot (version 2.4, Matrix Science) was set up to search casein database and Lactobacillus helveticus CICC6024 protein database which was established according to genome sequence.The database of casein amino acid sequence was established by searching BIOPEP[32]and Structure-function properties of food proteins[33],including αs1-casein(ID:1086–1089,variant A–D),αs2-casein (ID:1090, variants A1), β- casein (ID:1097-1103, variant A1-A3, B, C, E, F), κ- casein (ID:1117, variant A). The amino acid sequences obtained were sorted out and compiled into the first database, generating a fasta file. The amino acid sequence of Lactobacillus helveticus CICC6024 protein was obtained by RNA gene sequencing and genome sequencing, then translating into amino acid sequence of possibly expressed proteins.The second database was constructed with generating the second fasta file.
Tandem mass spectra were extracted, charge state deconvoluted and deisotoped by Compass Data Analysis version 4.1(Bruker Daltonics),generating mgf files.The Mascot 2.4 search engine was set up to search for the above two protein amino acid databases.Search parameters were: Carbamidomethyl (C) as fixed modifications, Oxidation (M) as variable modifications, no enzyme, two missed cleavages, 20 ppm mass tolerance for precursor ions and 0.5 Da for product ions, charge minimum +1. For reliable peptide identification, the following approach was applied: Significance threshold was set as 0.05 and all peptides with a score >15 were preselected.At least one unique peptide can be detected from one protein.
2.8. Selection and absolute quantification of target peptide
2.8.1. Target peptide
According to the analysis results above, select the common peptide DELQDKIHPF f(43–52) from β-casein var B among the extracellular and intracellular peptides,whose dupes was relativelyhigh in mass spectrometry results that means its content of the total peptides was same relatively high.Therefore,the synthesized peptide DELQDKIHPF was synthesized as a standard peptide to quantify the same peptide in the sample by the synthesizer Shanghai Toppeptide Co.,Ltd(Shanghai,China).

Table 1 MRM transitions for DELQDKIHPF.
The separation of peptides was carried out using the ACQUITY UPLC System equipped with ACQUITY UPLC Binary Solvent Manager,ACQUITY UPLC Sample Manager,and ACQUITY UPLC Column Manager (Waters, Milford, MA, USA). The Column was ACQUITY UPLC BEH300 C18 column (1.7 μm particle size, 2.1×100 mm)reversed phase analytical column(Waters,Milford,MA,USA),and equipped with a guard column of the same material.Mobile phase A was 100% H2O and mobile phase B was 100% acetonitrile. The injection volume was 5 μL.The flow rate of the mobile phase was 0.4 mL/min.The column temperature was 35◦C.The LC elution program was linear gradient as follows:0–0.5 min,18%B;0.5–3.5 min,18% B; 3.5–4 min, 85%B; 4 −6 min, 100% B; 6–6.5 min, 100% B;6.5–8 min, 18% B. The total injection cycling time was 8 min. Ion Source(Turbo Spray),Curtain Gas(35 Psi),Collision Gas(8 Psi),Ion-Spay Voltage(5500 V),Desolvation Temperature(500◦),Ion Source Gas1 (50 Psi) and Ion Source Gas2 (50 Psi) were optimized for MRM intensity derived from DELQDKIHPF. Precursor/product ion and their corresponding fragment for target peptide was created in the MRM transition list(Table 1).
Six-point calibration curve of the pure peptide DELQDKIHPF was constructed as follows. The synthetic peptide DELQDKIHPF were solubilized in ultrapure water, and then diluted with UP H2O, to obtain the 2, 5, 10, 15, 20 and 25 ng/mL solutions. The calibration curve was constructed from the peak area of the standard peptide versus concentration. The concentrations of DELQDKIHPF in two samples were calculated using a linear equation.
3. Results and Discussion
3.1. Peptide analysis by Nano LC–MS/MS
A detailed peptide analysis was obtained by direct Nano LC–MS/MS measurement in the m/z range from 100 to 1500 Da.Fig.1 shows specific Nano LC–MS/MS spectra for extracellular peptides and intracellular peptides.From the comparison with Fig.1A and B, we can see the peaks of Fig.1 B are larger in amount than Fig.1A. It means that intracellular peptides are much more complicated than extracellular peptides. Intracell is superior to extracell on the number of peptides producted by Lactobacillus helveticus. This result is strongly related to the type and amount of intracellular peptidases. The proteolytic system of lactic acid bacteria consists of three parts: cell wall proteases, different types of amino acid and oligopeptide transport systems, as well as a pool of intracellular peptidases (endopeptidase, aminopeptidase,proline-specific enzyme, tripeptidase and dipeptidases). To make use of casein in milk,lactic acid bacteria firstly hydrolyze casein into oligopeptides by cell wall proteases and then transport it into cells through a specific oligopeptide transport system,further degrading oligopeptides into smaller peptides and amino acids by intracellular peptidases to provide for the growth and utilization of bacteria[34,35].At the same time,intracellular peptides are not only from casein hydrolysis, but also from the Lactobacillus helveticus itself because of existing bacteria protein degradation.The peptide profiles recorded by Nano LC give a good overview of predominant peptides.
To identificate specific peptides,a MASCOT database search was applied for interpretation of the fragment spectra. All resulting assigned peptide sequences of casein and bacterial proteins were manually checked the p value less than 0.05 to achieve unambiguous identification. As a result, totally 241 different peptide sequences were confirmed from extracell and intracell as Table 2.Including peptide sequence, precursor protein, position, and origin. The sequences of the identified peptides ranged from five to twenty-six amino acids.
Our work eventually obtained all bioactive peptides by fermenting casein using Lactobacillus helveticus,which resulted in a specific¨peptide database¨that contained 241 peptide sequences derived from variants of casein and Lactobacillus helveticus proteins to be established, the casein-derived peptides including those derived from extracell and intracell of Lactobacillus helveticus.We published the ¨peptide database¨like a large ¨vault¨,not only including a variety of peptide fragments with biological activities,but containing proteins metabolism mechanism and potential research value during the evolution of human life. Bioactive peptides beneficial to the high-grade animals found from the lower organisms at bacterial level are whether to be reserved by ¨natural selection,survival of the fittest¨in the process of biological evolution from low-grade bacteria to high-grade animals,which play an important functional role in its life-sustaining physiological activities. Whether the original genetic information of some proteins with special functions is preserved in the process of biological evolution or not contains deep scientific significance,worthy of further exploration and research.
3.2. Peptide profile analysis
Fig.2A shows total 241 different peptide sequences were found,among which 212 peptides were from casein and 29 peptides came from Lactobacillus helveticus protein.These casein-derived peptides originated from β-casein (105), followed by αS1-casein (60), κcasein(26),and αS2-casein(21).And the proportion was 49.5%for β-casein, 28.3% for αS1-casein, 12.3% for κ-casein, and about 9.9%for αS2-casein,which shows that peptides from β-casein accounted for the majority. However, in milk proteins 34% of casein is αS1-casein,followed by 25%β-casein,9%κ-caseins,and 8%αS2-caseins[103].Therefore,the number of released peptides was not related to the abundance of the parent proteins but to the preferences of the proteases.β-casein must be prone to enzymatic degradation by Lactobacillus helveticus and release more active peptides that benefit to microbial survival.The hydrolysis of αS1-,αS2-and β-casein by highly proteolytic strains S.Thermophilus to release bioactive peptides was studied, generating the maximum number of peptides from β-casein,secondly αS2-casein and lastly αS1-casein[104].
Among these casein-derived biological active peptides, intracellular peptides were much more complex than extracellular peptides in terms of whether type or quantity considerations,and 69 extracellular peptides and 143 intracellular peptides were identified. The number of the intracellular peptides was more 2 - fold more than that of extracellular peptides,which is related to microbial unique proteolytic system.In addition,the average number of

Table 2 Intracellular and extracellular identified sequences in experimental groups.
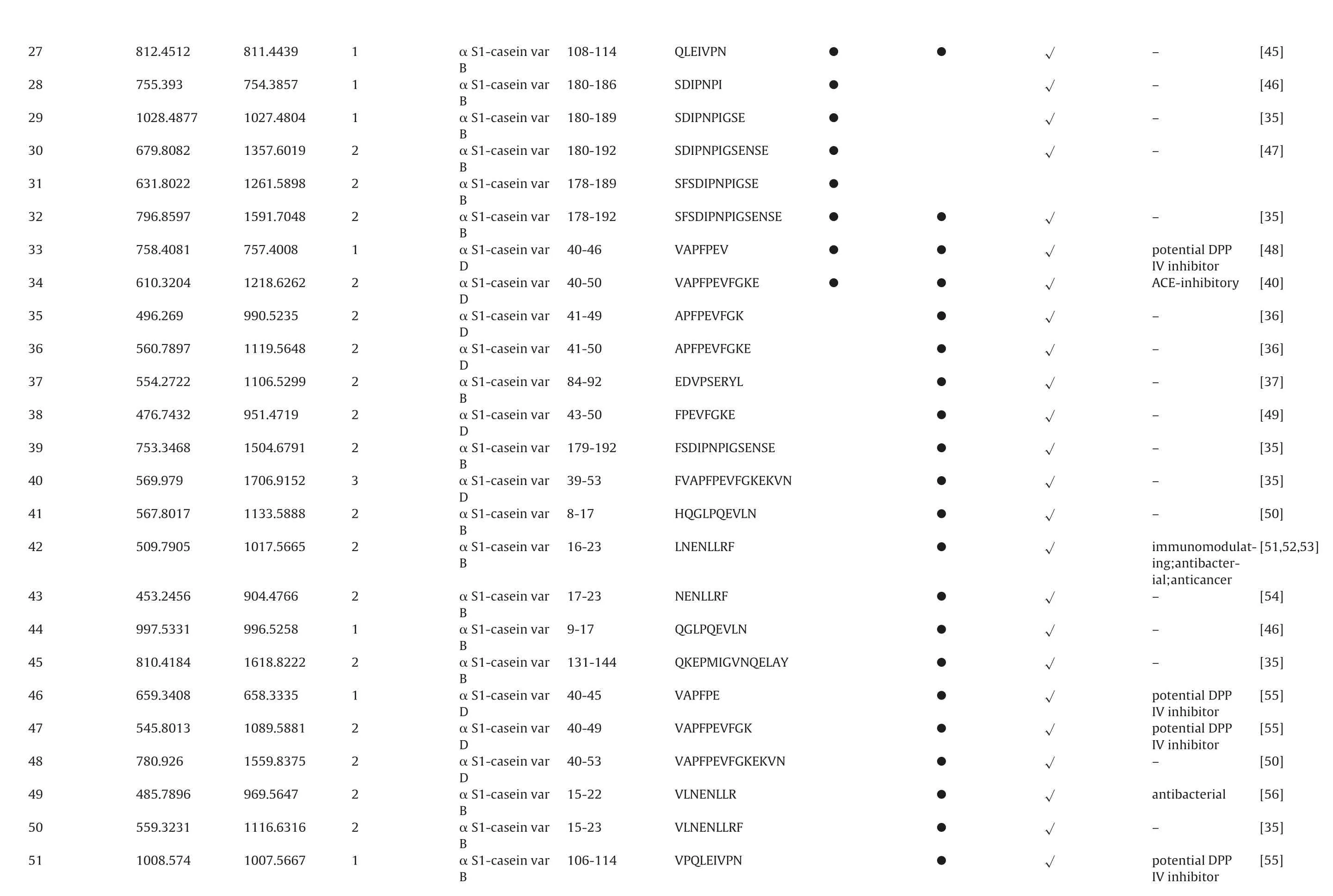
[45]–[46]–[35]–[47]–[35]–[48][40]potential DPP IVinhibitor ACE-inhibitory[36]–[36]–[37]–[49]–[35]–[35]–[50]–[51,52,53]immunomodulat-[54]ing;antibacterial;anticancer–[46]–[35]–[55][55][50]potential DPP IVinhibitor potential DPP IVinhibitor–[56]antibacterial[35]–[55]potential DPP IVinhibitor√√√√√√√√√√√√√√√√√√√√√√√√●●QLEIVPN 108-114●SDIPNPI 180-186 SDIPNPIGSE●180-189●SDIPNPIGSENSE 180-192 SFSDIPNPIGSE●178-189●●SFSDIPNPIGSENSE 178-192●●VAPFPEV 40-46●●VAPFPEVFGKE 40-50●APFPEVFGK 41-49 APFPEVFGKE●41-50●EDVPSERYL 84-92 FPEVFGKE●43-50 FSDIPNPIGSENSE●179-192●FVAPFPEVFGKEKVN 39-53 HQGLPQEVLN●8-17 LNENLLRF●16-23●NENLLRF 17-23●QGLPQEVLN 9-17 QKEPMIGVNQELAY●131-144 VAPFPE●40-45 VAPFPEVFGK●40-49 VAPFPEVFGKEKVN●40-53 VLNENLLR●15-22●VLNENLLRF 15-23●VPQLEIVPN 106-114 α S1-caseinvar B α S1-caseinvar B α S1-caseinvar B α S1-caseinvar B α S1-caseinvar B α S1-caseinvar B α S1-caseinvar D α S1-caseinvar D α S1-caseinvar D α S1-caseinvar D α S1-caseinvar B α S1-caseinvar D α S1-caseinvar B α S1-caseinvar D α S1-caseinvar B α S1-caseinvar B α S1-caseinvar B α S1-caseinvar B α S1-caseinvar B α S1-caseinvar D α S1-caseinvar D α S1-caseinvar D α S1-caseinvar B α S1-caseinvar B α S1-caseinvar B 1 811.4439 812.4512 27 1 754.3857 755.393 28 1 1027.4804 1028.4877 29 2 1357.6019 679.8082 30 2 1261.5898 631.8022 31 2 1591.7048 796.8597 32 1 757.4008 758.4081 33 2 1218.6262 610.3204 34 2 990.5235 496.269 35 2 1119.5648 560.7897 36 2 1106.5299 554.2722 37 2 951.4719 476.7432 38 2 1504.6791 753.3468 39 3 1706.9152 569.979 40 2 1133.5888 567.8017 41 2 1017.5665 509.7905 42 2 904.4766 453.2456 43 1 996.5258 997.5331 44 2 1618.8222 810.4184 45 1 658.3335 659.3408 46 2 1089.5881 545.8013 47 2 1559.8375 780.926 48 2 969.5647 485.7896 49 2 1116.6316 559.3231 50 1 1007.5667 1008.574 51

Table 2(Continued)

[40][65,66,67][40][68]ACE-inhibitory immunomodulating;ACE-inhibitor;antioxidative ACE-inhibitory potential antioxidant[36][36][35][35][69][70][71][36][72]–––––ACE-inhibitory––potential ACE-inhibitory[49][73]–ACEinhibitory;antioxidative[74]BitterPeptides[45]–[75][74][76][62][35][36][49][36][77][79]ACE-inhibitory BitterPeptides Opioidagonist potential Antihypertensive––––cytomodulatory effect immunomodulating[78]DPP IV inhibitor[80]–√√√√√√√√√√√√√√√√√√√√√√√√√√√√√●●●FPPQSVL GPVRGPFPI●GPVRGPFPIIV NIPPLTQT●●●●●●●●●QQQTEDELQDKIHPF QSLTLTDVE RELEELNVPG●●RELEELNVPGE SLTLTDVE●●SLTLTDVEN SLVYPFPG●SLVYPFPGPIHN●SQSLTLTDVE●TESQSLTLTDVE●●VVPPF VVPPFL●●VVPPFLQPE VYPFPGPIHN●AQTQSLVYPFPGPIHN●●●●LPPLL LVYPFPGPIHN QPEVMGVSKVAPKQKEMPFPKY KEAM QPVLGPVRGP●QPVLGPVRGPFP●●QQPVLGPVRGP SQSKVLPVPE●●YPFPGPIHN YPFPGPIHNS●●YPFPGPIHNSLPQ DELQDKIHPFAQTQ●●●DMPIQAF EDELQDKIHPF EELNVPGE●●ELQDKIHPFAQTQ EPVLGPVRGP●EPVLGPVRGPFP●●FPGPIPN FPGPIPNS●●●FPKYPVEPF FPKYPVEPFTE 157-163 199-207 β-caseinvarB β-caseinvarB 12 786.4285 938.5343 787.4357 470.2744 76 77 199-209 73-80 β-caseinvarB β-caseinvarB 21 1150.6916 882.476 576.3531 883.4833 78 79 38-52 123-131 1-10 1-11 124-131 124-132 57-64 83-87 β-caseinvarB β-caseinvarB β-caseinvarB β-caseinvarB β-caseinvarB β-caseinvarB β-caseinvarB β-caseinvarA1 57-68 β-caseinvarA1 122-131 β-caseinvarA1 120-131 β-caseinvarB 31221112121 1854.8745 1004.5053 1154.5978 1283.6434 876.4453 990.4861 878.4508 1339.701 1091.5309 1321.629 557.3204 619.2988 1005.5126 578.3062 642.829 877.4526 991.4934 879.4581 670.8578 1092.5382 661.8218 558.3276 80 81 82 83 84 85 86 87 88 89 90 83-88 83-91 β-caseinvarB β-caseinvarB β-caseinvarA1 59-68 112 670.4056 1024.5582 1139.578 671.4128 1025.5654 570.7963 91 92 93 β-caseinvarA1 53-68 β-caseinvarA1 135-139 β-caseinvarA1 58-68 β-caseinvarA3 89-114 2123 1767.9034 551.3703 1252.6653 2992.5062 884.959 552.3776 627.3399 998.5093 94 95 96 97 β-caseinvarA1 195-204 β-caseinvarA1 195-206 β-caseinvarA1 194-204 β-caseinvarA1 166-175 β-caseinvarA1 60-68 β-caseinvarA1 60-69 β-caseinvarA1 60-72 43-56 β-caseinvarB 22222223 1019.5824 1263.7019 1147.6341 1082.6008 1040.5152 1127.5469 1465.7391 1668.8221 510.7985 632.8582 574.8243 542.3077 521.2649 564.7807 733.8768 557.2813 98 99 100 101 102 103 104 105 184-190 42-52 4-11 44-56 195-204 β-caseinvarB β-caseinvarB β-caseinvarB β-caseinvarB β-caseinvarB 12122 836.3728 1369.656 885.4134 1553.7932 1019.5824 837.3801 685.8353 886.4207 777.9039 510.7985 106 107 108 109 110 195-206 62-68 62-69 111-119 111-121 β-caseinvarB β-caseinvarB β-caseinvarB β-caseinvarB β-caseinvarB 21122 1263.7019 740.3838 827.414 1124.5615 1352.6727 632.8582 741.3911 828.4213 563.288 677.3436 111 112 113 114 115
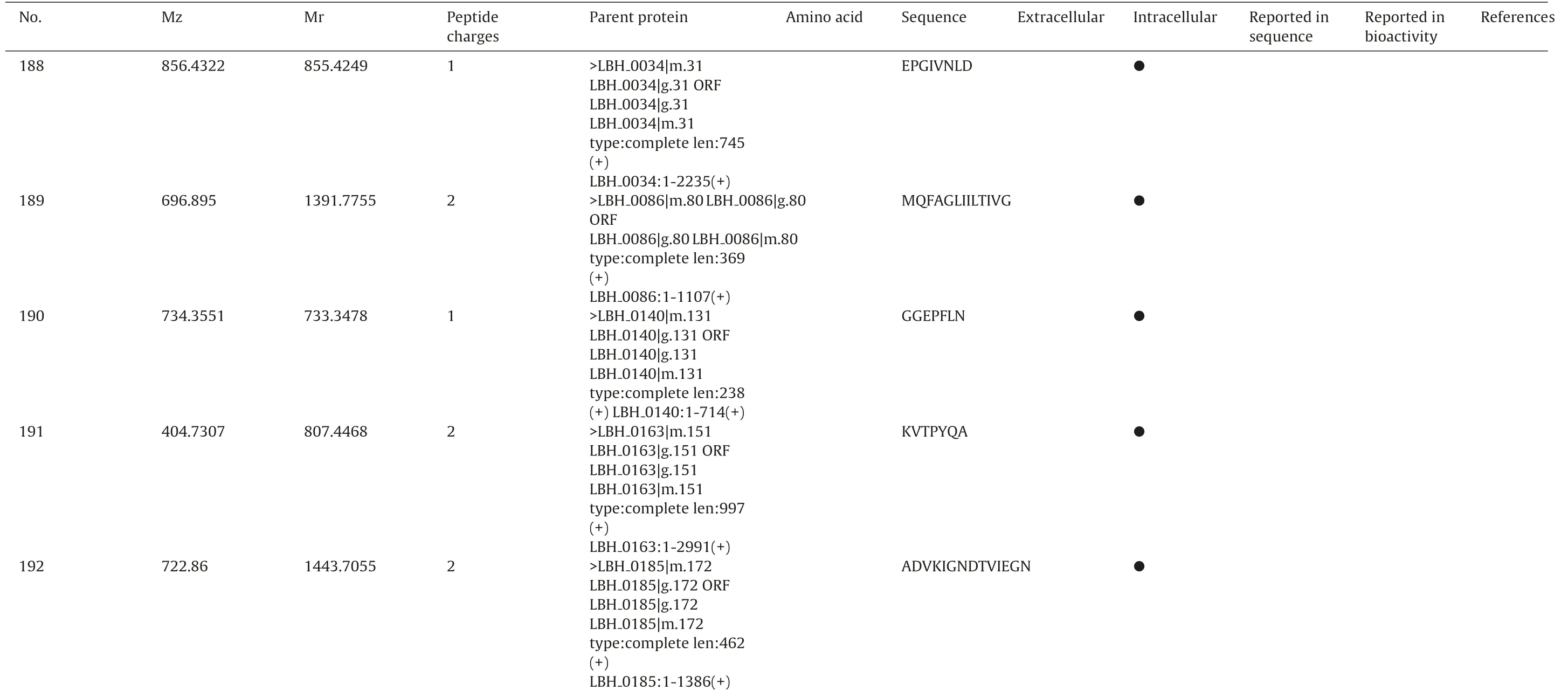
Table 2(Continued)
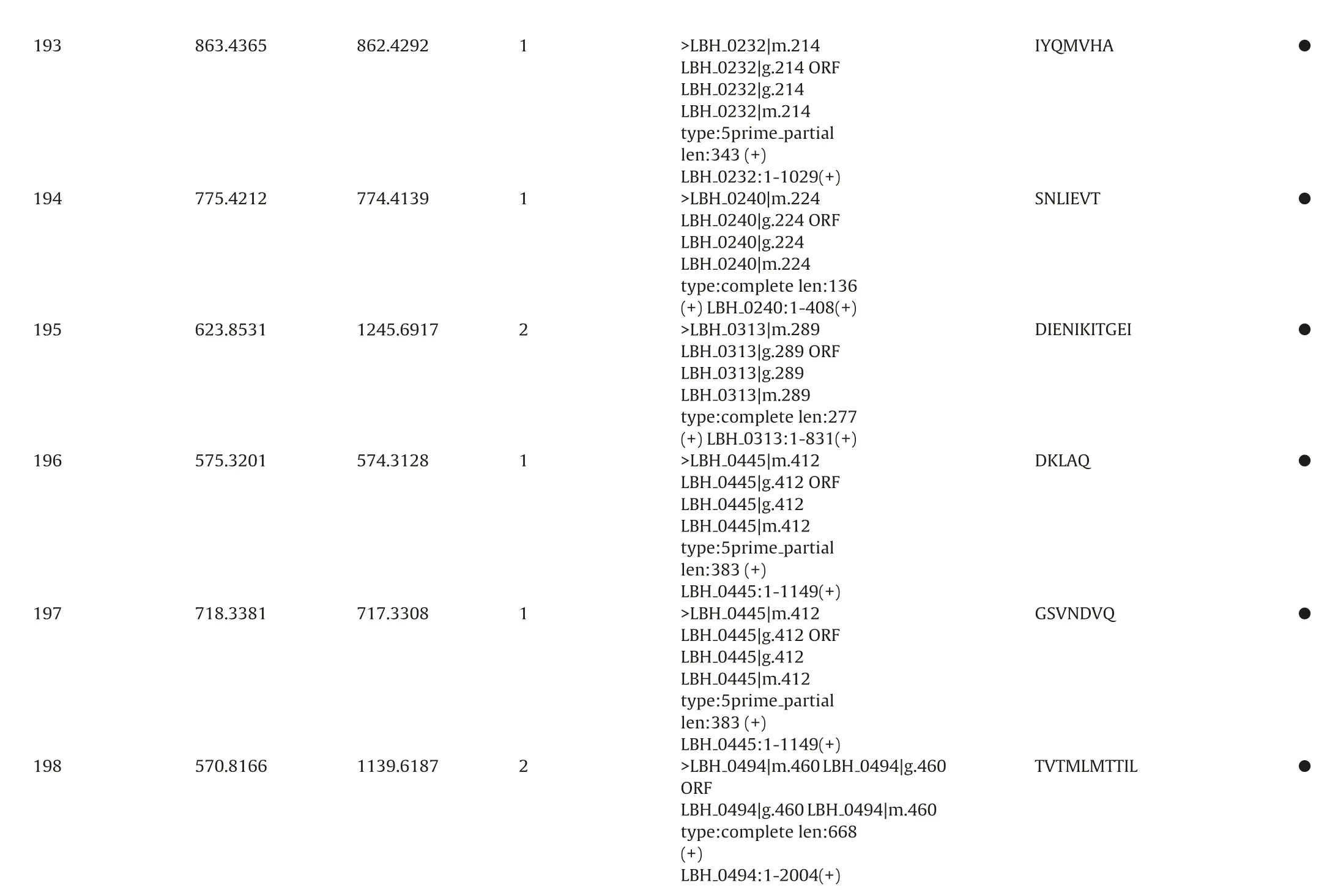
●IYQMVHA●SNLIEVT●DIENIKITGEI●DKLAQ●GSVNDVQ TTIL●TVTMLM 0494|g.460 0494|m.460 0232|m.214 partial 0240|m.224 0240:1-408(+)0313|m.289 0313:1-831(+)0445|m.412 partial 0445|m.412 partial 0494|m.460 LBH>LBH 0232|g.214ORF 0232|g.214 0232|m.214 0232:1-1029(+)0240|g.224ORF 0240|g.224 0240|m.224 0313|g.289ORF 0313|g.289 0313|m.289 0445|g.412ORF 0445|g.412 0445|m.412 0445:1-1149(+)0445|g.412ORF 0445|g.412 0445|m.412 0445:1-1149(+)0494|g.460 LBH 0494:1-2004(+)LBH LBH LBH type:5prime len:343(+)LBH>LBH LBH LBH LBH type:complete len:136(+)LBH>LBH LBH LBH LBH type:complete len:277(+)LBH>LBH LBH LBH LBH type:5prime len:383(+)LBH>LBH LBH LBH LBH type:5prime len:383(+)LBH>LBH ORF LBH type:complete len:668(+)LBH 1 862.4292 863.4365 193 1 774.4139 775.4212 194 2 1245.6917 623.8531 195 1 574.3128 575.3201 196 1 717.3308 718.3381 197 2 1139.6187 570.8166 198

Table 2(Continued)

●FDPTLHQ●DDVTEVM LDENDIIL●SVAPAAAGIN●●SRPETSG●MVIQH 1278|g.1190 1278|m.1190 1471|g.1370 1471|m.1370 1099|m.1026 partial 1154|m.1076 partial 1162|m.1083 1162:1-807(+)1221|m.1131 1278|m.1190 LBH 1471|m.1370 LBH 1471:1-981(+)>LBH 1099|g.1026 ORF 1099|g.1026 1099|m.1026 1099:1-618(+)1154|g.1076 ORF 1154|g.1076 1154|m.1076 1154:1-1662(+)1162|g.1083 ORF 1162|g.1083 1162|m.1083 1221|g.1131 ORF 1221|g.1131 1221|m.1131 1221:598-954(+)1278|g.1190 LBH 1278:1-1050(+)1471|g.1370 LBH LBH LBH LBH type:5prime len:206(+)LBH>LBH LBH LBH LBH type:5prime len:554(+)LBH>LBH LBH LBH LBH type:complete len:269(+)LBH>LBH LBH LBH LBH type:complete len:119(+)LBH>LBH ORF LBH type:complete len:350(+)LBH>LBH ORF LBH type:complete len:327(+)LBH 1 856.4196 857.4269 205 1 823.3407 824.348 206 1 943.4928 944.5001 207 1 869.4506 870.4579 208 1 733.3478 734.3551 209 1 642.3263 643.3336 210
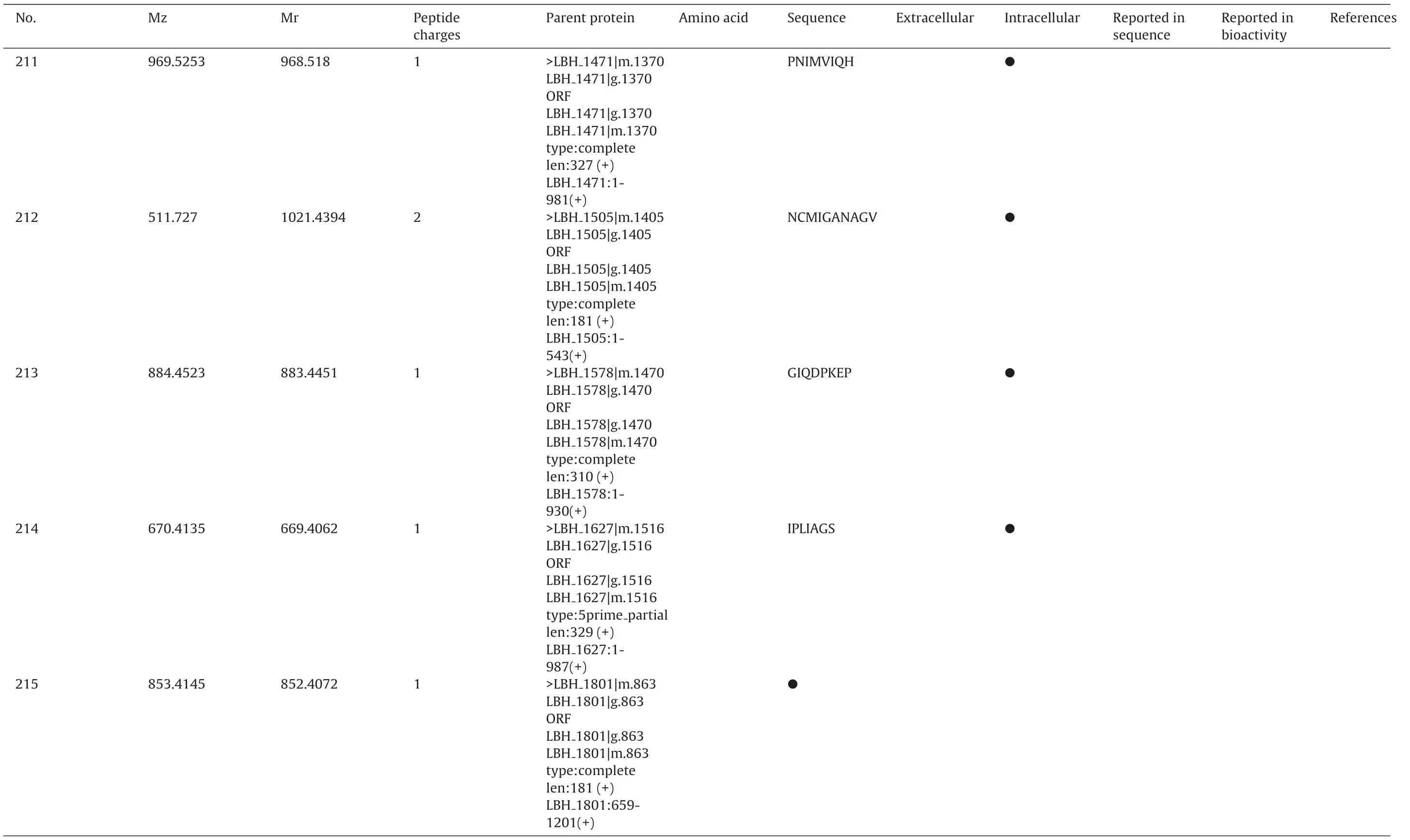
Table 2(Continued)
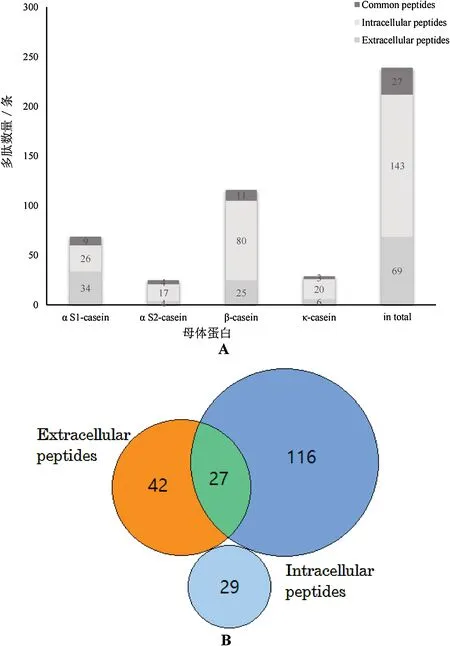
Fig.2. Distribution of identified peptides between extracell and intracell.Numbers show the total quantity of peptides identified either in extra or in intracell in total and for all parent proteins,i.e.αS1-,αS2,β-and κ-casein(A),and the comparison to the peptide profile of casein fermentation in a Venn diagram(B).
B extracellular peptides amino acids was 10.4,and less than or equal to 8 amino acids was 18,accounting for 26%;the average number of extracellular peptides amino acids was 10.3, and less than or equal to 8 amino acids was 40, accounting for 28%. Although the proportion of intracellular short peptides slightly larger than the extracellular peptides,on the whole,the two are basically consistent.In both,common peptides had a total of 27,just as the intersect of the orange and the blue circle in Fig.2B. These peptides not only existing in extracell, but also appearing in intracell were the result of peptides generated extracellularly being transported into the cells,or result from generation in intracell and transportion to extracell again, whose way is to be further tracked and explored.However, it has been described that the bacteria can absorb the extracellular hydrolyzed peptide fragments into the cell for make use of protein in the life of metabolic activities,and then make the next utilization that most of peptides are again hydrolyzed into small molecule peptides or even a single amino acid to provide for the growth[35].It is the found common peptides appearing in the cell that proved that it has not been degraded again and likely to be directly used by the bacteria. Z. Cheng et al. cultured lactic acid bacteria with 5-carboxyfluorescein (FAM) -labeled milk-derived peptide QEPV,made the bacteria washed three times and detected by flow cytometry, and finally the fluorescence intensity was significantly shifted, indicating that FAM-QEPV was absorbed by the cell.The qualitative and quantitative analysis by UPLC-MS/MS confirmed that QEPV could be directly absorbed from extracell to intracell for use by Streptococcus thermophilus. The smallest blue circle in the figure represents peptides derived from Lactobacillus helveticus protein, which is no coincidence on the sequence of casein-derived peptides. Whether the milk casein and the microbial protein have a such connection as same peptides or same core amino acid sequence remains to be explored.
The detected distribution of peptides amino acids in parent casein and cleavage sites of the parent casein were displayed in Fig.3,which indicated that the released extracellular peptides and intracellular peptides are distributed unevenly among the protein sequences. In β-casein var A1, peptides amino acids are focused on 57–70 sites. And it can be found that a relative high number of peptides are released from 178 site to 209 site of β-casein var B. However, none peptides were found at position 15–37 in the sequence of β-casein.What’s more,other popular distribution region are focused on the 178–199 in the sequence of αS1-casein var B, 39–50 in the sequence of αS1-casein var D, 72–86 and 170–182 in the sequence of κ-casein and 130–142 in the sequence of αS2-casein. Cleavage in β-casein are same most, and N68–S69 in var A1 and F52–A53, V178–P179, Q182–R183, A189–F190 and L192–Y193 in var B are more easily hydrolyzed by proteases.In αS1-casein,F23–F24,F179–S180,D181–I182,E189–N190,E192–K193,and L189–W199 in var B and F39–V40 in var D show a greater superiority than others. In κ-casein, only var A, A86–A87 and N181–T182 show a high susceptibility to proteolysis.And the resistance of N130–A131 and A131–V132 to hydrolysis is weaker in αS2-casein.E.R.S.Kunji et al.[105]demonstrated cleavage sites are related to the specificity of some microbiological proteinases. In the case of PrtP including E group protease (PI-type) and A group protease (PIII-type), the former PI-type preferentially hydrolyzes the β-casein and the latter PIII-type is capable of hydrolyzing the αS1-,β-and κ-casein,which can be further subdivided into a,b,c,d, e, f and g seven subtypes depending on different cleavage sites[106,107].
3.3. Functions confirmation and prediction of bioactive peptides
A detailed literature survey and a BIOPEP [108,109] database search were performed for the identification of known bioactive sequences in the generated peptide list in Table 2. The BIOPEP application contains a database, where sequences of biologically active peptides confirmed to have an activity either in vivo or in vitro based on literature reports are listed. A largest number of bioactive peptides are ACE inhibitors. Many research indicated that ACE can raise blood pressure by converting angiotensin I into angiotensin II,which strengthens the myocardial contractility,and at the same time makes the vascular smooth muscle contraction resulting in increased blood pressure.Consequently,ACE inhibitors can exert inhibition [110]. Twenty-one peptides were reported to have ACE-inhibitory ability. Secondly, five antioxidant peptides were identified. V.P. Shanmugam et al. [58] reported that the NAVPITPTL peptide possessed antioxidant activity of 1.49±0.10 TEAC (mmol TE mg1) and significantly differed between others(P <0.01).Five peptides with immunomodulation were identified.Five antibacterial peptides were reported by M.Hayes(2006)et al.[53,57,64,85,99]. Also, H. Uenishi [59] and M. J. Ojeda et al. [80]confirmed such three DPP-IV inhibitors. Dipeptidyl-peptidase 4(DPP-4)can rapidly degrade the rising incretin in the plasma after food intake so as to hinder the synthesis of insulin and bring about a high blood glucose concentration.Therefore,DPP-4 inhibitors can ameliorate glucose tolerance[111].Finally,there were other bioactive peptides, such as antiamnesic peptides, anticancer peptides,bitter peptides,and peptides with cell regulation ability,conducive to treat leptin-associated disorders and with lipoxygenase inhibition. Of course, there have been some peptides remained and not reported by scientists with regard to their bioactivities so far,that can be called as novel peptides and could have a potential of biological activities for the human body.

Fig.3. Amino acid sequences of the parent proteins αS1(var B,C,D)-,αS2(var A)-,β(var A1,A3,B)-,andκ-casein(var A)in single letter code.Arrows represent the identified released peptides.
Fig.4. Chromatographic peaks of DELQDKIHPF peptide in the extracellular(A)and intracellular(B)sample.
Therefore all the unknown peptides were then evaluated according to bioinformatic prediction by BIOPEP and literature survey.Thirteen peptides with the potential of some biological activity are obtained. The peptide PIGSENSEKTTMPLW is a potential antihypertensive peptide because of its fragment TTMPLW proved to have a ACE-inhibitory ability [112,113]. VAPFPE possessing the features of DPP-IV inhibitory peptides(i.e.,Leu,Ile or Phe at the Nterminus and/or Pro/Ala at position 2,and/or Pro at the C-terminus)according to known DPP-IV inhibitory peptides was considered to have a DPP-IV inhibitory potential[56].B.N.P.Sah et al.[69]purified antioxidant peptides from crude peptide extract of probiotic yoghurt using ultrafiltration and reversed phase-high performance liquid chromatography, followed by GI digestion, and identified several potential antioxidant peptides including NIPPLTQT in the digestate of P19.As for the remaining 47 novel peptides,there currently are not any experimental and scientific evidence for their sequences and possible biological activities, and their acitivities and potential application value remain to be explored. Currently,most of studies on the peptide activity are focused on the antihypertensive, hypoglycemic, antioxidative, immunomodulating and other medicinal application, and more and more scientists excavate and explore bioactive peptides in the use of microbial. And our ¨peptide database¨is the basis of our exploration of new bioactive peptides and as a database of exploring new valuable drugs through microbes or proteins in future.These new natural bioactive peptides are quite likely to be health care products for improving human health, and even become new drugs for the treatment of human diseases.
3.4. Target peptide absolute quantification
Selection of target peptide:In the Mascot search results,it was found that in a number of extracellular and intracellular peptides,a few peptides were hydrolyzed and ionized to identify more times,indirectly indicating that these peptides had a higher content and a higher ionization efficiency relative to other peptides.In this context, it was also found that there were a few peptides existing in both extracell and intracell,having the same sequences and having been identified several times.The valuable peptide most noteworthyly is DELQDKIHPF(amino acid number 43–52),which is closely related to the DELQ and the DELQDKIH found previously in our laboratory. DELQ was found to play an important role in promoting immune system activity.It was proved that the peptide has in vitro antioxidant activity by a[DPPH.](diphenyl picryl hydrazinyl radical) method and a FRAP (Ferric Reducing Ability Power) method,and in vitro proliferating lymphocyte function by MTT method[114].Our researchers also tested and veritied that DELQDKIH had an antioxidant activity in vitro using a [DPPH.] (diphenyl picryl hydrazinyl radical)method and a Total Antioxidant Capacity Assay Kit with ABTS method.Moreover,the peptide DKIHPF had been previously demonstrated to exhibit ACE inhibition with IC50 values as low as 256 M[115].So the peptide DELQDKIHPF is not only defined as the target peptide,but also the potential bioactive peptide.
The target peptide quantification was based on the calibration curve drawn using synthesised standard peptide. The calibration curve of the developed UPLC/MRM method for DELQDKIHPF within a concentration range of 2–25 ng/mL DELQDKIHPF was obtained,of which equation fitted by linear regression was y=1.26e+004x+-1.11e+004. Good linearity has been achieved with regression coefficient R2=0.9983.The acquired equation were used to quantify peptide samples according to peak areas,resulting to calculate final concentration of DELQDKIHPF peptide in both extracellular and intracellular samples.The final concentration of DELQDKIHPF was 23.1 ng/mL in extracell and 9.76 ng/mL in intracell, which laid the foundation for the subsequent research and production of bioactive peptides(Fig.4).
So far, many studies have shown that single or several bioactive peptide fragments found from fermented dairy products have low biological activity, simple function and some even no bioactivity. The application value of single peptide may not be high. It shows that these research results are not comprehensive and lack of system.The type and quantity of bioactive peptides derived from fermented dairy products vary greatly because of the effects of fermentation conditions,species of lactic acid bacteria and substrate concentration. In terms of functional characteristics, it is likely a series of peptides that play a role in the bioactivity as a comprehensive result; In terms of yield, the yield of single peptide in production applications is not high,and it can be said to be low,but the total yield of the mixed peptides is relatively higher.Therefore,in the future production of peptide drugs or health care products,it is combined peptides or mixed peptides that have higher value,more practical and greater prospects for development in the market in which health industry as the mainstream.
4. Conclusion
In this study,Lactobacillus helveticus CICC6024 was used to efficiently ferment milk-casein under relatively fixed fermentation conditions in order to achieve efficient production of bioactive peptides.241 bioactive peptides from the extracell and intracell of the fermentation broth were isolated and identificated by a series steps of centrifugation, ultrasonic cell disruption, ultrafiltration, solid phase extraction,gastrointestinal simulated digestion and NanoLCQ-TOFMS. The comprehensive analysis of the peptide profile and the summary of their functions were carried out.A specific ¨peptide database¨like a large ¨vault¨were published, where it was the types and quantities of bioactive peptides derived from casein and probiotics and their various biological activities that were explored,the potential application of extracellular and intracellular peptides also done. Also 47 novel bioactive peptides derived from casein were found,which laid the foundation for following investigation of new bioactive peptides. Finally, one of the potential peptide DELQDKIHPF was quantitatively analyzed by UPLC-U3Q method.The present work provides useful information that could be used as a basis of subsequent exploration and indicators for various production parameters of peptides capable of treating disease and protecting health,which may have a beneficial influence on human health industry.
杂志排行
食品科学与人类健康(英文)的其它文章
- Diet and medical foods in Parkinson’s disease
- Functional food products in Japan:A review
- Mathematical rules for synergistic,additive,and antagonistic effects of multi-drug combinations and their application in research and development of combinatorial drugs and special medical food combinations
- Biting force and tongue muscle strength as useful indicators for eating and swallowing capability assessment among elderly patients
- Influence of luteolin on the apoptosis of esophageal cancer Eca109 cells and its mechanism of action
- A value-added cooking process to improve the quality of soybean:Protecting its isoflavones and antioxidant activity
