Analysis of the autophagy gene expression profile of pancreatic cancer based on autophagy-related protein microtubule-associated protein 1A/1B-light chain 3
2019-05-13YanHuiYangYuXiangZhangYangGuiJiangBoLiuJunJunSunHuaFan
Yan-Hui Yang, Yu-Xiang Zhang, Yang Gui, Jiang-Bo Liu, Jun-Jun Sun, Hua Fan
Abstra c t BACKGROUND
Key words: Pancreatic cancer; Autophagy-related protein m icrotubule-associated protein 1A/1B-light chain 3; Perineural invasion; Gene Ontology analysis; Kyoto Encyclopedia of Genes and Genomes pathway analysis; Ubiquitin C
Core tip:In this study, we identified differentially expressed genes based on the autophagy-related protein m icrotubule-associated protein 1A/1B-light chain 3 (LC3) to analyze the gene expression profile of autophagy in pancreatic cancer. Three hundred and forty-seven genes that have no confirmed association w ith the autophagy process of human pancreatic cancer cells in previous studies were concentrated, and the key pathways involved in autophagy were enriched. Furthermore, a key gene ubiquitin C which is closely related to the occurrence of perineural invasion (PNI) was determined,suggesting that LC3 may influence the PNI and prognosis of pancreatic cancer through ubiquitin C.
Citation:Yang YH, Zhang YX, Gui Y, Liu JB, Sun JJ, Fan H. Analysis of the autophagy gene expression profile of pancreatic cancer based on autophagy-related protein microtubuleassociated protein 1A/1B-light chain 3. World J Gastroenterol 2019; 25(17): 2086-2098
URL: https://www.w jgnet.com/1007-9327/full/v25/i17/2086.htm
DOI:https://dx.doi.org/10.3748/w jg.v25.i17.2086
INTRODUCTION
Pancreatic cancer is a high ly invasive tum or of the d igestive system. It is of highm alignancy, its early d iagnosis is d ifficu lt, and it is not sensitive to rad iotherapy and chem otherapy. A t p resent, su rgical resection is the on ly relatively effective treatm ent.The m ajority of patients are at the late stage of the d isease w hen d iagnosed and thus have m issed the best treatm ent opportunity[1]. Therefore, identifying a new d irection for the treatm en t of pancreatic cancer has becom e the focus of pancreatic su rgery stud ies. As an im portant m echanism for tum or cells to escape apop tosis, au tophagy has both p rom oting and inhibiting effects on tum ors[2,3]. A t p resen t, m ore and m ore stud ies have found that au tophagy is closely related to the occu rrence, developm ent,d ifferentiation and p rognosis of pancreatic cancer[4,5].
Au tophagy-related p rotein m icrotubu le-associated p rotein 1A/1B-ligh t chain 3(LC3), as a key p rotein in the au tophagy p rocess, is involved in the form ation of the autophagosom e. A study by ou r research group found that high exp ression of LC3 in pan creatic can cer was positively co rrelated w ith neu ral invasion and p oor p rognosis[6]. On the basis of p revious stud ies and using LC3 as a guidance index, the au tophagy gene exp ression p rofile of pancreatic cancer was analyzed to gu ide the functional annotation of d ifferentially exp ressed genes and to enhance the reliability of bioin form atics p red iction and analysis. D ifferentially exp ressed genes involved in the au tophagy of pancreatic cancer w ere iden tified by a gene exp ression m icroarray technique. Protein in teraction netw o rks w ere constru cted and the fun ctional clustering of d ifferentially exp ressed genes was carried ou t. Key in teracting p roteins or genes betw een m odu les w ere screened and evaluated by statistical m ethod s, and the pathogenesis of pancreatic cancer was exp lored.
MATERIALS AND METHODS
Sample collection
Tw o groups of experim ental data w ere selected for the analysis of the w hole-genom e exp ression p rofile. Group 1 consisted of 16 norm al pancreatic tissue sam p les and 36 pancreatic cancer tissue sam p les. Group 2 consisted of 26 norm al pancreatic tissue sam p les and 26 pancreatic cancer tissue sam p les. A ll sam p les w ere obtained from su rgical excision specim ens o f pancreatic cancer patien ts and w ere d iagnosed,classified, graded and staged by pathology p rofessionals.
Gene sequencing
Fo llow ing the instructions o f the A ffym etrix gene m icroarray exp ression analysis m anual and using the A ffym etrix Hum an Genom e U133 Plus 2.0 A rray sequencing p latform (p latform num ber: GPL570)[7-10], gene exp ression in the sam p les was exam ined. A ll of the obtained data have been up loaded and subm itted to the Gene Exp ression Om nibus (GEO), a gene exp ression database.
Gene expression profile data acquisition and preprocessing
Based on the in form ation of the above tw o sets of sam p les and by exp loring the gene exp ression database of the National Center for Biotechnology In form ation (NCBI) of the United States[11], tw o sets o f pancreatic tissue gene exp ression p ro files w ere collected: GSE16515 and GSE15471. Data set-related in form ation is listed in Table 1.
The rm a function in the affy package of R language[12]was used to p rep rocess the raw data of the gene exp ression p rofiles, and the robust m u lti-array average (RMA)algorithm[13]was em p loyed to calcu late the am ount of gene exp ression from the raw data of exp ression p rofiles and to obtain the gene exp ression values of the p robes, that is, the signal strength values.
Gene annotation of probe data
Using the Annotate package o f R language com bined w ith the chip annotation docum ent, the p robe sets w ere labeled w ith the correspond ing genes (Entrez ID). In the situation of a single gene correspond ing to m u ltip le p robe sets, the average value of the m u ltip le p robe sets was used to rep resent the exp ression value of the gene. In the case of m u ltip le genes correspond ing to one p robe set, the p robe set data w ere deleted.
Screening and identification of differentially expressed genes
The significance analysis of m icroarrays (SAM) algorithm[14]was used to screen for d ifferentially exp ressed genes related to pancreatic cancer. The analysis p latform em p loyed the R language p latform.
Functional enrichment analysis of differentially expressed genes
Gene Ontology (GO) analysis of d ifferentially exp ressed genes was used to search for gene functions w hose changes m ight be correlated w ith the d ifferentially exp ressedgenes of d ifferent sam p les[15,16]. The pathw ay analysis of d ifferentially exp ressed genes was used to search for cellu lar pathw ays w hose changes m igh t be related to the d ifferentially exp ressed genes in d ifferent sam p les[17-19]. The significance thresholds of en richm en t analysis w ere 0.01 and 0.05 for GO analysis and pathw ay analysis,respectively. Descrip tions of the resu lt param eters of the functional annotations are show n in Supp lem entary Table 1.

Table 1 Relevant information of the genome expression profile data sets of pancreatic tissues
Protein interaction analysis of differentially expressed genes
Data screening and preprocessing:
Hum an p rotein in teraction netw ork data w ere dow n loaded from the STRING database[20], and p rotein interaction pairs w ith in teraction scores of m ore than 900 w ere selected. On ly p rotein interaction data containing d ifferentially exp ressed genes w ere screened from the above qualified p rotein interaction data. Modu les w ere m ined and d ifferentially exp ressed genes w ere annotated using the ClusterOne p lugin o f Cytoscape softw are.
Analysis of in form ation crosstalk betw een in teracting m odu les: The calcu lation m ethod of crosstalk significance is as follow s: in the context of a random netw ork, the num ber of cases in w hich the num ber of in teraction pairs betw een m odu les in N random netw orks (in this study, N = 1000) was greater than that in real netw orks was calcu lated and recorded as n. The form u la for calcu lating the P value is P = n/N; a P value less than or equal to 0.05 rep resents significant crosstalk betw een m odu les.
Pivot analysis: The definition o f p ivot requires satisfaction o f the fo llow ing tw o cond itions: (1) the pivot in teracts w ith tw o m odu les at the sam e tim e and has at least tw o in teraction pairs w ith each m od u le; and (2) the P value o f the significance analysis of the in teraction betw een the p ivot and each m odu le shou ld be less than or equal to 0.05. A ccord ing to the above descrip tions, the Python p rogram was w ritten to find the p ivots betw een the functional m odu les, and the hypergeom etric test m ethod was used for the significance analysis.
RESULTS
Preprocessing results of expression profile raw data
The d istribu tion of gene exp ression am oun t calcu lated by the RM A algorithm is show n in Figure 1A and B A fter data p rep rocessing, the gene exp ression p rofile data w ere redu ced from the original 54675 p robe exp ression values to 20502 gene exp ression values.
Extraction results of differentially expressed genes
A fter data standard ization and gene annotation, gene m icroarray significance analyses w ere perform ed on the tw o sets of data (GSE16515 and GSE15471) separately using the Sam function of the siggenes package of R language (Figu re 2A and B); a total of 6098 and 12928 d ifferentially exp ressed genes w ere obtained, respectively, and the first 40 genes w ere selected for d isp lay in Supp lem en tary Tab les 1 and 2,respectively. A total of 4870 core d ifferentially exp ressed genes w ere obtained from the in tersection o f the tw o sets o f d ifferen tially exp ressed genes for subsequen t functional annotation analysis.
Functional enrichment analysis of differentially expressed genes
In the p rocess of GO analysis of d ifferentially exp ressed genes, the involvem ent o f genes in bio logical p rocesses, m o lecu lar functions and cell com positions was annotated by setting d ifferen t param eters. The 14 functional item s related to apop tosis/autophagy are listed in Table 2.
The Kyoto Encycloped ia of Genes and Genom es (KEGG) analysis en riched the d ifferentially exp ressed genes into four pathw ays: hsa03050, hsa04144, hsa05412 and hsa04020. hsa04144 is related to endocy tosis, hsa04020 is related to the calcium signaling pathw ay, and both are, to a certain degree, related to au tophagy.
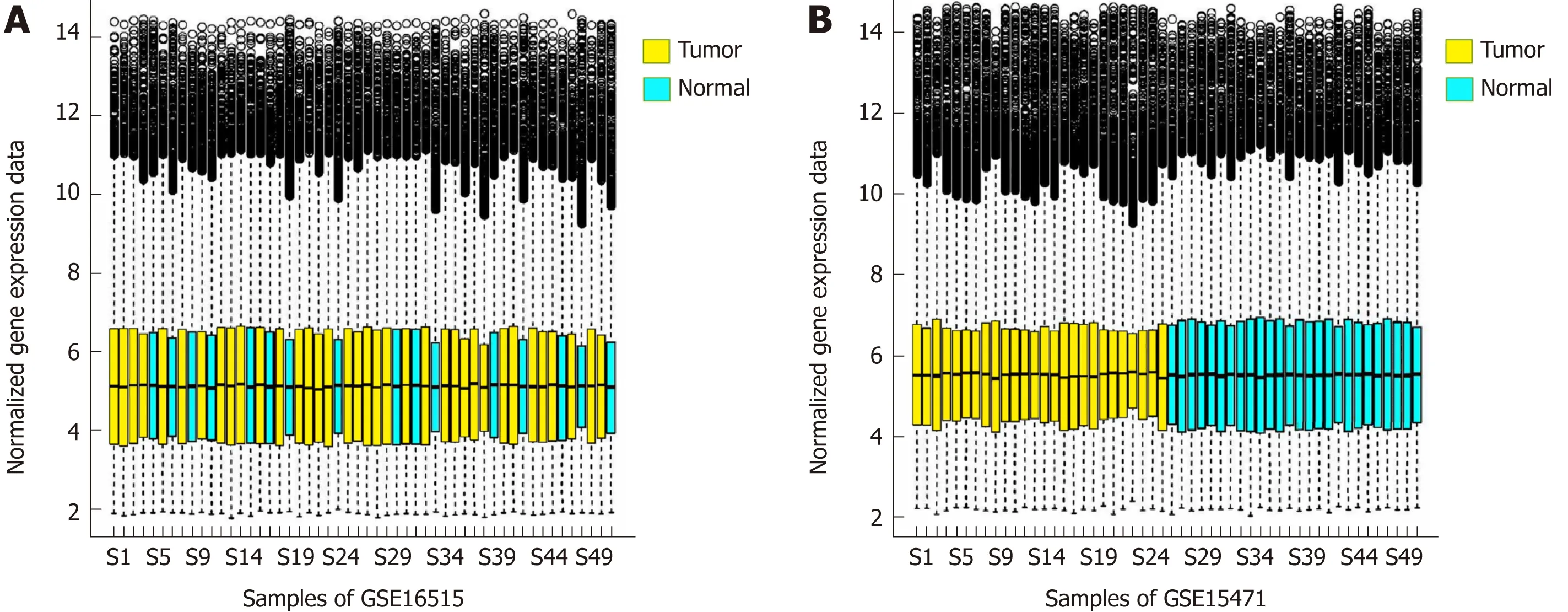
Figure 1 Box diagram of the gene expression distribution. A: Box diagram of the gene expression distribution of each sample after the standardization of set GSE16515. B: Box diagram of the gene expression distribution of each sample after the standardization of set GSE15471.
Searching the GENE functional annotation database o f the NCBI using th ree keyw ords, au tophagy, Homo sapiens and cancer (adop t and logic ru le), 851 genes w ere obtained. The 16 functional item s associated w ith au tophagy obtained by GO and KEGG analyses revealed a total of 986 d ifferentially exp ressed genes (Figu re 3). A fter rem oving genes that are clearly defined in the GENE database, 347 d ifferen tially exp ressed genes w ere obtained (Supp lem entary Table 3). The relationship betw een these genes and the autophagy of pancreatic cancer cells aw aits fu rther exp loration.
Interaction analysis of differentially expressed genes
Based on a p rotein in teraction netw ork database (STRING), a reliab le p rotein in teraction netw ork of 10524 p roteins and 196254 interaction pairs was obtained by selecting p rotein interaction pairs w ith an interaction score of greater than or equal to 900. Protein in teraction data con taining on ly d ifferentially exp ressed genes w ere subsequen tly screened from the above p rotein in teraction d ata, and a p rotein interaction netw ork of 2256 d ifferentially exp ressed genes and 12632 interaction pairs was ob tained. Figu re 4 show s the p rotein in teraction netw ork com p osed o f d ifferentially exp ressed genes. Red pivot dots ind icate that w hen m ining m odu les, the P values of the participating m odu les w ere less than 0.05, w hile yellow p ivot dots ind icate that w hen m ining m odu les, the P values of the participating m odu les w ere greater than 0.05. Grey p ivot dots ind icate that these genes w ere not involved in the m odu les. A total o f 65 clustering m odu les w ere screened by im porting the above in teraction d ata con tain ing d ifferen tially exp ressed genes in to Cy toscape (Supp lem entary Table 4).
GO and KEGG functional annotation of the d ifferentially exp ressed genes in the p rotein interaction netw ork was perform ed. The resu lts are show n in Supp lem entary Tables 5 and 6. GO analysis found that GO:0016236 was involved in au tophagy, w hile KEGG analysis revealed a high ly significan t (P = 1.06E-07) new pathw ay, hsa04216(ferrop tosis), a new cell death pattern associated w ith iron death.
Next, w e identified the M AP1L3A (LC3) gene in the excavated m odu le 32, w hich is the target gene of em phasis in this study. W e also found that this m odu le contains m u ltip le genes related to au tophagy (Supp lem entary Table 7). Through the functional enrichm ent analysis of m odu le 32, the genes of this m odu le w ere found to be m ain ly involved in pathw ays related to autophagy and iron-dependent cell death.
Fu rtherm ore, netw ork d iagram s w ere used to show the LC3 gene and genes that d irectly interact w ith LC3 (Figu re 5A) and to dem onstrate the functional m odu les in w hich the LC3 gene is involved (Figu re 5B).
Analysis of crosstalk between interacting modules
The com p lex in tracellu lar pathw ay netw orks w ere analyzed by crosstalk, and im portan t p rotein p ivots that affect signal transduction betw een the pathw ays w ere iden tified through p rotein interactions. The resu lts of the pathw ay crosstalk analysis of the aforem en tioned clustering m odu les are show n in Supp lem entary Table 8 and Figu re 6.
The above functional annotation analysis revealed that m odu les 33 and 40 w ere both associated w ith au top hagy; the crosstalk betw een the tw o m od u les is dem onstrated using a netw ork d iagram by Cytoscape (Figu re 7). The genes involved in the tw o m odu les and their functional descrip tions are listed in Supp lem en tary Table 9.
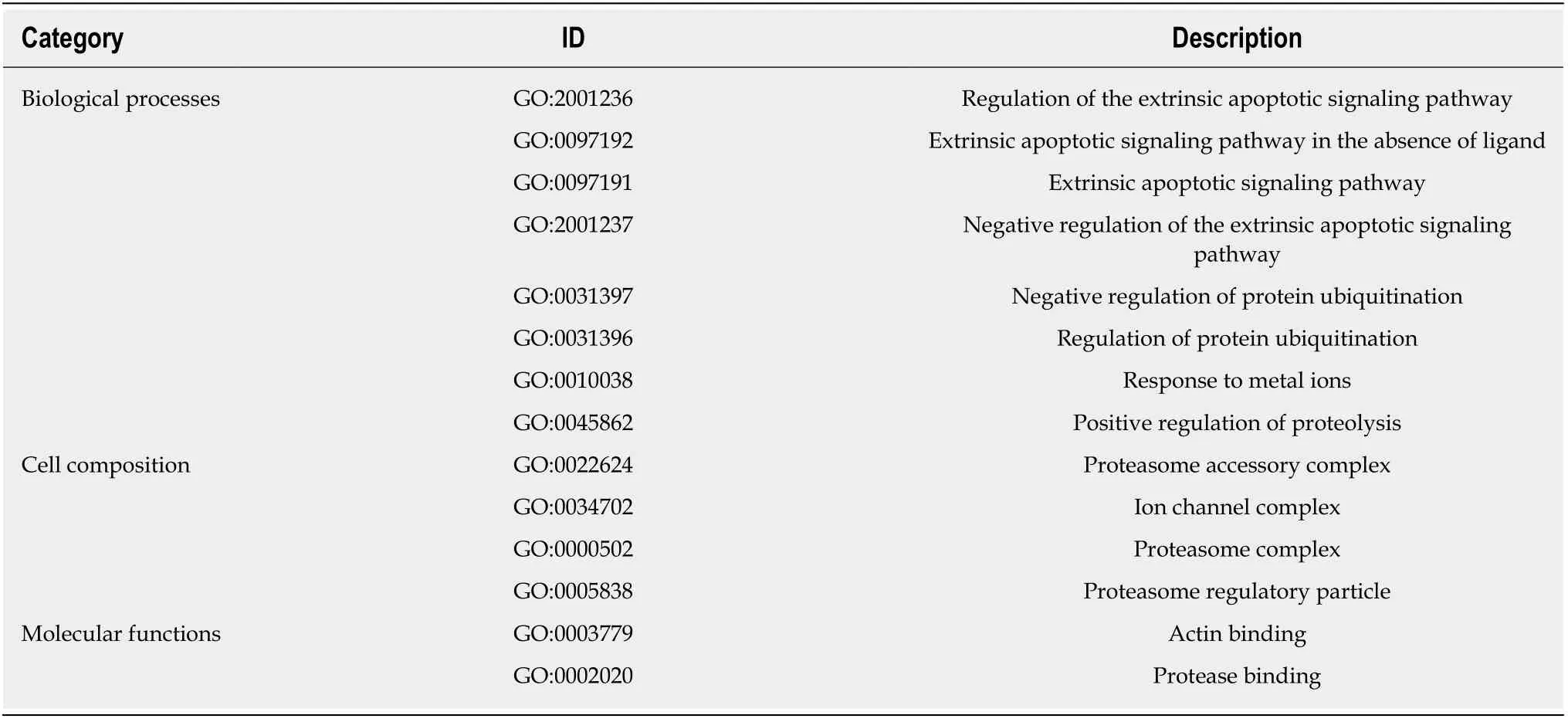
Table 2 Items related to apoptosis/autophagy in the Gene Ontology enrichment of differentially expressed genes
Pivot analysis
Pivot analysis revealed that the gene 7316, ubiqu itin C (UBC), was associated w ith several m odu les related to cancer and au tophagy. A fter rem oving the redundan t m odu les, seven m odu les interacting w ith this p ivot w ere obtained (Supp lem en tary Table 10). These m odu les all contain genes that d irectly interact w ith gene 7316.Functional en richm ent analysis show ed that m ost of these 7 m odu les are related to hum an cell death, especially the au tophagy p rocess. In m odu le 32, the p ivot d irectly interacts w ith gene 84557 (M AP1LC3A) and is associated w ith m u ltip le m odu les(Figure 8).
DISCUSSION
Pancreatic cancer has a high m alignant degree, a low early d iagnosis rate, and a less than 20% chance of rad ical resection. The incidence and m ortality rate of pancreatic cancer are high. From 2000 to 2014, the overall 5-year su rvival rate of pancreatic cancer in d ifferen t regions of the w orld was betw een 5% and 15%[21]. In China,app roxim ately 90100 new cases of pancreatic cancer w ere reported in 2015, ranking ninth am ong all m alignant tum ors, w hile m ortality reached 79400 cases, ranking sixth am ong all m alignan t tum ors; the 5-year su rvival rate was 9.9%[22]. Un til now, an effective d iagnosis and treatm ent for pancreatic cancer have been lacking, and the exact causes and pathogenesis of pancreatic cancer are not w ell understood. In this stud y, bioin fo rm atics analysis based on the LC3 gene was used to iden tify d ifferen tially exp ressed genes in pancreatic cancer and the resu lts w ere comp rehensively analyzed to p rovide a new experim ental basis for the d iagnosis and treatm ent of pancreatic cancer.
First, the GO analysis resu lts of the d ifferentially exp ressed genes associated w ith pancreatic cancer w ere p relim inarily stud ied, and 347 genes that had not been clearly defined by the GENE database as d irectly related to the au tophagy of hum an cancer cells w ere iden tified, p rovid ing new in form ation and a novel d irection for fu tu re cancer and autophagy research. Previous stud ies have show n that the occu rrence and developm ent of pancreatic cancer are the resu lt of the in teraction and in fluence o f m u ltip le genes and factors[23,24]. In this study, GO functional annotation analysis revealed that functional annotations w ere associated w ith m u ltip le d ifferentially exp ressed genes, ind irectly con firm ing the earlier resu lts. This study was not lim ited to an exp ression abnorm ality or a m u tation of a sing le gene or p rotein bu t rather analyzed the gene m icroarray exp ression p ro file data to obtain the suspected d ifferen tially exp ressed genes associated w ith tum ors, p rov id ing usefu l data for fu tu re stud ies and p rovid ing a reliab le and detailed experim en tal basis for the d iagnosis and treatm ent of com p lex d iseases. Second, the KEGG analysis resu lts of pancreatic cancer-related d ifferen tially exp ressed genes w ere analyzed. The d ifferen tially exp ressed genes w ere en riched in fou r pathw ays, and the com p lex m olecu lar m echanism s involved in these pathw ays are related to the occu rrence o ftum ors, in flamm ation, imm une d isorders, and id iopathic d iseases[25].
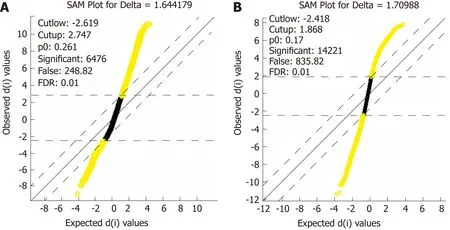
Figure 2 Distribution diagram of the statistical analysis of gene expression. A: Distribution diagram of the statistical analysis of gene expression after the extraction of differentially expressed genes in set GSE16515; B:Distribution diagram of the statistical analysis of gene expression after the extraction of differentially expressed genes in set GSE15471. Yellow for differentially expressed genes and black for non-differentially expressed genes.
In this study, w e com bined the gene exp ression database and the STRING p rotein interaction database to m ine m odu les in the p rotein interaction netw ork involving the d ifferentially exp ressed genes and found that m odu les 33 and 40 w ere both associated w ith au tophagy. In view of the functional descrip tions, the genes involved in these tw o m odu les are closely related to cellu lar p rocesses such as cancer, au tophagy and apop tosis. Th rough functional enrichm ent analysis o f the genes invo lved in these m od u les, KEGG en richm en t fu rther iden tified pathw ay hsa04216, that is,ferrop tosis[26], w hich is characterized by the p roduction of reactive oxygen species(ROS) from the peroxidation of accum u lated iron and lip ids. Kang et al[27]review ed the relationship betw een au tophagy and ferrop tosis and noted that the activation of ferrop tosis depends on its induction by au tophagy.
In the m odu le in teraction stud y, w e found that UBC, as a p ivot o f m odu le interactions, connected several m odu les related to cancer and autophagy and p lays an im portan t role in m u ltip le cell death m odu les related to au tophagy (Figu res 1-8),consistent w ith cu rrent know ledge and ou r understand ing of cancer and cell death.UBC, as a p recu rsor of ubiquitin, p lays a key role in the occu rrence and developm ent of d iseases such as au tophagy, cancer, and in flamm ation th rough the ubiquitin p roteasom e system (UPS)[28]. A large num ber o f stud ies have show n a close relationship betw een UPS and au tophagy[29,30]. Previously, UPS and au tophagy have been considered com p lem entary degradation system s w ith no intersection. How ever,som e m onoubiquitinated p roteins can also be degraded by au tophagy[31]. Pandey et al[32]perform ed in vitro experim ents and show ed that m onoubiquitination and histone deacety lase 6 (HDAC6) are the key signals linking the tw o system s of autophagy and UPS, consistent w ith the resu lts of the cu rrent study that the ubiquitin p recu rsor UBC connects m u ltip le autophagy-related m odu les.
A p revious study show ed that the signaling pathw ay adap ter p rotein P62 contains an LC3 recognition sequence that form s oligom ers to recru it ubiqu itinated p roteins th rough a ubiqu itin bind ing dom ain and interacts w ith LC3 to form degradation substrates for the au tophagosom e[33]. Ubiqu itin is also one o f the substrates o f m o lecu lar chaperone-m ed iated au tophagy[34]. In this study, a d irect in teraction betw een LC3 and the p ivot gene UBC was identified, w hich has certain significance for the stud y o f the m echanism by w h ich au tophagy scavenges ubiqu itinated substrates. Ubiqu itin and UPS are also closely related to the occu rrence and developm ent of tum ors. Tang et al[35]con firm ed that ubiquitin is high ly exp ressed in m any types of tum or tissues. Liu et al[36]show ed that UPS cou ld selectively degrade the p roducts o f oncogenes and tum or supp ressor genes, as w ell as apop tosisregu lating p roteins, thus regu lating cell m u tation and tum origenesis.
In recent years, the perineu ral invasion (PN I) of pancreatic cancer has becom e a research hotspot in academ ia and in the clinic; how ever, its m echanism has not yet been elucidated. A com p licated tum or m icroenvironm en t, au tophagy, and neu ral p lasticity[37]exert im portant effects on the nerves around and inside the pancreas. It has been reported[38]that pancreatic cancer tissue w ith high exp ression of ubiquitinspecific p rotease 9X (USP9X), a m em ber of the ubiquitin-specific p rotease subfam ilyo f the fam ily o f deubiqu itinating enzym es, m ay be m ore invasive, and h igh exp ression of USP9X is closely associated w ith the p rognosis of pancreatic cancer. In UPS, the reversible p rocess of p rotein ubiquitination and deubiquitination catalyzed by ubiqu itinating enzym es (UBEs) and deubiqu itinating enzym es (DUBs) is also closely related to PN I o f the tum or[39]. UBC-term inal hyd ro lases (UCHs) are a subfam ily of DUBs. Previous stud ies have show n that the UCH fam ily p lays d ifferent roles in the p rogression of different tum ors[40]. Ubiquitin carboxyl term inal esterase L1,a fam ily m em ber of UCHs, is not on ly overexp ressed in neu ral tissue[41]bu t is also used as a m arker of nerve fibers to study PN I[42].
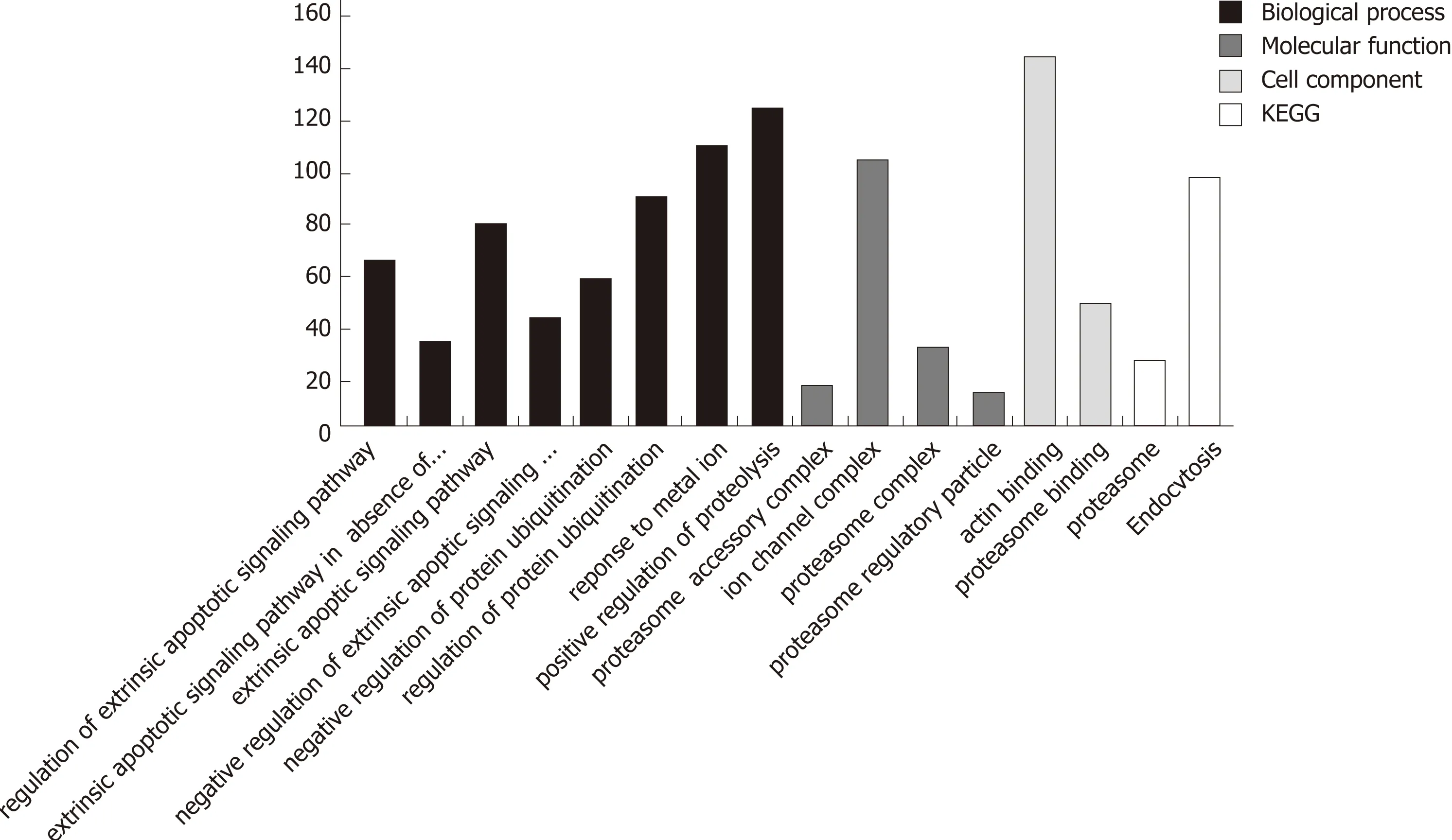
Figure 3 Distribution of the numbers of differentially expressed genes in functional items associated with apoptosis/autophagy.
In the study o f m odu lar clustering based on p rotein in teractions, UBC was clustered w ith m u ltip le au tophagy-related genes in m odu le 32 and d irectly interacted w ith LC3 and NBR1 (Figu res 2-7). UCH uses ubiquitinated p roteins as substrates to catalyze the rem oval of ubiquitin m olecu les. Therefore, UBC and UCH also d irectly in teract. M oreover, the exp ression level of UCH is closely related to the p rocess o f PN I. Based on ou r p revious study[6]in w hich the positive rate of PN I was significantly positively correlated w ith the high exp ression of LC3 in pancreatic cancer patients and that PN I and LC3 levels are independen t risk factors for the poor p rognosis o f pancreatic cancer, w e postu late that the autophagy-related p rotein LC3 m ay establish a close association w ith pancreatic cancer PN I th rough UBC and its associated ubiquitin p roteasom e system.
In summ ary, the p resent study com bined the gene exp ression p rofile m icroarray technique w ith bioin form atics analysis technology to analyze and m ine a large quantity of data and identified new significantly d ifferentially exp ressed genes related to the occu rrence of au tophagy in pancreatic cancer, w hich expands the au tophagyrelated gene p ro file of pancreatic cancer and is help fu l for the search of cand idate suscep tible genes and rare m u tations that m ay be associated w ith the occu rrence and developm ent of au tophagy in pancreatic cancer. The iden tification and review o f UBC, a key gene that d irectly in teracts w ith LC3, suggest that it m ay be a key factor that leads to a poor p rognosis of pancreatic cancer m ed iated by PN I and suggests a new d irection for further research.
In this study, w e iden tified d ifferen tially exp ressed genes betw een pancreatic cancer cells and norm al cells at the w hole-genom e level using the w hole-genom e exp ression p ro filing techn ique, iden tified 347 genes that have no con firm ed association w ith the au tophagy p rocess of hum an pancreatic cancer cells in p revious stud ies, and d iscovered and clarified in form ation abou t the pathw ays invo lved in autophagy. Fu rtherm ore, w e identified UBC, w hich p lays an im portant role in several m odu les related to cell death, through gene exp ression m icroarray analysis based on au tophagy and LC3 and found that UBC is w idely involved in tum or cell grow th,invasion and m etastasis. It was also found that ubiquitin is closely related to PN I. It is believed that LC3 m ay affect the PN I and p rognosis of pancreatic cancer th rough
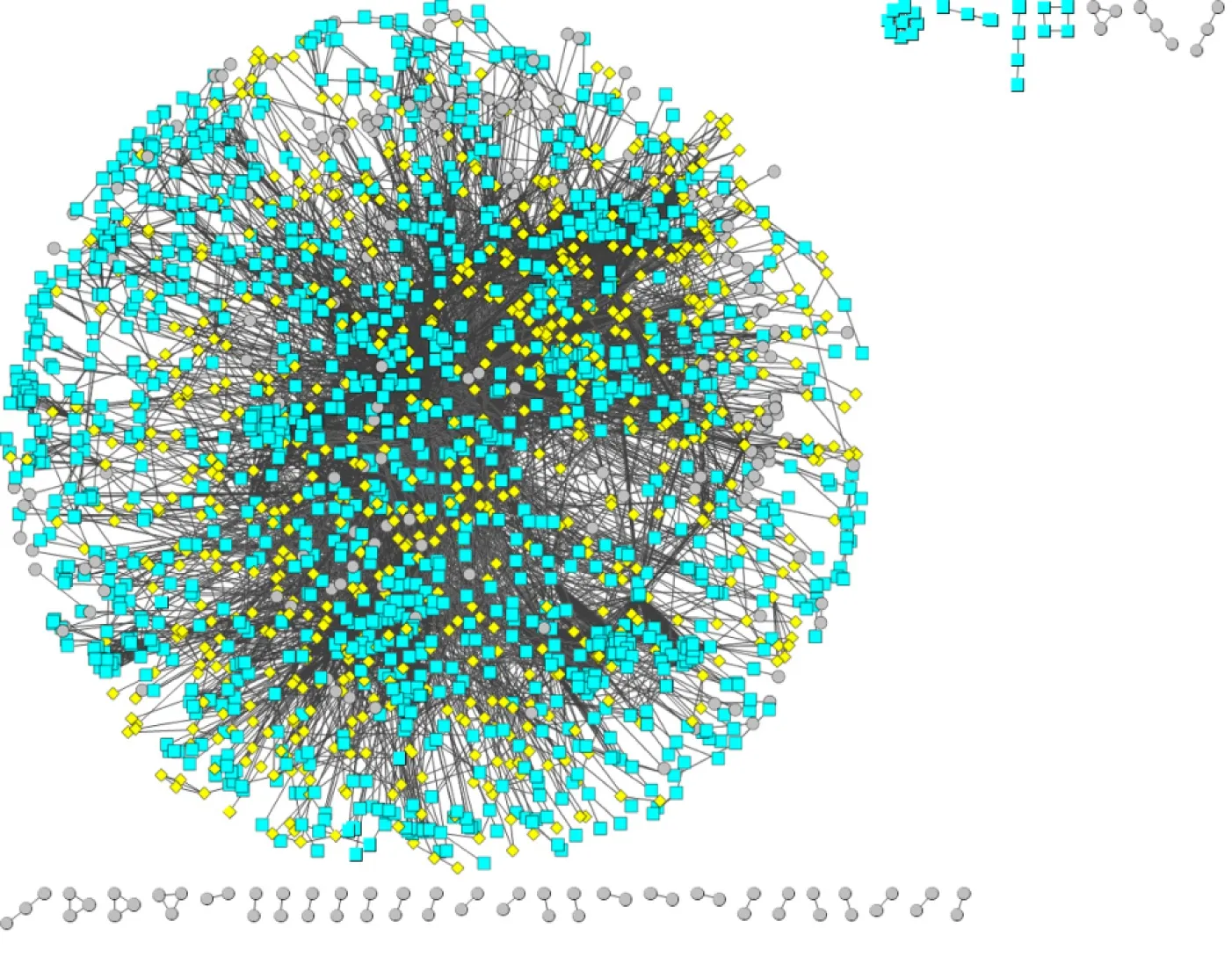
Figure 4 Protein interaction network composed of differentially expressed genes.
UBC, w hich is help fu l for the study and treatm ent of pancreatic cancer and p rovides an im portant clue to the pathogenesis of pancreatic cancer.
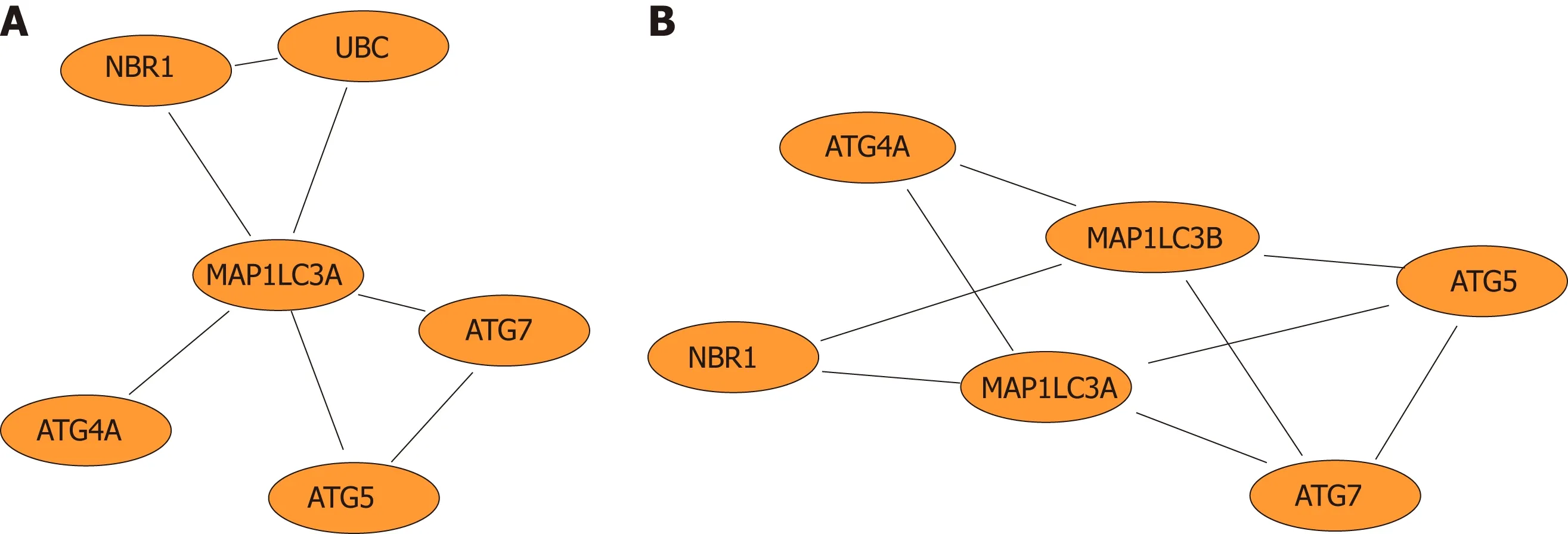
Figure 5 Network diagram. A: Network diagram of genes that directly interact with the autophagy-related protein microtubule-associated protein 1A/1B-light chain 3(LC3) gene; B: Network diagram of functional modules in which the LC3 gene is involved.
Figure 6
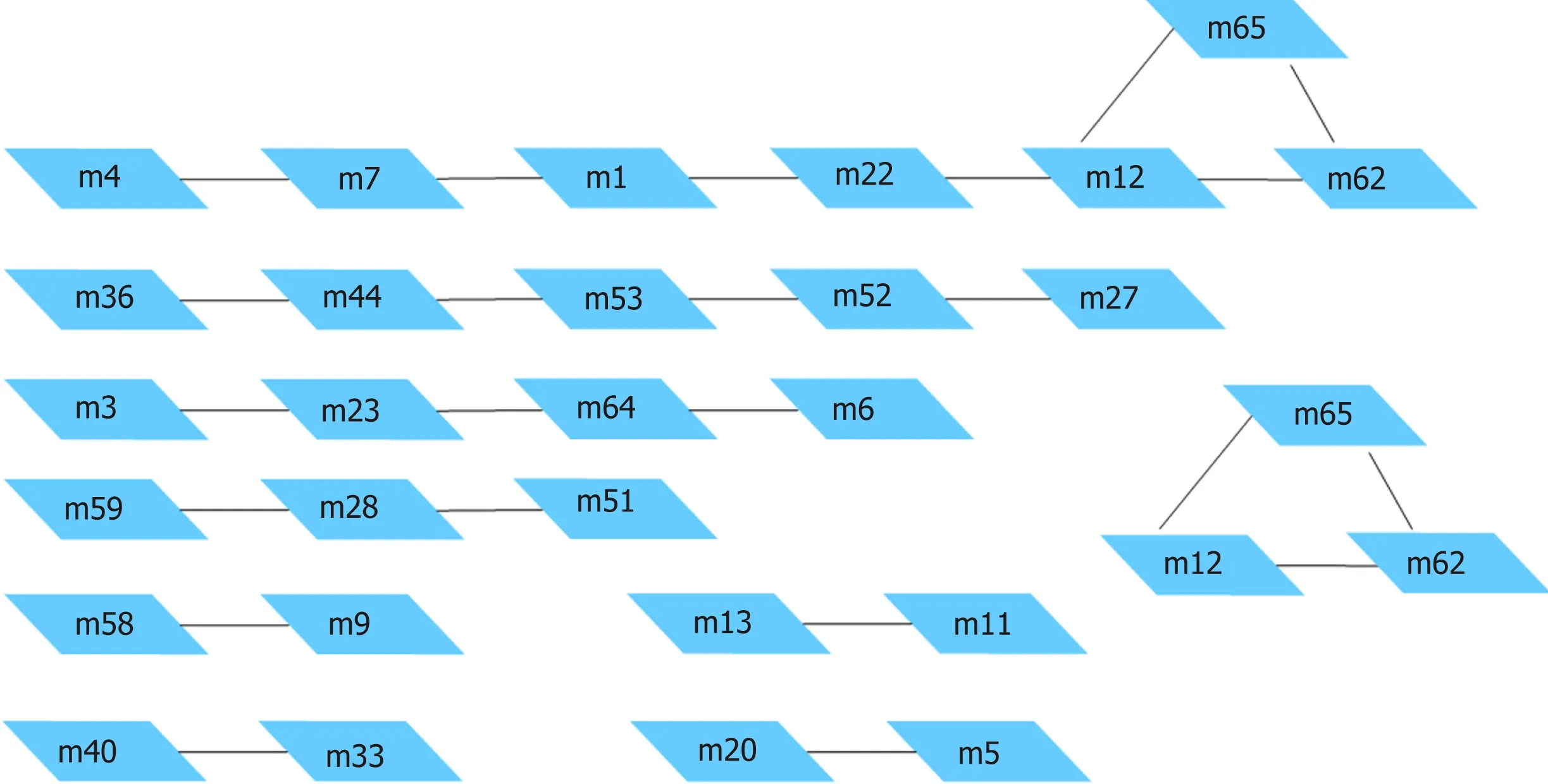
Figure 6 Module pairs with significant crosstalk.
Figure 7
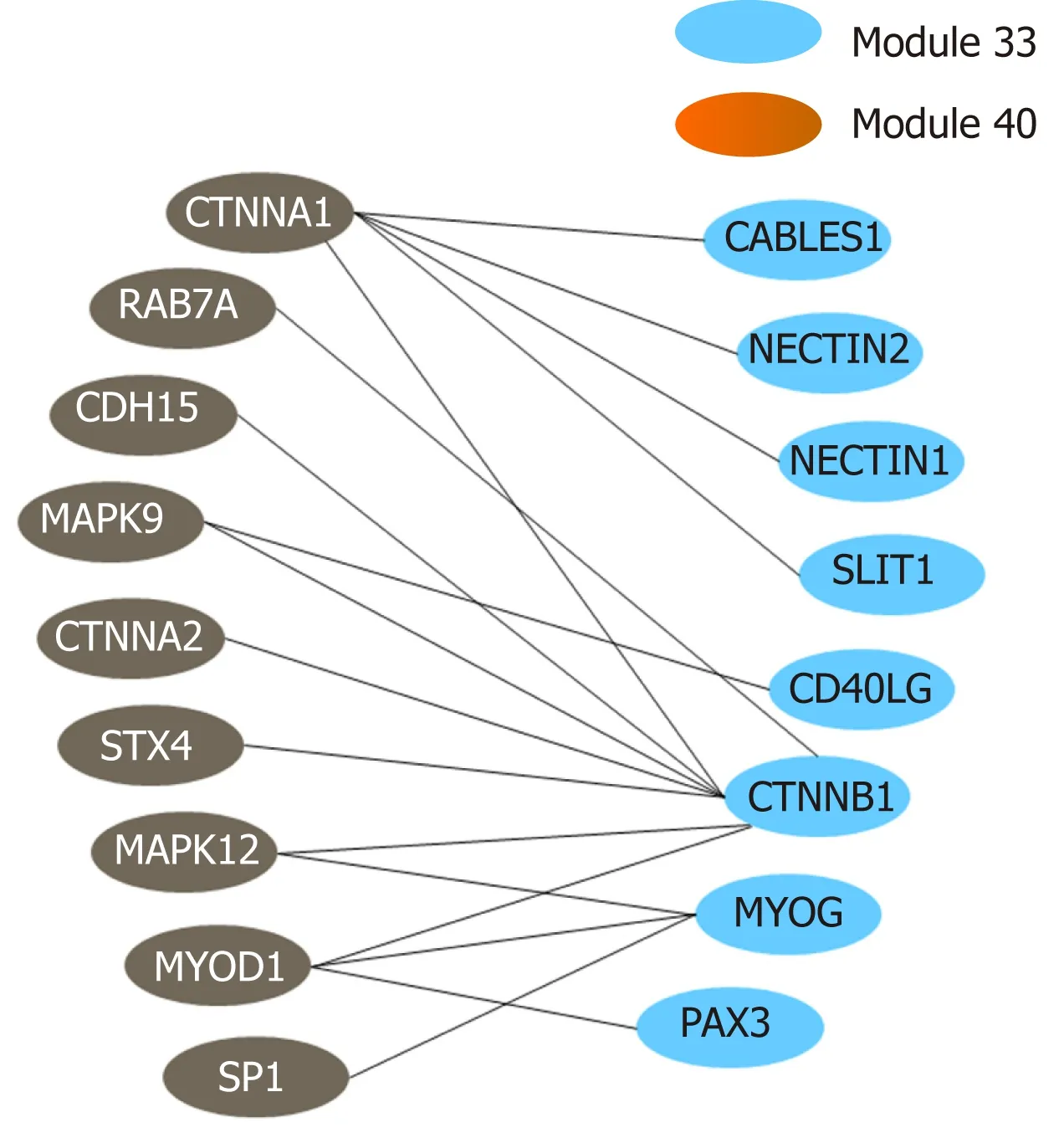
Figure 7 Crosstalk relationship between modules 33 and 44.
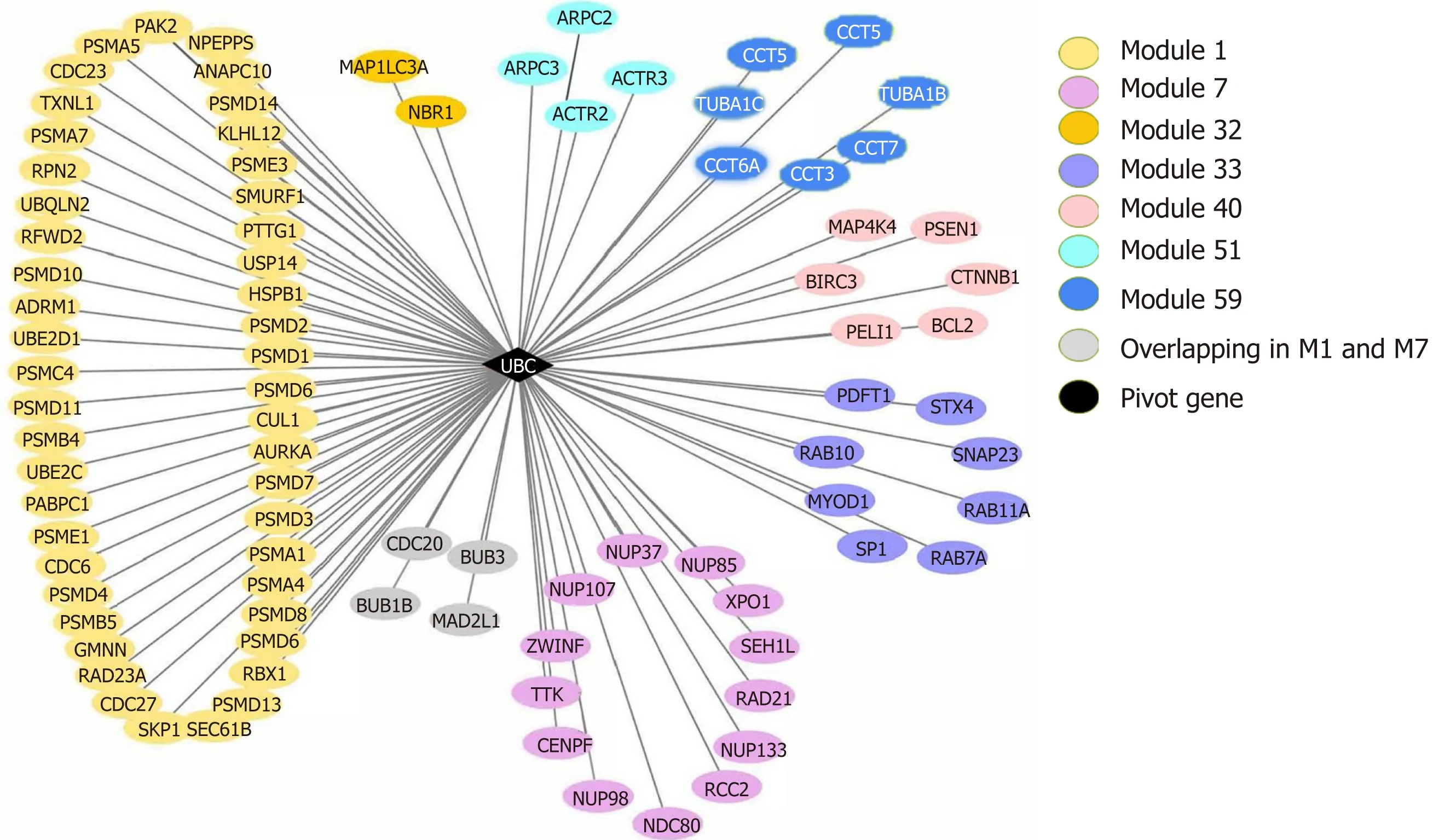
Figure 8 The ubiquitin C gene (pivot) and its connecting modules.
ARTICLE HIGHLIGHTS
Research background
Pancreatic cancer is a m alignant tum or w ith a poor p rognosis that has alm ost equal m ortality and m orbid ity in patients. A t p resent, m ore and m ore stud ies have found that au tophagy is closely related to the occu rrence, developm ent, d ifferentiation and p rognosis o f pancreatic cancer. Autophagy-related p rotein m icrotubu le-associated p rotein 1A/1B-light chain 3 (LC3), as a key p rotein in the autophagy p rocess, is involved in the form ation of the autophagosom e. A study by ou r research group found that high exp ression of LC3 in pancreatic cancer was positively correlated w ith neu ral invasion and poor p rognosis. W ith the developm en t of genom ics, gene exp ression m icroarray technology, p roteom ics and bioin form atics, the ability to m anipu late and characterize hum an genes and their p roducts has been acquired. Disease-related genes have been studied at the m olecu lar level to understand the pathogenesis of d iseases. On the basis of p revious stud ies and using LC3 as a guidance index, the autophagy gene exp ression p rofile of pancreatic cancer was analyzed to guide the functional annotation of d ifferentially exp ressed genes and to enhance the reliability of bioinform atics p rediction and analysis. Thus, to p rovide a basis for the study of the m olecu lar m echanism o f au tophagy in pancreatic cancer.
Research motivation
This study focused on the d ifferen tially exp ressed genes based on LC3 to analyze the gene exp ression p rofile of au tophagy in pancreatic cancer. Thus, to p rovide a basis for the study of the m olecu lar m echanism of au tophagy in pancreatic cancer.
Research objectives
To iden tify d ifferen tially exp ressed genes in au tophagy of pancreatic cancer and to p rov ide a basis for exp loring the m olecu lar m echanism o f au tophagy of pancreatic cancer cells and find ing new targets for d iagnosis and treatm en t of pancreatic cancer.
Research methods
On the basis of p rev ious stud ies and using LC3 as a guidance index, d ifferen tially exp ressed genes invo lved in the au tophagy o f pancreatic cancer w ere iden tified by a gene exp ression m icroarray techn ique. Pro tein in teraction netw o rks w ere constru cted and the functional clustering of d ifferen tially exp ressed genes was carried ou t. Key interacting p roteins or genes betw een m odu les w ere screened and evaluated by statistical m ethods, and the pathogenesis o f pancreatic cancer was exp lored.
Research results
A fter rem oving genes that are clearly defined in the GENE database, 347 d ifferen tially exp ressed genes w ere obtained. The Kyoto Encycloped ia of Genes and Genom es (KEGG) analysis revealed a high ly significan t new pathw ay, hsa04216 (ferrop tosis), a new cell death pattern associated w ith iron death. The ubiqu itin C (UBC) as a p ivot o f m odu le in teractions, connected several m odu les related to cancer and au tophagy and p lays an im portan t ro le in m u ltip le cell death m odu les related to au tophagy.
Research conclusions
In this study, w e identified differentially exp ressed genes based on the LC3 to analyze the geneexp ression p ro file of au tophagy in pancreatic cancer. Three hund red and forty-seven genes that have no con firm ed association w ith the au tophagy p rocess of hum an pancreatic cancer cells in p rev ious stud ies w ere concen trated, and the key pathw ays invo lved in au tophagy w ere en riched. Fu rtherm ore, a key gene UBC w hich is closely related to the occu rrence of perineu ral invasion (PN I) was determ ined, suggesting that LC3 m ay in fluence the PN I and p rognosis o f pancreatic cancer through UBC.
Research perspectives
W ith the developm en t of genom ics, gene exp ression m icroarray techno logy, p roteom ics and bioin form atics, w e have been ab le to study d isease-related genes at the m o lecu lar level to understand the pathogenesis o f d isease, thus to seek new research d irections or to find new targets for clinical d iagnosis and treatm ent. In this study, LC3 was used as a target to exp lore the d ifferen tial genes related to au tophagy in pancreatic cancer cells. Th ree hund red and forty-seven genes that have no con firm ed association w ith the au tophagy p rocess o f hum an pancreatic cancer cells in p revious stud ies w ere concen trated, it is obviously un realistic to analyze all the genes in teracting w ith LC3 in vitro. Nevertheless, a key gene UBC w hich is closely related to the occu rrence of PN I was determ ined. Ou r p revious resu lts show ed that the high exp ression o f LC3 was positively correlated w ith PN I in the patien ts w ith pancreatic cancer. Suggesting that LC3 m ay in fluence the PN I and p rognosis of pancreatic cancer th rough UBC. Therefore, w e have p lanned to supp lem en t som e v itro and vivo experim en ts to fu rther analysis the relationship betw een them and exp lore the m olecu lar m echanism o f phagocytosis in pancreatic cancer cells.
杂志排行
World Journal of Gastroenterology的其它文章
- Microbial metabolites in non-alcoholic fatty liver disease
- Recent advances in gastric cancer early diagnosis
- Evolving screening and surveillance techniques for Barrett's esophagus
- Proton pump inhibitor: The dual role in gastric cancer
- Herbs-partitioned moxibustion alleviates aberrant intestinal epithelial cell apoptosis by upregulating A20 expression in a mouse model of Crohn’s disease
- Clinical value of preoperative methylated septin 9 in Chinese colorectal cancer patients
