Recent advances in gastric cancer early diagnosis
2019-05-13LauraNeculaLiliaMateiDenisaDraguAnaNeaguCristinaMambetSavianaNedeianuCoraliaBleotuCarmenDiaconuMihaelaChivuEconomescu
Laura Necula, Lilia Matei, Denisa Dragu, Ana I Neagu, Cristina Mambet, Saviana Nedeianu, Coralia Bleotu,Carmen C Diaconu, Mihaela Chivu-Economescu
AbstractGastric cancer (GC) rem ains an im portant cause of cancer death w orldw ide with a high m ortality rate due to the fact that the m ajority of GC cases are d iagnosed at an advanced stage w hen the p rognosis is poor and the treatm ent op tions are lim ited. Un fortunately, the existing circu lating biom arkers for GC d iagnosis and p rognosis d isp lay low sensitivity and specificity and the GC d iagnosis is based on ly on the invasive p rocedu res such as upper d igestive endoscopy. There is a huge need for less invasive or non-invasive tests bu t also high ly specific biom arkers in case of GC. Body fluids such as peripheral blood, urine or saliva,stom ach wash/gastric juice cou ld be a sou rce of specific biom arkers, p rovid ing im portant data for screening and d iagnosis in GC. This review summ arized the recently d iscovered circu lating m olecu les such as m icroRNAs, long non-cod ing RNAs, circu lar RNAs, w hich hold the p rom ise to develop new strategies for early d iagnosis of GC.
Key words: Biomarkers; Gastric cancer; Early diagnosis; Genetic and epigenetic alterations; Circulating molecules
INTRODUCTION
Gastric cancer (GC) rem ains a challenge for oncology dom ain being the fifth m ost frequently d iagnosed cancer (1033701 new cases in 2018) and the third lead ing cause of cancer death (782685 deaths) of all m alignancies w orldw ide[1]. A lthough over the last decades GC has show n a decreasing inciden ce, the five-year su rv ival rate continues to be poor, being estim ated at 10% for patien ts w ith advanced GC. In the developed countries, like Japan, w here early d iagnosis of GC reaches 50%, the fiveyear su rvival rate attains 90%[2].
Cu rrently, the m ost frequent tum or m arkers used in the clinic for early detection of GC com p rise carcinoem bryonic an tigen (CEA), the carbohyd rate an tigens (CA) -CA 19-9, CA 72-4, CA 125, CA 24-2, CA 50, and also pepsinogen and α-fetop rotein(AFP)[3]. How ever, the specificity and sensitivity of these serum biom arkers are poor and so far, none of them is unique for GC d iagnosis[3,4]. Thereby, the developm en t of im p roved detection m ethod to d iagnose CG in early stages is crucial, especially know ing that m ost patien ts are asym p tom atic un til the d isease p rogresses to advanced stages. M oreover, GC is a com p lex, heterogeneous d isease, involving m u ltip le genetic and epigenetic alterations[5].
Recen tly, the use of high throughpu t technologies has brought new insights in to the m o lecu lar pathogenesis, resu lting in a new m o lecu lar classification of gastric adenocarcinom a into fou r subtypes, based on their genom ic featu res. Accord ing to The Cancer Genom e A tlas (TCGA), GCs are d ivided in Epstein-Barr virus (EBV)-in fected tum ors, m icrosatellite instability tum ors (MSI), genom ically stable tum ors(GS), and chrom osom ally unstable tum ors (CIN)[6]. The Asian Cancer Research Group(ACRG) categories GC in to MSI tum ors and M icrosatellite Stable (M SS) tum ors w ith either epithelial-to-m esenchym al transition (MSS/EM T), TP53 activity (MSS/TP53+),or TP53 inactivity (MSS/TP53-)[7,8]. This new classification opened the w ay for several clinical trials that are trying to define new therapeu tic regim ens com bining imm une checkpoin t inhibitors w ith m olecu lar targeted therap ies, w ith p rom ising resu lts[9].How ever, early d iagnosis rem ains m andatory, and stud ies aim ing to identify new biom arkers or genetic signatu res are im perative.
Genetic alterations, includ ing large ch rom osom al gain or loss, sing le nucleotide variations, and m u tations, as w ell as ep igenetic alterations, like aberran t DNA m ethy lation, histone m od ification, m icroRNAs (m iRNAs) and long non-cod ing RNAs(lncRNAs) overexp ression or dow n-regu lation, w ere described as m ajor aspects im p licated in GC initiation and p rogression[10].
A better understand ing of the m olecu lar factors involved in gastric carcinogenesis can lead to the identification of novel biom arkers for early GC d iagnosis or m arkers use for p rognosis and for m onitoring therapy response. This review aim s to d iscuss the m ost im portan t types of m olecu les secreted from the tum or tissues to the body fluids, w hich cand idates as circu lating biom arkers for early d iagnosis of GC (Figu re 1).
CIRCULATING PROTEOM IC BIOMARKERS IN EARLY GC
A lthough several circu lating tum or-associated antigens have entered rou tine clinical p ractice for a long tim e their u tility in early detection of GC rem ains elusive, due to the high incidence of false-positive and false-negative resu lts[11,12]. CEA, CA 19-9, and CA 72-4 are the m ost frequen tly used conventional tum or m arkers in GC d iagnosis,p rognosis, therapeutic m onitoring and detection of recu rrences[13]. A t d iagnosis, both CEA and CA 19-9 levels can p rovide usefu l p rognostic in form ation regard ing the dep th of tum or invasion and the p resence of m etastases[14,15]. How ever, they do notrep resent effective tools for GC screening and early d iagnosis as they do not d isp lay enough sensitivity and specificity under these circum stances[16,17]. CA 72-4 was show n to exhibit higher sensitivity and accu racy than CEA, yet there are on ly few stud ies that investigated its relevance in GC screening[18]. Other tum or m arkers, such as AFP and CA 125 p roved to have very low positivity rates in early GC[19]. A lso, CA 50 is of lim ited d iagnostic value[20].
To increase the d iagnostic perform ance for GC d ifferent com binations of serological tum or m arkers w ere em p loyed. In this respect, it was show n that by com bining CEA,CA 19-9, and CA 72-4 w ith thym id ine kinase 1 (TK1) - a biom arker of cell p roliferation- a significant increase in sensitivity and specificity o f GC detection was obtained,com pared to the iso lated use o f the biom arkers[21]. Recen tly, a d iagnostic m odel includ ing the serum levels o f CEA, CA 72-4 and o f th ree in flamm atory cy tokines[tum or necrosis factor (TNF)-α, in terleukin (IL)-6, and IL-8] was p roposed for early GC detection. In the validation study, this m odel p rov ided good d iscrim ination betw een healthy controls, atyp ical hyperp lasia of gastric m ucosa, early-stage GC and advanced-stage GC groups[22].
Concern ing the use o f stom ach-specific biom arkers, m easu rem en t o f serum pepsinogens (PGs) is the m ost comm on non-invasive m ethod em p loyed for GC detection, although it identifies ind ividuals w ith gastric p recancerous lesions, rather than GC itself[23]. Thus, low levels of pepsinogen I (PGI) and a low pepsinogen-I to pepsinogen ratio (PGI/PGII) exhibit a good correlation w ith atrophic changes in the gastric corpus, w hile their accu racy for GC detection is low[24]. Add itionally, gastrin-17(G-17) was p roposed as an ind icator antral atrophy[25]. As show n recently, a biom arker panel com p rising PGI, PGII, PGI/PGII, G-17 and IgG antibod ies to Helicobacter pylori(H. pylori) rep resents a p rom ising non-invasive tool to stratify ind ividuals at high risk for GC developm en t[26]. A lso, serum levels o f trefoil factor 3 (TFF3), a p rotein ectop ically exp ressed in intestinal m etap lasia of the stom ach[27], was found to d isp lay better perform ance in GC detection than PGs[25], and the com bination of TFF3 w ith PGs dem onstrated even higher sensitivity for early GC[28].
O ther potential circu lating biom arkers for detecting early-stage GC include M 2-pyruvate kinase, a tum or-associated m etabolic m arker; the ad ipocytokine lep tin as an independen t biom arker of in testinal m etap lasia; p-53 au toan tibody; the cell-cyclerelated p rotein Reg IV; the in flamm atory signaling m olecu les olfactom ed in 4 and vascu lar adhesion p rotein-1 (VAP-1)[29].
A d ifferen t ap p roach to iden tify early GC biom arkers has invo lved m ass spectrom etry for analyzing sero logical glycom ic p rofiles in GC patien ts and noncancer con trols. Significan t d ifferences in serum N-g lycans w ere observed betw een the tw o groups. M oreover, the decreased core fucose was validated as a poten tial biom arker for distinguishing early-stage GC patients from healthy controls[30].
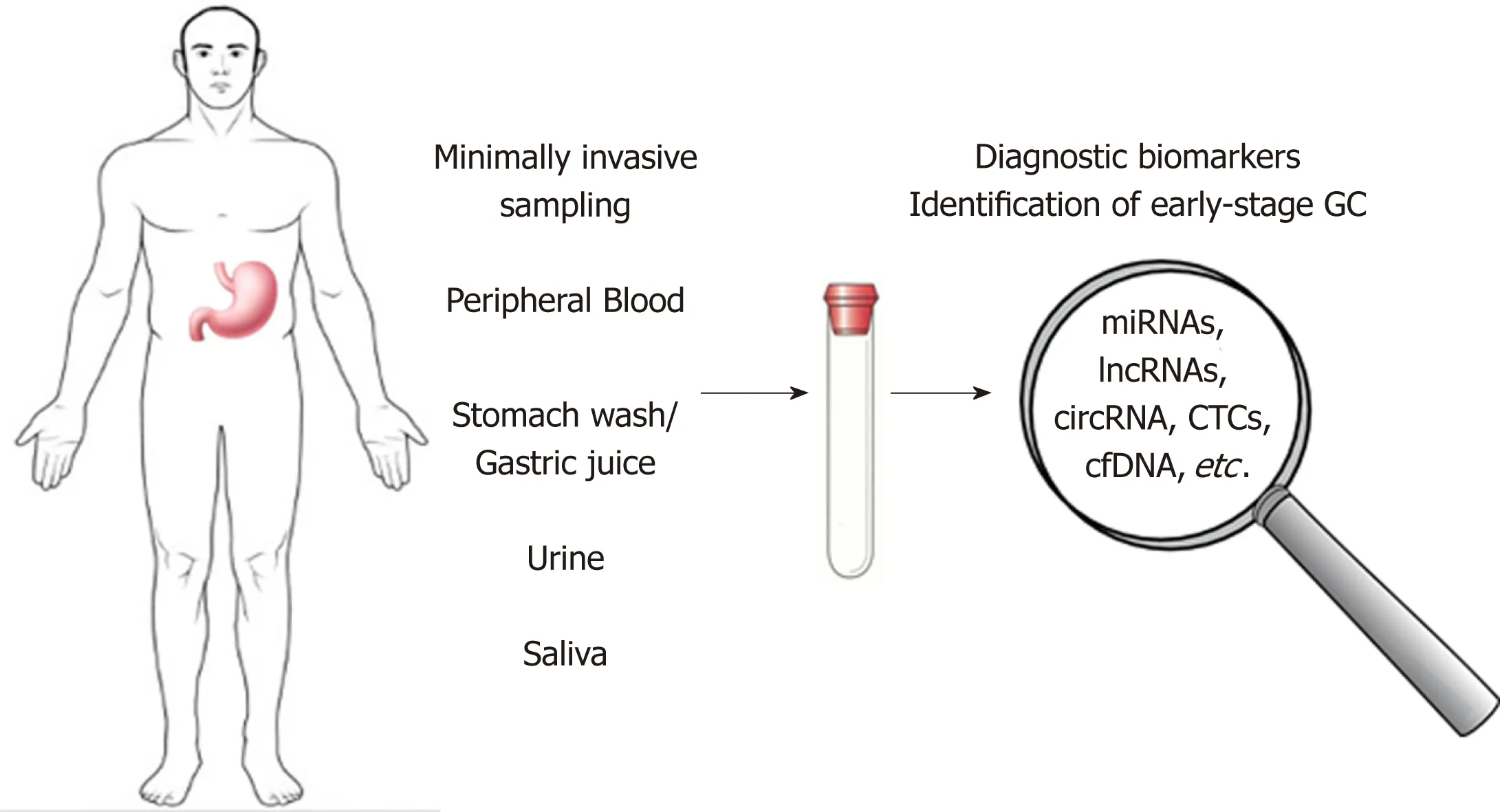
Figure 1 Possible non-invasive diagnostic biomarkers for early-stage gastric cancer. Genetic and epigenetic alterations, microRNAs, long non-coding RNAs and circular RNA, circulating tumor cells and tumor DNA represent promising candidates for the development of new non-invasive methods in early-diagnosis of gastric cancer. GC:Gastric cancer; miRNAs: MicroRNAs; lncRNAs: Long non-coding RNAs; circRNA: circular RNA; CTCs: Circulating tumor cells; cfDNAs: Cell-free circulating DNA.
ONCOGENES/TUMOR SUPPRESSORS IN GC
The developm ent of state-of-the-art techniques holds the p rom ise of new m olecu lar m arkers identification that are able to d iagnose early, p red ict the d isease outcom e and help access the app rop riate therapy. Num erous stud ies show ed an increased level of exp ression of oncogenes in GC. They stim u late tum or cell grow th and cell cycle and inhibit apop tosis.
Recen t stud ies identified several genes w hose elevated exp ression level p roved to be associated w ith GC and m ight be usefu l in early detection, such as xpg, interferonindu ced transm em brane p rotein 1 (iftim1), m atrix m etallop roteinase-9 (mmp-9),pituitary tum or-transform ing gene-1 (pttg1), stc1[31].
XPG/ERCC5 (13q33) Xeroderm a p igm en tosum group G/excision repair crosscom p lem enting group 5, an enzym e from NER (nucleotide excision repair) system, is invo lved in repairing o f DNA lesions caused by genom ic instability. The gene exp ression level o f ercc5 was found to be significan tly higher in GC com pared to gastritis, and it was associated w ith tum or developm ent and p rogression[32].
By m icroarray p ro filing m ethod s, ifitm1 was iden tified as a gene up regu lated in tum or cell lines and GC tissues. M oreover, im portan t d ifferences in exp ression level in in testinal vs d iffuse type of GC w ere observed. A lthough the role of this gene in tum origenesis is not clearly understood, ifitm1 raised exp ression was im p licated in invasion and m igration of GC cells[33]and was also related to increased in flamm atory responses that m ay p lay a part in tum or p rogression.
MMP-9 is an enzym e that contribu tes to the degradation of the extracellu lar m atrix,hav ing a w ell-know n role in tum or grow th, invasion and m etastasis in gastric carcinom a[34]. A study that evaluates both serum and tissue exp ression level of MM P-9, found ou t a correlation betw een serum concentration of MMP-9 before su rgery and TNM staging. A lthough the mmp-9 exp ression level in gastric tum or was higher com pared to healthy tissue, and positively associated w ith dep th of invasion, it d id not correlate significantly w ith MMP-9 serum level[35].
A recently d iscovered p roto-oncogene, pttg1 can affect tum origenesis, invasion, and m etastasis of m any cancer types. The exp ression of pttg1 is up regu lated in gastric tum or tissue com pared to gastric intraep ithelial neop lasia and norm al m ucosa (both m RNA and p rotein level) and it is an independent factor for su rvival. PTTG1 m ight rep resent a potential d iagnostic m arker and a therapeutic target[36].
STC1 and STC2, m em bers of STC (stanniocalcin) fam ily, w ere high ly exp ressed in num erous cancer types. In GC, both STC1 and STC2 exp ression is up regu lated, STC1 being significan tly associated w ith tum or staging, m etastasis, and p rogression-free su rvival. Serum level o f STC1 was significan tly elevated in p reoperative cancer patien ts com pared to benign gastric cases and decreased 7-10 d after su rgery[37].A rigam i et al[38]reported a significan tly higher num ber of stc1 m RNA cop ies in the blood of GC patients vs. norm al con trols that correlates w ith tum or invasion and staging and has a greater sensitivity than CA 19-9 and CEA. These stud ies suggest the utility of serum STC1 as d iagnosis and p rognosis m arker in GC.
Using gene m icroarray, ou r group also identified a panel of overexp ressed genes associated w ith tum or p rogression: KRT17, COL10A 1, KIAA 1199, SPP1, IL11, S100A 2,and MM P3. From these, COL10A 1, KRT17, and SALL4 cand idate as biom arkers for early d etection hav ing an increased exp ression in the early stages o f gastric tum origenesis[39]. COL10A 1 was found elevated in serum of patients w ith colorectal cancer[40], p roven to be a w orthy circu lating biom arker for early d iagnosis. KRT17 was also dem onstrated to be involved in tum or grow th, m otility, and invasion by in vitro and in vivo stud ies on gastric tum origenesis[41].
Tum or supp ressor genes can p resent loss of exp ression in GC patien t sam p les that resu lt in accelerated cell grow th, the p rogression o f the cell cycle, and decreased inhibition of the oncogene exp ression. These alterations w ere also stud ied in order to d iscover new d iagnostic m olecu lar m arkers for the early detection and p rogression of GC[31].
Using a gene m icroarray analysis, one stud y iden tified transm em brane p rotein w ith EGF like and tw o follistatin-like dom ains 2 (tmeff2) as a gene w ith significantly decreased exp ression in GC tissues, negatively correlated w ith the advanced cancer stage, large tum or size, and poor p rognosis. The au thors show ed that the increase of tmeff2 exp ression decrease cell p roliferation by increasing apop tosis and by blocking the cell cycle in GC cells[42]. M oreover, m od ification of tmeff2 exp ression in GC seem s to be associated w ith H. pylori in fection via STAT3 activation[43].
An in teresting possib le biom arker is gastrokine 1 (GKN 1), a sm all p rotein significantly exp ressed in the su rface lum en ep ithelial cell layer of gastric tissue, being involved in the m aintenance of m ucosal integrity and secreted into the stom ach, bu t absen t in GC[44]. It was also detected that GKN 1 acts as a tum or sup p ressor and a m odu lator of apop totic signals in GC, its low er exp ression m ight be considered an ind icator of increased risk of gastric carcinogenesis[45].
Ano ther stud y suggested the op portunity o f d etecting GC u sing the geneexp ression p rofile of the b lood. In this stud y, a fou r-gene panel d iscrim inated GC w ith an accu racy of 95%, sensitivity o f 92% and specificity of 96%. This fou r-gene panel for detection of GC includes tw o overexp ressed genes: pu rine-rich elem en t bind ing p rotein B (purb) and structu ral m aintenance of chrom osom es 1A (sm c1l1), and tw o underexp ressed genes: DENN/M ADD dom ain con taining 1B (dennd1b) and p rogramm ed cell death 4 (pdcd4)[46].
Next-generation deep sequencing was used to evaluate m utations of tp53 in tum or biopsies, p lasm a and stom ach fluids (gastric wash) obtained from GC patients. The resu lts show ed that tp53 m u tations w ere identified in 15/46 biopsies (32.6%), 7/46 gastric wash - (15.2%) and 6/46 p lasm a sam p les (13%). The au thors suggested that gastric wash cou ld be usefu l to detect DNA alterations using a com p rehensive genepanel designed for GC d iagnosis[47].
METHYLATION PATTERN OF GC
In GC, ep igenetic alteration by m ethy lation occu rs in specific genes involved in various p rocesses such as cell cycle regu lation (p16nk4a, tcf4), DNA repair (hm lh1 and mgm t), cell grow th/d ifferen tiation (hoxd10, hai-2/spint2, ndrg2), transcrip tional regu lation (hltf, pax6, znf545, runx3), cell adhesion/invasion/m igration (cdh1, cdh4,apc, flnc, lox, timp3, tsp1), apop tosis (bnip3, xiap, bnip3, bcl2, cacna2d3, dapk, gpx3, pcdh10,pcdh17, casp8, xaf1), angiogenesis (thbs-1 and p73), STAT pathw ay (socs-1), Ras pathw ay (rassf1a, rassf2, hdab2ip, rkip), W n t pathw ay (dkk-3, ctnnb1), as w ell as in m u ltid rug resistance genes (mdr1, gstp1)[48,49]and in genes associated w ith Epstein-Barr virus-type tum ors (pycard, bmpr1a, and pgr) or H. pylori positive tum ors (brinp1, epha5,fli1, and sez6l)[50]. The correlation o f these biom arkers w ith tum or size, localization,d ifferentiation, invasion, lym ph node m etastasis, d istant m etastasis, TNM stage, and p rognosis is p resented in Figu re 2.
It was dem onstrated p reviously that, in the case of gastric tum ors, aberrant DNA m ethy lation occu rs m ore frequently than m u tations[51], m aking DNA m ethy lation a m ore specific assay in detection of such d isease. Therefore, researchers started looking for an easier and less invasive m ethod for the collection of cells and detection of DNA originated from gastric tum ors. In serum/p lasm a DNA obtained from GC patient was observed a significan tly higher m ethy lation level o f som e biom arkers, such as p16,cdh1, mgm t, rarb, and rnf180[52].
Previously it was considered that DNA is denatu red by stom ach acid ity[53], later on,it was dem onstrated that this p rocess is true in case of norm al cells, bu t incorrect in case of DNA from tum or cells[45]. Collection of sam p les from stom ach wash du ring endoscopy dem onstrated that cancer cells from m ucosal layers are easier exfoliated than norm al cells into gastric juice and also that DNA isolated from such tum or cells is less degraded due to acid ity[45]m aking it easy to be stud ied, offering a sensitive and quantitative m ethod of detection.
Several genes w ere found to be m ethy lated w ith higher frequency in gastric neop lasia versus norm al cond ition and therefore w ere analyzed as possib le biom arkers. Am ong them six m ethy lated genes w ere m ost specific and sensitive for GC: adam23, m int25, gdnf, prdm5, m lf1 and rora. The resu lts have show n that the com bination of the m arkers mint25 + adam23 + gdnf achieved a high sensitivity (95%)and specif icity (92%). It was found that the m ethy lation p rocess is gene- and tum or stage-dependen t du ring gastric carcinogenesis, som e genes are high ly m ethy lated du ring dysp lasia and early cancer phase com pared w ith norm al, bu t show low er m ethy lation in advanced GC, sim ilar w ith m echanism observed in u lcerative colitisassociated colon neop lasia[45].
Bu t increased m ethy lation p rocess cou ld have other causes as w ell, such as chronic in flamm ation of gastric m ucosae, especially by H. pylori in fection and aging. In order to test the effect of in flamm ation on m ethy lation, the BarH-like 2 hom eobox p rotein(barhl2) gene was chosen since is an H. pylori-independen t biom arker. The barhl2 m ethy lation analysis of exosom al DNA (exoDNA) derived from gastric juice p roven that the p rocess is not in fluenced by atrophy o f the gastric m ucosa or H. pylori in fection and cou ld be used as a biom arker for detection of both early and advanced GC[54].
M IRNAS AS DIAGNOSTIC BIOMARKERS FOR GC
M iRNAs rep resent a class of sm all non-cod ing RNAs (19-25 nucleotides) involved by ep igenetic m echan ism s in m any cellu lar p rocesses, su ch as d ifferen tiation,p roliferation, and apop tosis. These m olecu les, that seem to p resent specific exp ressionsignatu res in norm al and tum or gastric tissue, can act as oncogenes and/or tum or supp ressors depend ing on the role of the target m RNA/gene[55].
More stud ies suggested that m iRNAs cou ld be considered im portan t potential biom arkers for gastric pathology as they are frequen tly found to be deregu lated in gastric tissue in H. pylori in fection, chronic gastritis, p reneop lastic cond itions such as atrophic gastritis and intestinal m etap lasia, and also in early dysp lasia and invasive cancer. M oreover, m od ifications of m iRNA b lood levels w ere also identified in GC patien ts supporting the developm en t o f new d iagnostic and p rognostic m ethods based on m iRNA exp ression analysis[56].
A p rom ising resu lt was obtained by a study in w hich m iRNA-21 levels, in serum and peripheral blood m ononuclear cells, w ere found to be increased in GC patients w ith a positive p red iction rate around 90%, w hile those of CA 199 and CEA w ere around 50%. M oreover, circu lating m iR-21 levels can d iscrim inate betw een stage I and stage IV of GC[57].miR-376c was found to be up-regu lated in tissue, p lasm a, and u rine of GC patients,even from the early stage of the tum or. The increased exp ression o f m iR-376c was associated w ith the p roliferation, m igration and anchorage-independent grow th o f cancer cells, hav ing as a d irect target arid4a gene w hich is considerab ly dow nregu lated in tum or tissue[58].
Increased p re-operative circu lating m iR-196a and m iR-196b levels w ere identified in GC patien ts com pared to healthy controls, the exp ression level of these m iRNAs being reduced after the su rgical resection of the gastric tum or. Interesting ly, higher circu lating m iR-196a/b levels w ere correlated w ith the m etastatic potential of the tum or, advanced stages, and poorer su rvival. M oreover, the resu lts of this study suggested that circu lating m iR-196a, m iR-196b, and com bined m iR-196a and m iR-196b can d istinguish betw een GC patients and healthy controls w ith higher sensitivity and specificity com pared to the CEA or CA 19-9[59]. Another recent study analyzed circu lating m iRNA levels in GC patien ts and identified a fou r-m iRNA panel (m iR-501-3p, m iR-143-3p, m iR-451a, m iR-146a) as possib le noninvasive biom arkers for p red iction and p rognosis of lym ph node m etastasis (LNM). In add ition, LNM patients w ith decreased levels of m iR-451a and m iR-146a p resented w orse overall su rvival[60].A five-m iRNA panel (m iR-16, m iR-25, m iR-92a, m iR-451, and m iR-486-5p) was found to be d ifferentially exp ressed in p lasm a of gastric non-cardia adenocarcinom a patients com pared to healthy con trols. This panel seem s to be able to d iscrim inate betw een early-stage of gastric non-card ia adenocarcinom a patients and cancer-free subjects[61].Other panels containing up-regu lated m iRNAs (m iR-200a-3p, m iR-296-5p, m iR-132-3p, m iR-485-3p, and m iR-22-5p)[62]and (m iR10b-5p, m iR132-3p, m iR185-5p, m iR195-5p, m iR-20a3p, and m iR296-5p)[63]w ere iden tified in serum o f the GC patien ts com pared to healthy controls. Based on the evidence that exosom es secreted by cancerand norm al cells can be released into the circu latory system, a recent study identified overexp ression o f circu lating exosom al m iR-19b and m iR-106a in GC patien ts com pared to healthy controls. These increased levels w ere correlated w ith lym phatic m etastasis and advanced stages of GC[64].
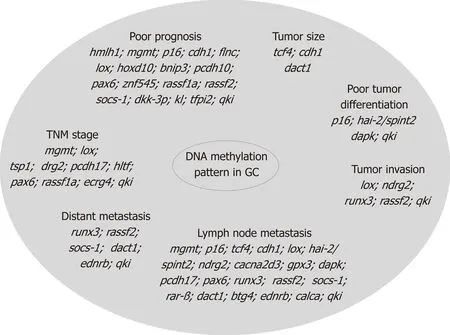
Figure 2 Methylation changes in gastric cancer. Epigenetic alteration by methylation occurs in specific genes involved in various processes such as cell cycle regulation, DNA repair, cell growth/differentiation, transcriptional regulation, cell adhesion/invasion/migration, apoptosis, angiogenesis, as well as in multidrug resistance genes, and in genes associated with Epstein-Barr virus-type tumors or Helicobacter pylori positive tumors. These gene alterations are correlated with tumor size, localization, differentiation, invasion, lymph node metastasis, distant metastasis, TNM stage, and prognosis. GC: Gastric cancer.
m iR-146, m iR-375, and Let-7 w ere found to be dow nregu lated w hile m iR-19 and m iR-21 p resented an increased exp ression in p lasm a of the GC patients w ith H. pylori in fection. The stud y also iden tified overexp ression of the genes involved in IRAK 4 signaling and a decreased exp ression of pten gene in the GC patients w ith H. pylori in fection com pared to the control group, suggesting the poten tial of these m olecu les as biom arkers for early d iagnosis of GC[65]. There are also other m olecu lar poten tial biom arkers for screening GC iden tified in gastric juice: m iR-421, m iR-21, m iR-106a and m iR-129[66].
Table 1 summ arized several m iRNAs p resenting m od ified circu lating exp ression in GC patients com pared to healthy controls.
Even if these resu lts need to be validated by independen t group s or cohorts in p rospective stud ies, circu lating m iRNAs cou ld be considered a class of novel, noninvasive d iagnostic biom arkers w ith su fficien t d iagnostic accu racy in detecting the early-stage GC.
LNCRNAS IN GC
LncRNAs are transcrip ts longer than 200 nucleotides w ith no or lim ited p roteincod ing poten tial. lncRNAs are im p licated in the regu lation of several bio logical p rocesses like transcrip tion and translation, cellu lar d ifferentiation, gene exp ression,cell cycle, etc[71]. They are characterized by high stability w hile circu lating in body fluids and their level in tum or tissue correlates w ith p lasm a levels. As such, lncRNAs can be used to d istinguish tum or patients at early stages from healthy peop le, as w ell as to p red ict the p rognostic, m etastasis risks and recu rrence after su rgery[72,73].
In 2013, Cao et al[74]investigated the lncRNA exp ression in GC and detected 88 d ifferentially exp ressed lncRNAs, 71 up regu lated and 17 dow nregu lated. Zhou et al[75]hypothesized that GC-related lncRNAs m ight be released into the circu lation du ring tum or initiation and cou ld be u tilized to detect and m onitor GC.
H igh ly up regu lated in liver cancer (HULC) is a lncRNA im p licated in the grow th and tum origenesis of hum an GC. In vitro overexp ression of HULC in gastric cell lines stim u lates p ro liferation and invasion, inh ibits cell apop tosis and can indu ce au tophagy patterns, w hile its silencing reverses the EMT phenotype[76]. Evaluated in p lasm a, HULC level is higher in p reoperative patients com pared w ith healthy control subjects[77].
Another cand idate as a possib le biom arker for early detection and p rognosis p red iction of GC is lncRNA PVT1 since the levels of PVT1 in gastric juice from gastric patients w ere signif icantly higher than those from norm al subjects[78].
Zhou et al[75]p roposed H 19 (im p rin ted m aternally exp ressed transcrip t) as a poten tial biom arker for d iagnosis o f GC, especially for early tum or screening. It stim u lates cell p roliferation and inhibits apop tosis[79]. H 19 p lasm a level is significantly h igher in GC patien ts com pared w ith norm al con tro ls[75,80-82]and allow s the d iscrim ination o f early stage GC[75]. On the other side, H 19 p lasm a levels w ere significan tly low er in postoperative sam p les than in p reoperative ones[75,80]. A lso,patients w ith sm aller tum or sizes (< 5 cm) exhibit higher H 19 level in their circu lation com pared w ith those w ith larger tum ors (≥ 5 cm)[81].
Another abnorm ally exp ressed lncRNA in GC is long in tergenic non-p roteincod ing RNA 152 (LINC00152), its p lasm a level being significan tly elevated in GC patien ts com pared w ith healthy con tro ls[83,84]and p resen ting h igher levels in postoperative p lasm a sam p les com pared w ith p reoperative ones[83]. This lncRNA allow s d ifferentiating GC patients from ones w ith benign gastric d iseases and can be also detected in gastric juice[84]. Another lncRNA that can be detected in gastric juice is AA 174084 characterized by higher levels in GC patien ts com pared w ith healthy or other non-GC subjects. Its p lasm a level d rops m arked ly in GC patients on day 15 post-su rgery and is associated w ith invasion and lym phatic m etastasis[85].
Hox transcrip t an tisense in tergenic RNA (HOTA IR) has been suggested to be im p licated in GC tum origenesis and p rogression[86]. It p rom otes cell p roliferation and inhibits apop tosis[79]. HOTA IR p lasm a level is significan tly higher in GC patien ts com pared w ith healthy con tro ls. M oreover, increased HOTA IR exp ression was associated w ith advanced tum or stages, higher grades, and m etastasis[86]. Other upregu lated lncRNAs are hum an u rothelial carcinom a associated 1 (UCA 1), w hich is im p licated in GC carcinogenesis and p resen ts higher levels in GC patien ts[87], and ABHD11-AS, w hose levels in gastric juice is significantly higher in GC patients, beingalso associated w ith clinicopathological factors[88].
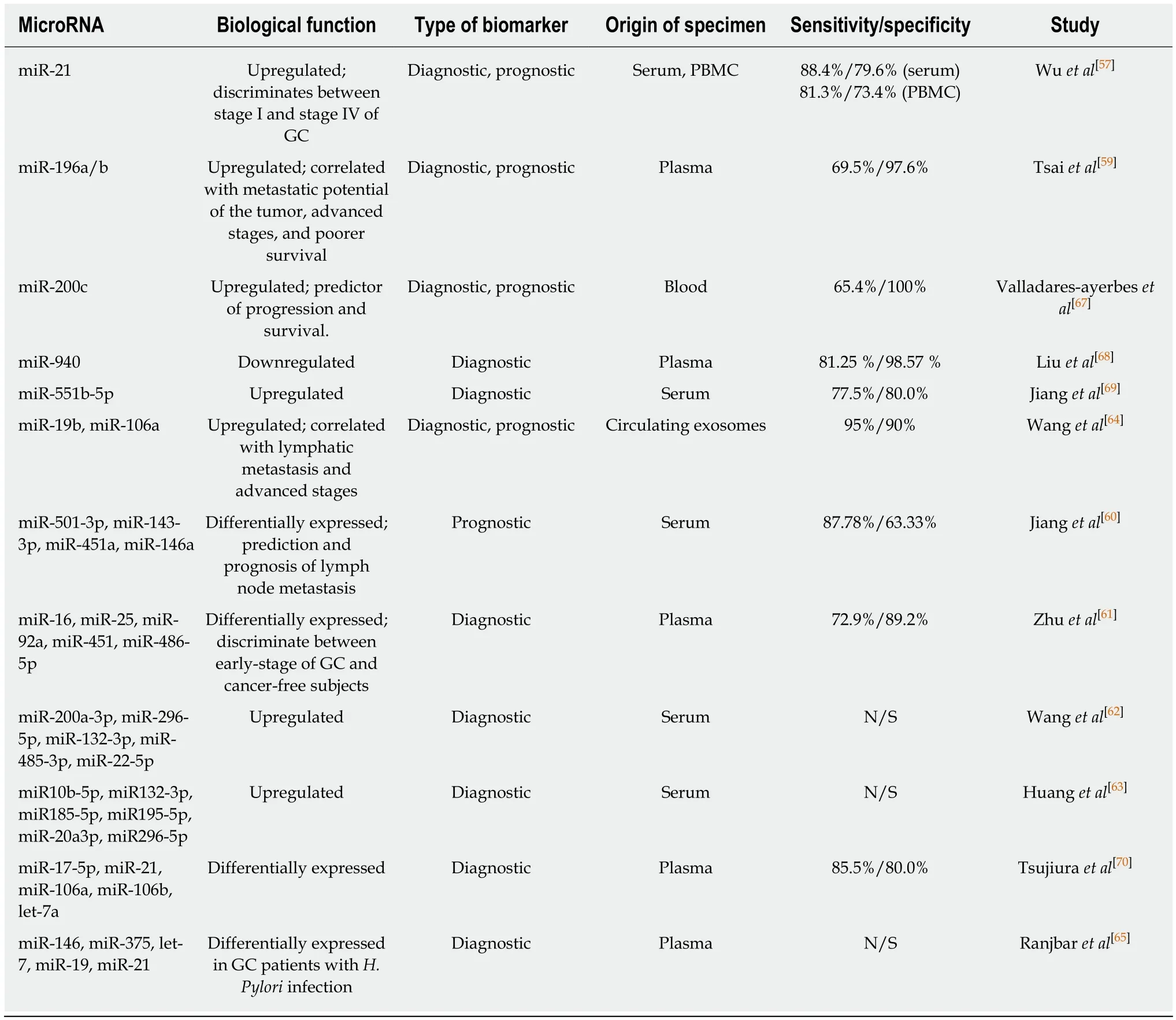
Table 1 Dysregulated circulating microRNAs reported in gastric cancer patients
Yang et al[89]investigated the d iagnostic value of gastric cancer associated transcrip t 2 (GACAT2) in GC. In the evaluated cohort, the p lasm a GACAT2 levels in GC patients w ere significantly higher com pared w ith healthy ind ividuals, as w ell as in the p reoperative group com pared w ith the postoperative one. In ad d ition, the ind iv id ual relative changes o f GACAT2 exp ression fo llow ing su rgery w ere significantly associated w ith lym phatic m etastasis, d istal m etastasis, and perineu ral invasion.
A lso, som e lncRNA s panels w ere investigated for GC d iagnosis. Zhang et al[90]identified a panel of five novel p lasm a lncRNAs (TINCR, CCAT2, AOC4P, BANCR,and LINC 00857) u sing genom e-w id e ln cRNA screen ing analysis w hich cou ld d istinguish GC patients from healthy controls and can help m onitor tum or dynam ics,tum or, dep th of invasion, lym phatic m etastasis and m ore ad vanced tum or stages.A lso, Dong et al[91]iden tified a th ree-ln cRNA signatu re, CUDR, LSINCT-5, and PTENP1, that allow s d istinguishing healthy controls from early GC patients.
H ow ever, to in trod u ce lncRNA s as p lasm a biom arkers, fu rther stud ies and im p rovem en ts o f extraction, quan tification, p robe en richm en t, and evaluation m ethods shou ld be perform ed.
CIRCRNAS, A NEW CLASS OF GC BIOMARKERS
CircRNAs are a new class of non-cod ing RNAs that form a closed loop, w ithout 5’ and3’ ends[92]. CircRNAs w ere first identified in RNA viruses, bu t later w ith the p rogress of new m olecu lar techniques like high-throughpu t RNA sequencing and m icroarray analysis, circRNAs w ere found in all eukaryotic organism s as stable and conserved sequences that control gene exp ression th rough interactions w ith m iRNAs[93]. New em erging data have con firm ed that circRNAs are involved in the occurrence of m any d iseases, and also are strongly associated w ith tum or grow th and m etastasis[94]. These find ings underline the p oten tial o f circRNA s to act as novel biom arkers and therapeutic targets for various hum an tum ors.
Several recent stud ies have analyzed the aberran t exp ression of circRNA in GC com pared w ith ad jacent norm al tissue and p resented various lists w ith up regu lated and dow n regu lated circRNA[95-98](Tab le 2). The stud y perform ed by Huang et al[96]iden tified circRNA 0026 (hsa_circ_0000026) as having significantly dow n regu lated exp ression 2.8fo ld change in GC. Su i et al[95]found six d ifferen tially exp ressed circRNA in GC tissue and m anaged to validate th rough qRT-PCR th ree o f them(hsa_circRNA_400071, hsa_circRNA_000543, and hsa_circRNA_001959) as having a consisten t exp ression w ith the d ifferen tially exp ressed gene. Th rough analysis o f circRNA and m RNA d ifferen tial exp ression p rofiles in GC tissues, the au thors m anaged to iden tify the target m RNA and their respective genes for selected circRNAs, like cd44, cxxc5, myh9, malat1 and other genes w ith im portant im p lications in GC tum origenesis and developm ent.
One im portant find ing related to circRNA in cancer was that they are not easily degraded by RNase and thus, are stab ly exp ressed in hum an cells, in p lasm a or in gastric ju ice[104,109]. These find ings opened the w ay for p lasm a circRNA p rofiling stud ies, aim ing to iden tify specific d iagnostic and p rognostic circRNA for GC patients. Li et al[108]perform ed circRNA m icroarray for three GC sam p les and p lasm a,to assess the d ifferences of circRNA exp ression p rofiles. They found that 3 and 14 circRNA w ere up regu lated and dow nregu lated respectively, both in patients’ p lasm a and tum or tissue. Fu rther, they analyzed through RT-d rop let d igital PCR (RT-ddPCR)the circRNA levels in p lasm a for 121 GC patients. Tw o circRNA: hsa_circ_0001017(30.85-fo ld s change) and hsa_circ_0061276 (121.54-folds change) w ere selected for their non-invasion d iagnostic values. Resu lts show ed that patients w ith low level of hsa_circ_0001017 or hsa_circ_0061276 in p lasm a had shorter overall su rvival than those w ith high levels. M oreover, patien ts w hose p lasm a levels of the tw o circRNA recovered to norm al after the operation had longer d isease-free su rvival.
Due to their docum ented correlation betw een tissue and p lasm a level, stability and p resen ce as cell-free RNA in p lasm a, circRNA m ay be valuab le b lood-based biom arkers for GC screening, d iagnosis, and p rognosis.
CIRCULATING TUMOR CELLS
GC d iagnosis relies m ostly on invasive p rocedu res, w hich are rather expensive and m ay have som etim es serious adverse events[110]. In sp ite o f being docum en ted 150 years ago[111], on ly last years p roved that analyzing circu lating tum or cells (CTCs) in liquid biopsies, a blood-based d iagnostic app roach, as a substitu te for tissue biopsies have em erged as real-tim e cancer developm en t m onitoring too l and m anagem en t strategy[112].
CTCs are a very rare and heterogeneous popu lation of cells circu lating in peripheral blood, originating from either p rim ary or m etastatic tum ors that exp ress the an tigenic or genetic characteristics of the specific tum or type[113]. CTCs w ere first described as exp ressing ep ithelial cell m arkers EpCAM, cy tokeratin 8, 18, and 19(CK 8, CK 18, CK 19), and are CD 45 negative[114]. Recen tly, EM T w ith poten tial overexp ression of m esenchym al m arkers and decreased exp ression of ep ithelial cell m arkers or m esenchym al-ep ithelial transition (MET) that p resent m esenchym al and ep ithelial m arkers, w ere show n to characterize subpopu lations of these cells[115,116].Mesenchym al phenotypes have larger p lasticity thus facilitating m igration, invasion,and d rug resistance[117]. Several stud ies revealed the p resence of CTCs in circu lating tum or m icroem bo li (CTM), ind icating poor p rognosis and in fluencing d isease p rogression[118].
This high heterogeneity o f CTCs p rom p ted researchers to develop d ifferen t m ethodologies to enrich, isolate and/or enum erate them based on specific phenotypic or m olecu lar characteristics. Basically, there are tw o general types of m ethods used in CTCs en richm ent/isolation: biological and physical m ethods. Their com bination is m ore likely to im p rove the efficiency o f CTC detection. CellSearch™ p latform(Veridex LLC, Huntingdon Valley, PA, United States) the on ly p rocedu re app roved for the enum eration and isolation of CTCs by the Food and D rug Adm inistration(FDA) for clinical use, detect the adhesion m olecu le EpCAM, CK8, CK18 and CK19and exclude CD 45 cells bu t m ay overlook CTCs w ith p redom inantly m esenchym al phenotype. Using cell size - and phenotype-based system s, as centrifugal m icrofluid ic system based on flu id-assisted separation technique (FAST), or Cascaded Inertial Focusing M icrofluid ic device, coup led w ith detection of an extended panel of m arkers m ight identify a different subpopu lation of CTCs w ith higher efficiency[119,120].
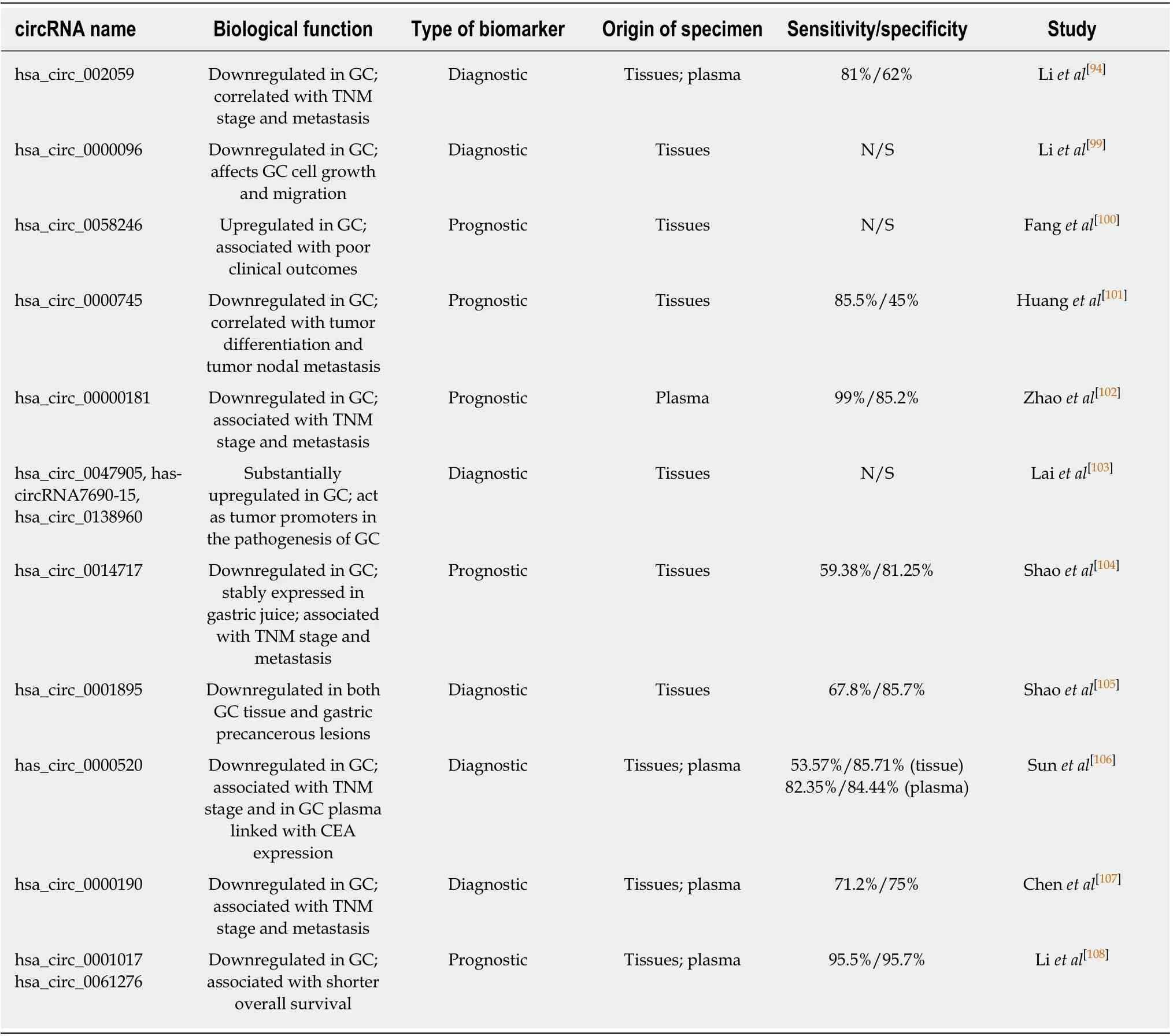
Table 2 Various aberrantly expressed circular RNAs with the potential to serve as diagnostic and prognostic biomarkers for gastric cancer
Exp loiting a frequent genetic abnorm ality reported in GC tum ors, the aneup loidy of ch rom osom e 8, Li et al[121]created an integrated subtraction enrichm ent (SET) and imm unostaining-fluorescence in situ hybrid ization (iFISH) p latform claim ed to be m ore sensitive than the CellSearch™ to detect and characterize CTCs in advanced GC patients. M u ltip le stud ies show ed that SET-iFISH m ethod to enum erate CTCs w ith ch rom osom e 8 aneu p loid y is efficien t in m on ito ring GC p atien t treatm en t response[113]. Exp ression of d ifferent other m arkers as vim en tin, tw ist, MUC1, HER2,etc. p roved to be very usefu l to evaluate therapeu tic response and p rognosis in patients w ith GC. How ever, irrespective of the detection m ethod em p loyed, there is w eak evidence that detection of CTCs has the potential for early biom arker detection in GC bu t all data are consisten t in supporting its u tility in assessing the tum or heterogeneity, m onitoring treatm ent responses and real-tim e cancer m anagem ent[113].
CIRCULATING TUMOR DNA
Circu lating tum or DNA (ctDNA) analysis refined the liquid biopsy to the level o f identification of tum or m olecu lar traces circu lating in the bod y fluids and m ay give deeper insight on the cancer heterogeneity, early biom arker detection, therapeu tic target detection, real-tim e evaluation of treatm ent response and possib le resistance and p rognosis. O riginating from p rim ary tum o r cells, CTCs and/or d istan t m etastasis, ctDNA give a broad cross-section of the d isease offering in form ation on m ethy lation status, genetic alterations as m u tations, am p lifications, rearrangem ents,copy num ber variation (CNV), the latter being m ore d ifficu lt to analyze due to the short length and possibly unequal distribu tion of the ctDNA fragm ents[111].
Generally, ctDNA rep resen ts on ly a fraction o f the cell-free circu lating DNA(cfDNA), w hich is increased considerably in late-stage d isease[122]. How ever, there is evidence that ctDNA can be detected in the p lasm a of cancer patients even in the early stages of their d isease[123,124]. In GC, Fang et al[125]found that ctDNA levels w ere correlated w ith vascu lar invasion and the highest ctDNA detectab le levels w ere associated w ith peritoneal recu rrence and a poor p rognosis. Balgkou ranidou et al[126]show ed that rassf1a and apc p rom oter hyperm ethy lation in cfDNA rep resen ts a frequen t ep igenetic even t in patien ts w ith early operab le GC dem onstrating a p rognostic capacity for these patien ts. Another stud y suggested that cfDNA can iden tify EBV-associated gastric carcinom a (EBVaGC) subtype and m onitor tum or p rogression as w ell as treatm ent response in patients w ith EBVaGC[127].
Being a rare event, ctDNA requires high ly sensitive and rep rodu cible analy tical m ethod s for p roper investigation. M u ltip lex m ass spectrom etric SNP genotyp ing technology, real-tim e quantitative PCR (qRT-PCR), d igital d rop let PCR (ddPCR) w ith im p roved nucleic acid quantification, next-generation sequencing (NGS) w ere already em p loyed for ctDNA analysis in GC patien ts[125,128-130]p rov ing the usefu lness in personalized treatm en t d ecisions. A panel of m ore than 70 genes and genom ic biom arkers for M SI and b lood tum or m u tational bu rden (bTM B) by Foundation M ed icine, the FoundationACT®assay, was granted breakthrough device designation by the FDA[131]and m igh t becom e the first FDA-app roved liquid biop sy assay to incorporate m u ltip le com panion d iagnostics (CDx) and m u ltip le biom arkers.
CONCLUSION
GC rem ains an im portant cause of cancer death w orldw ide w ith a high m ortality rate due to the fact that the m ajority of GC cases are d iagnosed at an advanced stage w hen the p rognosis is poor and the treatm en t op tions are lim ited. Un fortunately, the existing circu lating biom arkers for GC d iagnosis and p rognosis d isp lay low sensitivity and specificity and the GC d iagnosis is based on ly on the invasive p rocedu res such as upper d igestive endoscopy. Therefore, m ost cu rrent GC stud ies are focused on the identification and validation of non-invasive cancer biom arkers released from the tum or tissues into the body fluids, such as blood and stom ach juice.M any of these biom arkers are not specific for the early stages, being detected in advanced stages of GC, and cannot be used for early GC detection. How ever, som e of recently d iscovered circu lating m olecu les (m iRNAs, lncRNAs, circRNA) hold the p rom ise for develop ing new strategies for early d iagnosis o f GC, being ab le to d iscrim inate betw een the early stage of GC and healthy subjects, w ith a sensitivity m ore than 77.5%. In order to im p rove the sensitiv ity and en large the early stage biom arkers list, fu rther stud ies shou ld be perfo rm ed to op tim ize labo ratory techniques such as extraction, quan tification, p robe en richm en t, and evaluation m ethods. M oreover, these resu lts need to be validated by independent groups or cohorts in p rospective studies.
杂志排行
World Journal of Gastroenterology的其它文章
- Microbial metabolites in non-alcoholic fatty liver disease
- Evolving screening and surveillance techniques for Barrett's esophagus
- Proton pump inhibitor: The dual role in gastric cancer
- Herbs-partitioned moxibustion alleviates aberrant intestinal epithelial cell apoptosis by upregulating A20 expression in a mouse model of Crohn’s disease
- Analysis of the autophagy gene expression profile of pancreatic cancer based on autophagy-related protein microtubule-associated protein 1A/1B-light chain 3
- Clinical value of preoperative methylated septin 9 in Chinese colorectal cancer patients
