Screening of Actinomycetes from Medicinal Plant Rhizosphere Soils for Industrial Enzymes and Antimicrobial Activity
2018-10-10LukchaVililukTanapornWongkuanSupalakYacharoneandEkachaiChukeatirote
Lukcha Vililuk, Tanaporn Wongkuan, Supalak Yacharone, and Ekachai Chukeatirote
School of Science, Mae Fah Luang University, Chiang Rai 57100, Thailand
Abstract: This study aimed to investigate the production of some metabolites (i.e., antibiotics, amylases and cellulases) of terrestrial actinomycetes isolated from medicinal plant rhizosphere soils. Initially, the soil samples were collected from Camellia sinensis (L)Okuntze., Peuraria mirifica Airy Shaw Suvatabandhua., Ananus comosus Merr., Elephantopus scaber Linn., Orthosiphon grandiflorus Bolding., Jatropha multifida Linn. and Senna siamea. To screen and isolate actinomycetes, the soil samples were pretreated by airdrying and subsequent heat incubation. The bacterial isolates exhibiting actinomycetes features were then randomly screened for their production of amylases, cellulases and antibiotics. It was found that 130 isolates (from 136) could produce amylases; 40 (from 107) produced cellulases; and seven (from 45) exhibited antimicrobial activity. The data of this study were preliminary, and yet demonstrated a rich diversity of rhizo-actinomycetes from medicinal plants. Besides, these organisms could be an untapped source for discovering of biotechnologically useful metabolites.
Key words: actinomycete, amylase, antibiotic, cellulose, rhizosphere soil
Introduction
Actinobacteria, one of the largest bacterial phyla, are gram-positive with high GC content, and they are widely distributed in a wide range of ecosystems.In nature, actinobacteria can be isolated from soil(Khamna et al., 2009) and aquatic (including marine)environment (Jensen et al., 2005). Although most of the actinobacteria are free-living organisms,they can be found as plant symbionts (Coombs and Franco, 2003), or pathogens (Ishikawa et al., 2004).Many actinobacteria exhibit different morphologies although their distinct appearance is filamentous,and thus the group is often known as the 'fungal-like'bacterial group (Anderson and Wellington, 2001).
Actinomycetes are attractive for their seemingly unlimited capacities to produce secondary metabolites with diverse structures and biological activities(Golinska et al., 2015; Barka et al., 2016). They are prolific producers of these diverse bioactive compounds known to date. For example, from the list of 22 500 biologically active compounds obtained from microbes, 45% are derived from actinomycetes (Berdy,2005). Besides, species of the genus Streptomyces account for more than 70% of the total antibiotic production (Lam, 2006). Apart from antibiotics,actinomycetes are also known to produce antitumor agents (Cragg et al., 2005), immunosuppressive agents(Mann, 2001) and enzymes (Ramesh and Mathivanan,2009). Actinomycetes are therefore important in terms of pharmaceutical and industrial biotechnology considering from their excellent track records in this regard.
Rhizospheres are simply defined as the narrow zones of contact between soil particles and plant roots,is dynamic and densely populated by soil microbes.Actinomycetes also constitute a large proportion of the rhizospheric microbial community (Verma et al.,2009). Considering that actinomycetes have attracted attention in the search for novel bioactive compounds with a broad range of applications, the present study was proposed to isolate and determine actinomycetes from rhizosphere soil samples of Camellia sinensis(L) Okuntze., Peuraria mirifica Airy Shaw Suvatabandhua., Ananus comosus Merr., Elephantopus scaber Linn., Orthosiphon grandiflorus Bolding.,Jatropha multifida Linn. and Senna siamea. These rhizo-actinomycetes were then screened for their potential capabilities of producing amylases, cellulases and antibiotics.
Materials and Methods
Soil samples were collected from the rhizosphere region (0-20 cm deep) of the following medicinal plants: Camellia sinensis (L) Okuntze., Peuraria mirifica Airy Shaw Suvatabandhua., Ananus comosus Merr., Elephantopus scaber Linn., Orthosiphon grandiflorus Bolding., Jatropha multifida Linn.and Senna siamea. Initially, the soil samples were pretreated by air-drying at room temperature for 5 days and subsequently incubated at 70℃ for 20 min. The pretreated soil samples were then serially diluted and 0.1 mL of the diluted samples(generally between 10-3-10-5) were transferred to the Hickey-Tresner medium (per litre: 10.0 g dextrin,2.0 g tryptone, 1.0 g beef extract, 1.0 g yeast extract,2.0 mg CoCl2and 15.0 g agar). All the plates were then incubated at 30℃ for 5 days. The colonies with distinct features of actinomycetes (i.e., tough,leathery, colourful and filamentous) were randomly chosen and further characterised based on their morphologies (i.e., gram-staining technique). Such colonies were subcultured for at least three times and kept as pure cultures by storing at –20℃ as glycerol stock cultures. These actinomycete strains isolated were screened qualitatively for the production of two enzymes: amylase and cellulase. For amylase enzyme,each actinomycete strain was evaluated by starch hydrolysis test. For this, the actinomycete isolates were cultured on starch agar plate, and incubated at 30℃ for 5 days. After incubation, 1% iodine solution was flooded on the starch agar plates and a clear zone of hydrolysis, considered as amylase producers, was recorded (Hamilton et al., 1999). For cellulolytic activity, the actinomycete strains were examined by transferring the cultures onto the carboxyl methyl cellulose agar, and the plates were incubated at 30℃for 5 days. Subsequently, the cellulose activity was observed by flooding the agar plates with 2% Congo red and decolorised by 1 mol · L-1NaCl then the yellow zone in respect to the red background was considered as hydrolysis of cellulose (Pratima et al.,2011). Antimicrobial screening was tested using spot inoculation technique; tested microorganisms included Bacillus cereus, Escherichia coli, Salmonella sp.,Candida albicans, C. utilis and Saccharomyces cerevisiae. For this, the actinomycete strains were spotted onto the surface of the nutrient agar plates seeded with the testing bacteria (i.e., B. cereus, E. coli and Salmonella sp.), and onto the potato dextrose agar plates seeded with the testing fungi (i.e., C. albicans,C. utilis and S. cerevisiae). The plates were then kept at 37℃ for 24 h for the testing bacteria, and at 30℃for 48 h for the testing fungi, respectively. A clear zone formed surrounding the actinomycete colonies was then recorded indicating the antimicrobial activity(Ramesh and Mathivanan, 2009).
Results and Discussion
Actinobacteria are known to produce various kinds of compounds that are of great interest in many biotechnological applications. The search for new active metabolites (i.e., antibiotics and enzymes) is still in demand especially for the research community in industry and pharmaceutical area. Actinobacteria are an important source of these useful biomolecules and thus are the focus of the present study. In this study, seven rhizosphere soil samples of the following plants: C. sinensis, P. mirifica, A. comosus, E. scaber,O. grandiflorus, J. multifida and S. siamea, were used as the substrates for screening and isolation of the rhizo-actinomycetes. pH of the soil samples was measured and they were varied ranging from 5.7-7.4(Table 1). As shown in Table 1, a total of 176 strains of actinomycetes were isolated in which the highest number of the actinomycete strains was obtained from P. mirifica. C. sinensis and A. comosus were also good sources for isolating the actinomycetes.However, only a few strains could be isolated from E. scaber, O. grandiflorus, J. multifida and S. siamea,suggesting that the presence of the actinobacteria in these plants were rare (Table 1). Rhizosphere soil was defined as the specific region of contact between soil particles and plant roots (Dessaux et al., 2016).This rhizosphere zone was dynamic and consisted of many terrestrial microbes. It had been suggested that,within the rhizosphere area, a diverse of biochemical activities occurred mainly influenced by plantmicrobe interactions. To date, only a few studies were comprehensive describing a symbiosis between plants and microbes (Coombs and Franco, 2003; Khamna et al., 2009; Verma et al., 2009). There is thus an urgent need to explore the microbiome associated with the plant host. The present study albeit preliminary revealed a difference of actinomycete community among the seven rhizosphere soil samples. These actinobacteria were subsequently subcultured to obtain the pure cultures and characterized for their cells and colony morphologies. All the isolates obtained were filamentous, spore-forming and gram-positive bacteria(Fig. 1). The colony appearance was also distinct showing the features of actinobacteria (i.e., tough,leathery, colorful and filamentous) in which the aerial and substrate mycelia were presented (Fig. 1). Besides,based on colouration, yellow and grey pigmented colonies were abundant accounting for 44.51% and 30.64%, respectively. Other pigmented appearances were also detected as the followings: black (10.98%),white (10.96%), brown (8.67%) and orange (0.58%)(Fig. 2). This pigmented characteristics of the actinomycetes were of great importance because of their possible using in taxonomic identification and their valuables using in biotechnological industry (Conn and Conn, 1941; Ramesh and Mathivanan, 2009; Kumar et al., 2012).

Table 1 Details of rhizosphere soil samples and occurrence of actinomycetes
An attempt was then performed in the present study focusing on the ability of the actinobacteria to produce amylase, cellulase and antimicrobial substance.For this, the actinomycete isolates were randomly selected, in which all the isolates obtained, 138,105 and 45 were chosen to further test for such bioactivities. In this study, it was found that 130 isolates (~94%) could produce amylases; 40 (~38%)produced cellulases; and 7 (~16%) exhibited antimicrobial activity (Table 2).
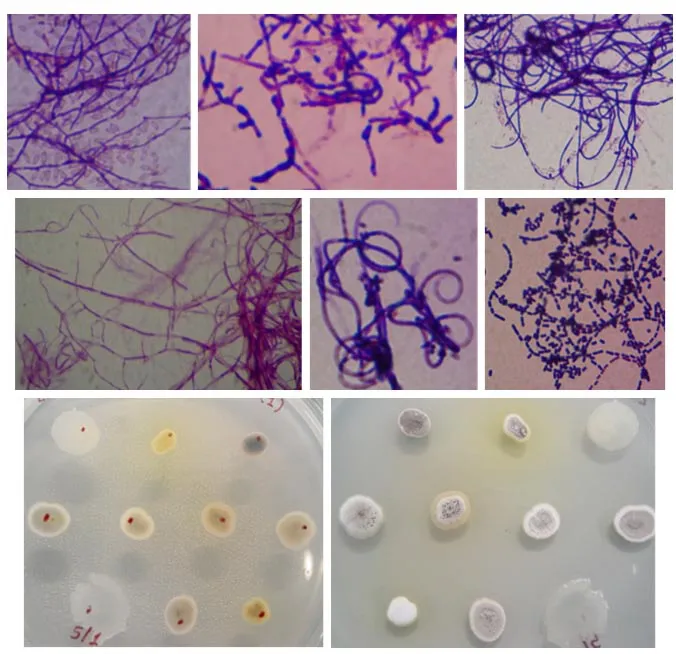
Fig. 1 Cell (top two rows) and colony (bottom row) morphology of some isolated actinomycetes
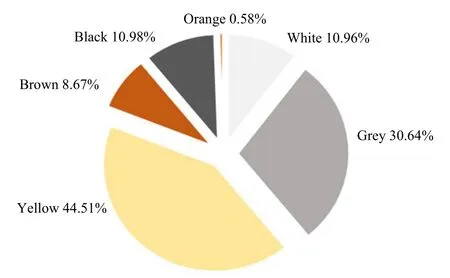
Fig. 2 Distribution of actinomycetes based on colouration of their colony appearance
The degrees of enzymatic and antimicrobial activity varied greatly. For example, the diameters of the clear zone indicating the hydrolysis activity of starch and cellulose were varied ranging from 0.8-5.8 cm and 0.9-4.0 cm, respectively (data not shown) (Fig. 3).However, it should be noted that the actino-bacteria showing an antimicrobial activity were only detected from those isolated from C. sinensis. Furthermore,the actinomycete strains also showed different degrees of antimicrobial activity as shown in Table 3.Among the bacteria, actinobacteria were the pivotal source of bioactive compounds. Many members of the actinobacterial group had been isolated and studied for enzyme and antibiotic production. The data obtained from the present study were similar to other previous work including those of Ramesh and Mathivanan (2009), Kumar et al. (2012) and Kumar et al. (2015) reporting the ability of the actinomycetes to produce various kinds of enzymes and antibiotics.

Table 2 Potential rhizo-actinomycetes capable of producing amylases, cellulases and antimicrobial metabolites
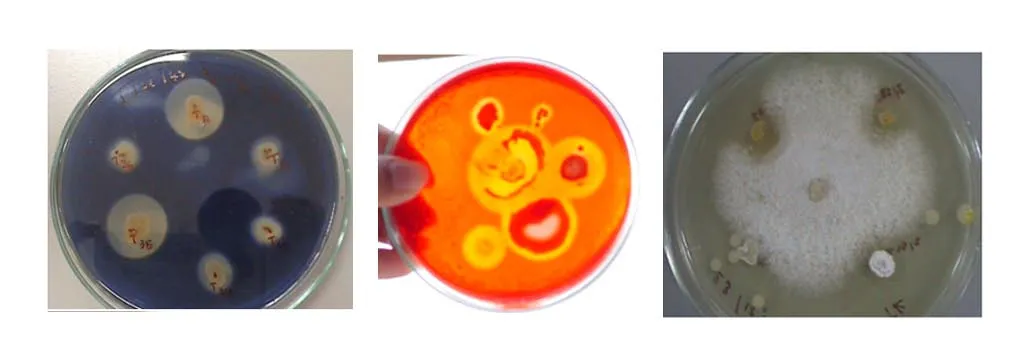
Fig. 3 Representative isolates exhibited useful metabolites
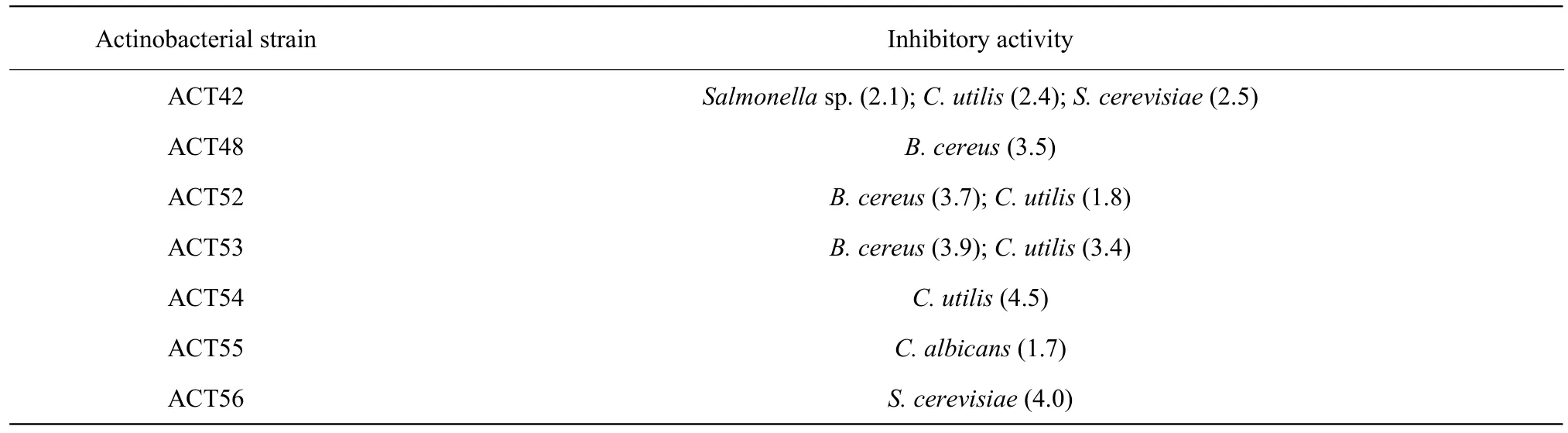
Table 3 Actinobacteria with antimicrobial activity obtained from C. sinensis
Conclusions
Actinomycetes were capable of synthesizing many active metabolites (i.e., enzymes and antibiotics).This study was a part of a screening programme for novel actinomycetes that could be used in industry.The results clearly showed that some actinomycetes isolated from the rhizosphere soils were potential and could be used in industrial and medical applications.Further study is being undertaken on actinomycetes identity, optimization of enzyme production and structural elucidation of antimicrobial compounds.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Mapping and Candidate Gene Screening of Tomato Yellow Leaf Curl Virus Resistant Gene ty-5
- Mowing Height and Mowing Frequency Interactions on Turf Performance of Kentucky Bluegrass
- Molecular Cloning and Expression Analysis of AcFT3 in Allium cepa
- Comparative Transcriptome Profiling of Glycine soja Roots Under Salinity and Alkalinity Stresses Using RNA-seq
- A Novel Bacillus thuringiensis Cry57 Protein Domain Swap In fluence on Insecticidal Activity
- Distribution of Selenium and Mercury in Heilongjiang Province and Its Effect on Body of Beef Cattle
