Mapping and Candidate Gene Screening of Tomato Yellow Leaf Curl Virus Resistant Gene ty-5
2018-10-10RenJingLiJingfuandYangHuanhuan
Ren Jing, Li Jing-fu, and Yang Huan-huan
College of Horticulture, Northeast Agricultural University, Harbin 150030, China
Abstract: To identify the inheritance pattern and perform fine mapping of ty-5 gene, P1, P2, F1, BC1 and F2 generations were obtained through a cross between CLN32120a-23 (containing ty-5 gene, P1) and S. lycopersicum Moneymaker (fully susceptible, P2). The results showed that resistance of ty-5 gene was determined by a recessive effect. Meanwhile, it was presumed that another resistance gene might be involved in mediating the resistance to tomato yellow leaf curl virus (TYLCV). In this study, fine mapping was used to map TYLCV resistance locus to an interval between NAC1 and TES2461 on the short arm of chromosome 4 with genetic distances of 0.5 and 0.8 cM, respectively. qRT-PCR results showed that four candidate genes, SlNAC1; LOC104229164; LOC101260925 and LOC101261508 having resistance-related expression patterns, were the likely target genes of ty-5. In addition, it was found that the codominant marker TES2461 could be used in marker-assisted selection (MAS) breeding. The findings of this research provided the basis for future cloning of ty-5 gene as well as MAS breeding and plant resistance mechanism studies.
Key words: tomato, tomato yellow leaf curl virus (TYLCV), genetic analysis, qRT-PCR, gene mapping
Introduction
Tomato yellow leaf curl virus (TYLCV) is considered as a major disease in cultivated tomato (Navot et al.,1991; Scott et al., 1996) and has threatened tomatogrowing areas throughout the world (Lapidot et al.,1997; Polston and Anderson, 1997). To date, five major TYLCV resistance genes have been identified in wild tomato species, including S. habrochaites,S. pimpinellifolium, S. chilense, S. cheesmaniae and S. peruvianum (Lapidot and Friedmann, 2002).Ty-1 originates from S. chilense and is mapped on chromosome 6 in tomato (Zamir et al., 1994).Moreover, Hanson et al. (2000) found that B6013 resistance is under the control of a single dominant gene. Ty-3 derived from LA2779 accession has a close genetic distance with Ty-1 gene and is mapped to chromosome 6 (Ji et al., 2007). Ty-3 is located near cLEG-31-P-16 and T1079. Further, Ty-1 and Ty-3 are mapped to a small genomic region (approximately 70 kb) on chromosome 6, which are recently identified as allelic and that they code for a RdRP (Verlaan et al., 2013). In later studies, Ty-2 is found to be a resistance locus (Hanson et al., 2006). Currently, Ty-2 is known to be located near two markers, UP8 and M1, with a genetic distance of 0.4 cM (Yang et al.,2014). Later, Ty-4 from S. chilense accession LA1932 was mapped to chromosome 3 (Ji et al., 2009). The plants containing both Ty-2 and Ty-3 or Ty-2 and Ty-4 are all shown to be resistant to TYLCV, whereas plants containing both Ty-3 and Ty-4 are all tolerant.Different genes show an additive effect, and plants containing Ty-2, Ty-3 and Ty-4 genes exhibit the highest resistance to TYLCV.
Anbinder et al. (2009) identified a resistance locus,Ty-5, which accounts for about 46.6% of the variation in disease severity index (DSI), characterizes by 34<LOD score<35 and is mapped at the vicinity of SlNAC1 marker (Anbinder et al., 2009). However,the genetic distance between Ty-5 and SlNAC1 marker is vague. Hutton et al. (2012) indicated that Ty-5 associated resistance is recessive and should thus be named ty-5 instead of Ty-5, which is cosegregated with SINAC1. Previous studies showed that plants carrying ty-5 gene show effective resistance to TYLCV in the field and can be used in breeding and resistance mechanism studies as new resistance materials. In this study, several SSR primers close to SINAC1 were selected for ty-5 gene fine mapping.The comprehensive study explored the inheritance of ty-5 gene as well as gene mapping to gain a deeper understanding of the genetic characteristics of ty-5 gene as well as the mechanism of disease resistance in response to ty-5 gene. This study provided the basis for ty-5 gene cloning and the application of ty-5 gene in tomato breeding (Hanson et al., 2000).
Materials and Methods
Plant materials and disease evaluation
CLN32120a-23 (resistant female, P1) containing ty-5 gene (kindly provided by the Asian Vegetable Research and Development Center, AVRDC) was crossed with the susceptible line Moneymaker(susceptible male, P2) (kindly provided by the Chinese Academy of Agricultural Sciences). The resulting F1plants were self-crossed to produce F2seeds, and F1plants were backcrossed with CLN32120a-23 to obtain BC1. In August, 2014, all the seedlings grew in a greenhouse under favorable conditions. A total of 1 013 individuals of F2population were used for inheritance analysis of ty-5 gene.
Whiteflies were kindly provided by the Jiangsu Academy of Agricultural Sciences. In mid-August,when P1, P2, F1, F2and BC1seedlings grew to 3-4 true leaves stage, inoculation was performed according to the method of Navot (Kalloo and Banerjee, 2006).Disease evaluation was conducted four weeks after TYLCV inoculation, and plants were scored for disease severity (DS) according to the method of Scott et al. (1996) based on the severity index. Intermediate scores (0.5, 1.5, etc.) were incorporated to allow for more precise disease severity ratings (0-1 for resistant,1.5-2.5, for intermediate resistance and ≥3 for susceptible) (Friedmann et al., 1998).
DNA extraction and marker screening
DNA samples were extracted from P1, P2, F1and F2individuals using the method of Fulton et al(1995). ty-5 gene was initially located in the interval between J04-1 and TG182 approximately 30 cM on chromosome 4 in tomato (Anbinder et al., 2009). This study selected 355 pairs of SSR molecular markers between J04-1 and TG182 for mapping of ty-5 gene(Polston and Hiebert, 2009) (http://solgenomics.net/) (http://marker.kazusa.or.jp/). The markers were screened using CLN32120a-23, Moneymaker and F2plants. Linkage analysis was then performed for these markers in 1 013 F2individuals. JoinMap 4.0 was used for fine mapping of ty-5 gene, with a minimum logarithm of odds (LOD) threshold of 3.0. A genetic map of chromosome 4 in tomato was calculated in centi-Morgans (cM). Resistance loci were placed on the linkage maps using QTL Icimapping (Chinese Academy of Sciences, CAS) (Kosambi, 2011).
Gene prediction and sequence analysis
To obtain detailed DNA sequence information, primers were designed for the candidate gene loci SlNAC1,LOC104229164, LOC101260925 and LOC101261508,using Primer 5.0 software (Table 1). The template sequences were derived from the reference genome sequence of Solanum genome network, SGN (http://solgenomics.net/). The obtained DNA sequences were submitted to NCBI database and analyzed using the Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Open Reading Frame Finder (ORF Finder,http://www.ncbi.nlm.nih.gov/gorf/gorf.html) tools of NCBI. Gene structure analysis was performed using the online tool SMART (http://smart.embl-heidelberg.de/). The statistical analyses were performed using SPSS (Version 16.0, SPSS Institute).
Quantitative real-time PCR analysis
Candidate gene expression analysis was performed using qRT-PCR. CLN32120a-23 and Moneymaker were inoculated at 3-4 true leaves stage (Rotenberg et al., 2006). Young leaves were collected at 5-day intervals (0, 5, 10 and 15 days) after TYLCV inoculation and were stored at –80℃. About three leaves of tomato plant were selected for sampling. The leaves were all the young leaves. The total RNA was extracted with three biological repeats using TRIzol reagent method. Reverse transcription was performed using the reverse transcriptase M-MLV (RNase H-)reverse transcription kit of TaKaRa, according to the operating instructions.
qRT-PCR reaction mixture contained 10 μL of 2xTrans Start Top Green qPCR Super Mix (Trans Gen,China), 1 μL of each primer, 2 μL of cDNA templates(1 : 5 dilution), and sterile distilled water to make up a total volume of 20 μL. The thermal conditions were as the followings: 95℃ for 10 min and 40 cycles of 95℃ for 5 s, 59℃ for 15 s, and 72℃ for 30 s. To detect primer demonization or other artifacts of amplification, a melting-curve analysis was performed immediately after completion of qRT-PCR (95℃ for 15 s,55℃ for 15 s, followed by slowly increasing the temperature by 0.5℃ per cycle to 95℃ with continuous measurement of fluorescence). Data analysis was performed using 2–ΔΔCTmethod (Livak and Schmittgen,2001) with EFa1, (F: 5'-CCACCAATCTTGTAC ACATCC-3' R: 5'-AGACCACCAAGTACTACT GCAC-3' ) as a reference gene for normalization(Table 1).

Table 1 Primers used for qRT-PCR analysis and candidate loci sequencing
Results
Inheritance studies of ty-5
CLN32120a-23 was found to be resistant to TYLCV,whereas the Moneymaker and F1plants were susceptible, demonstrating that ty-5 was a recessive resistance gene. Of all the 1 013 F2plants, 416 were regarded as resistant, while the remaining 597 plants were susceptible. The frequency distribution of disease scales in F2populations is shown in Fig. 1. The segregation ratio between resistant and susceptible plants in F2population corresponded to the ratio of 7R : 9S (χ27 : 9=1.85, P=0.17). The segregation ratio between resistant and susceptible plants in BC1 plants corresponded to the ratio of 1R : 3S (χ21 : 3 =2.16, P=0.14) (Table 2).
In this study, TYLCV resistance locus was mapped to the marker interval from NAC1 to TES2461 on the short arm of chromosome 4. The locus accounted for approximately 50% of the variance in TYLCV resistance among the total F2populations, which had almost equal dominance and additive value(Table 3).

Fig. 1 Frequency distribution of disease scales in F2 populations
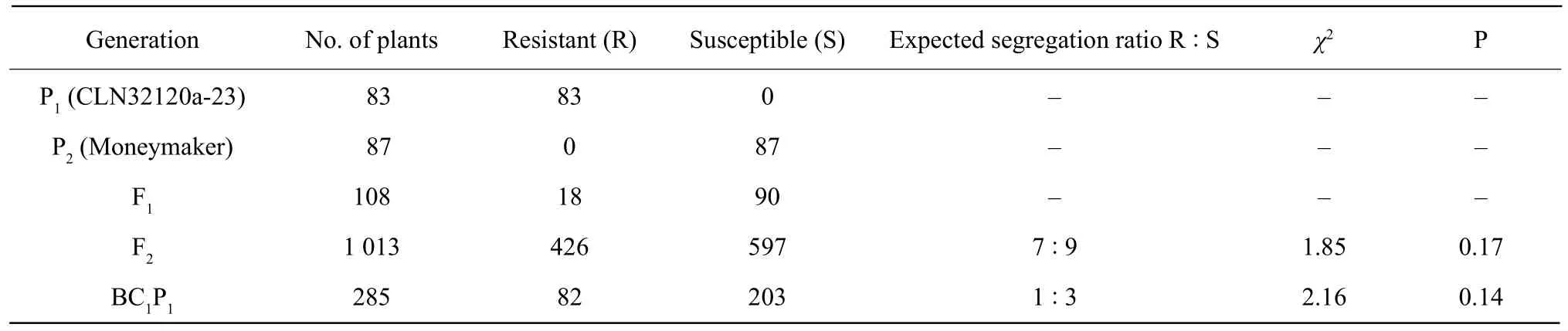
Table 2 Genetic analysis of ty-5 disease-resistance in different generations
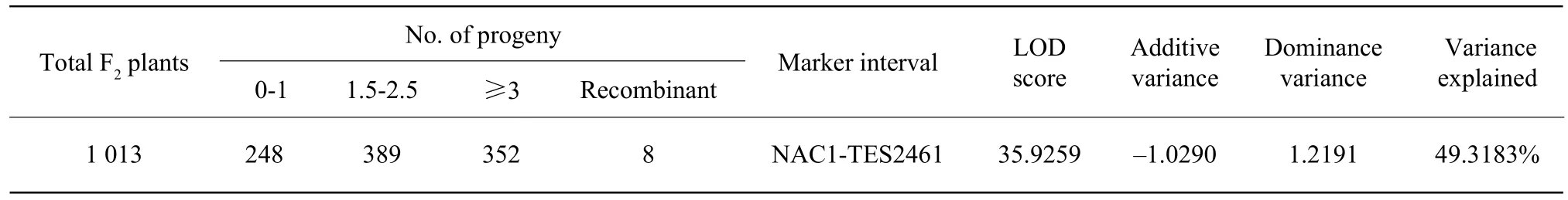
Table 3 Analysis of resistance locus in F2 mapping progeny
Fine mapping of ty-5
In this phase of the experiment, seven pairs of SSR markers that were closely linked to ty-5 on chromosome 4 were screened out. A fine genetic map of ty-5 gene was constructed. NAC1 and TES2461 were the closest flanking markers of ty-5 gene, as shown in Fig. 2a, and the genetic distances were 0.5 and 0.8 cM, respectively.
In addition, SSR marker TES2461 (forward primer:5'-GACTGCATTGGATTTGGCTT-3'; reverse primer:5'-CAATCGATGCACAAAACACC-3') was found to be linked with the resistance trait. As shown in Fig. 3,a fragment of 550 bp was amplified in CLN32120a-23,a fragment of 780 bp was amplified in Moneymaker,and both of these fragments were amplified in F1samples. Among the total F2plants, the susceptible plants showed the Moneymaker or F1genotype, all of the resistant plants showed CLN32120a-23 genotype,and eight susceptible plants showed CLN32120a-23 genotype. The inoculation and molecular marker identification results of all the F2line plants showed a near consensus. There were 33 materials that showed resistance to disease in the field, the resistance genotypes ty-5/ty-5 of molecular marker detection were 30. Therefore, the molecular detection was about 90%coincident with the results of the field identification.
Functional annotation of candidate genes
The candidate genes were screened out from the target region based on the annotations in NCBI.These candidate genes were defensin-like protein 1(LOC104229164), serine/threonine-protein kinase Aurora-3 (LOC101261508), stress-related transcription factor SINAC1 (NAC1) and histidinol-phosphate aminotransferase (LOC101260925) (Fig. 2b).

Fig. 2 Molecular marker linkage and chromosomal mapping of TYLCV resistant gene ty-5 in tomato

Fig. 3 Marker TES2461 amplification in different generations
Gene structure analysis was performed using the online tool SMART (http://smart.embl-heidelberg.de/). Results showed that SINAC1 locus proteins contained the same NAM (no apical meristem) protein that belonged to NAC domains. LOC104229164 locus proteins contained Knot1 protein, which belonged to defensin-like protein 1, LOC101261508 belonged to the protein kinase and LOC101260925 locus proteins contained aminotrane protein, which belonged to histidinol-phosphate aminotransferase.
Expression patterns of candidate genes
The relative expression level of candidate genes in CLN32120a-23 and Moneymaker was conformed by using qRT-PCR. Results showed three candidate genes: SlNAC1, LOC104229164 and LOC101261508 showed expression patterns related to the resistance response process (Fig. 4), and all the primer sequences of the candidate genes were reported in Table 1. The three candidate genes were expressed at a low level before inoculation and increased slightly after inoculation. This expression level was then increased rapidly 10 days after inoculation (TYLCV) and continued to increase during the following days. In particular, compared with 0 day, the expression levels of all the three resistance-related genes were increased on the 10th and 15th days, after inoculation (TYLCV). Nevertheless,the gene expression level of LOC101260925 was probably incompatible with being resistance related.In conclusion, qRT-PCR results in the study indicated that the expression levels of SlNAC1, LOC104229164 and LOC101261508 were compatible with being resistance related, and the three candidate genes were probably the target genes of ty-5.

Fig. 4 qRT-PCR analysis of expression of four candidate genes in susceptible (Moneymaker) and resistant (CLN32120a-23)lines
Discussion
A recessive gene ty-5
In this study, disease severity scores of CLN32120a-23, Moneymaker and F1showed that the resistance of CLN32120a-23 was controlled by a recessive resistance gene. Further, the segregation ratio between resistant and susceptible plants in F2population and BC1plants corresponded to the expected ratio of 7R : 9S and 1R : 3S, respectively. Consequently, it was presumed that ty-5 gene resistance to TYLCV was controlled by two genes, suggesting that another resistance gene might be involved in mediating the resistance to TYLCV.
Meanwhile, TYLCV resistance locus was mapped to the marker interval from NAC1 to TES2461 on the short arm of chromosome 4. This locus accounted for approximately 50% of the variance in TYLCV resistance among the total F2populations, which had almost equal dominance and additive value. Similarly,Anbinder et al. (2009) found an additional resistance locus from TY172 line that was different from any of the resistance genes; the resistance locus was named Ty-5 and was near SINAC1, which was characterized by R2=39.7% (33<LOD<34). Furthermore, Hutton et al. (2012) indicated that Ty-5 associated resistance was recessive and was named ty-5 instead of Ty-5,which was co-segregated with SINAC1.
Fine mapping of ty-5 gene
ty-5 gene was previously assigned to chromosome 4 near CAPS marker SINAC1. To our knowledge, no subsequent studies of this gene had been performed.The lacking of understanding of this gene restricted its application. In this study, 1 013 individuals of F2plant population were used in an attempt to map ty-5 gene by means of SSR molecular markers for fine mapping.Finally, seven polymorphic markers were screened out and closely linked to ty-5 gene on chromosome 4 in tomato. Furthermore, NAC1 and TES2461 were the closest flanking markers of ty-5 gene, and the genetic distances were 0.5 and 0.8 cM, respectively.This study provided the basis for the prediction of ty-5 candidate genes.
Prediction of ty-5 candidate genes
A total of 17 genes were found in this target region,according to the tomato genome sequence in NCBI.Further, irrelevant genes were excluded by annotating structure and function of the genes. Finally, four candidate genes related to the resistance were found.Based on the linkage map and tomato genome sequence, protein genes were predicted in the target region with the tomato genome annotations in NCBI.A defensin-like protein 1 (LOC104229164), an serine/threonine-protein kinase Aurora-3 (LOC101261508), a stress-related transcription factor SINAC1 (NAC1) and a histidinol-phosphate aminotransferase (LOC101260 925) were involved in this region. The results of qRTPCR in the study showed that SlNAC1, LOC104229164 and LOC101261508 exhibited the expression patterns of being resistance related. This result provided evidence that the expression patterns of the three candidate genes might depend on the occurrence of ty-5 gene resistance response to TYLCV. SlNAC1 locus proteins contained the same NAM (no apical meristem) and belonged to NAC domain protein family, which were accompanied by diverse C-terminal transcriptional activation domains. Again, studies had shown that NAC domain proteins played an important role in plant hormonal control and defense (Huh et al.,2012; Wang and Culver, 2012). Results showed that SlNAC1, a member of NAC domain protein family,played an important role in the replication of tomato leaf curl virus (TLCV) and possibly TYLCV replication (Selth et al., 2005). LOC104229164 locus proteins contained Knot1 protein, belonging to defensin-like protein 1, which participated in the regulation of the defense response to viruses.
Interestingly, it was previously suggested that resistance in TY172 was controlled by three genes.Two of these genes were regarded as additive, one was described as partially dominant, and the other was thought to be recessive; both of them were reported to be controlled by a third recessive gene (Friedmann et al., 1998). Anbinder et al. (2009) reported that SlNAC1 and SlSUMO appeared to be excellent candidate genes for TYLCV resistance and might be implicated in viral DNA replication and accumulation.In a recent study, Hutton et al. (2012) implied that an additional resistance gene might be involved. In this study, the results suggested that ty-5 candidate genes, which were a class of NAC domain proteins,might regulate the resistance of TYLCV together with a defensin-like protein. Through genetic mapping,expression pattern analysis and functional analysis,it was shown that ty-5 candidate gene was novel and completely different from any other TYLCV resistance genes.
In tomato, five TYLCV resistance genes were discovered; Ty-1 and Ty-3 were allelic and had been cloned, and they coded for an RNA-dependent RNA polymerase (RDR) belonging to RDRγ type, which had an atypical DFDGD motif in the catalytic domain.To date, other resistance genes (Ty-2, Ty-4 and ty-5)had not yet been cloned, and the findings of this research provided the basis for the cloning of ty-5 gene. Based on the three candidate genes involved in this target region, it was aimed to verify the function of SINAC1 by means of virus-induced gene silencing(VIGS). Functional verification of candidate genes in the resistant tomato line CLN32120a-23 was currently ongoing in our laboratory.
Using of Marker TES2461 in marker-assisted selection (MAS) breeding
TES2461 was a codominant marker that closely linked to the resistance trait. This marker was located near ty-5 resistance trait at genetic distance of 0.8 cM,making the products in CLN32120a-23 and Moneymaker different in size. This marker was tested in F2individuals. However, five F2individuals showed an inconsistent genotype. These five F2individuals were resistant in the inoculation test, but showed the susceptible genotype in TES2461 test. In other words, ty-5 gene, might together with another resistant trait, might display a vital resistance effect on TYLCV.Moreover, ty-5 gene might also be slightly affected by incompletely recessive inheritance, leading to a higher disease severity score for some heterozygous plants,and these plants were divided into the susceptible bulk. Although not all the samples yielded consistent results in the inoculation and TES2461 tests, the veracity of TES2461 in genotype identification was sufficient for MAS breeding work (Czosnek and Laterrot, 1997).
Conclusions
In the study, the results showed that the novel TYLCV resistance locus was recessive effect. It was presumed that ty-5 gene resistance to TYLCV might be controlled by two genes, suggesting that another resistance gene was involved. TYLCV resistance locus was mapped to the marker interval from NAC1 to TES2461 on the short arm of chromosome 4. The genetic distances between ty-5 and the closest flanking markers, NAC1 and TES2461, were 0.5 and 0.8 cM,respectively. The findings of this research provided the basis for future cloning of ty-5 gene as well as markerassisted selection (MAS) breeding.
Acknowledgments
Thank Prof. Li for his efforts in revising manuscript.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Design and Experiment of four Lines Trailed Potato Fertilization Seeder
- Predictive Modeling for Growth and Enterotoxin Production of Staphylococcus aureus in Milk
- Evaluated Characteritics of Chicken Bone Marrow-derived Dendritic Cells Following LPS Induced Maturation
- Evolution and Expression Patterns of Forkhead Box N1 in Pig
- Distribution of Selenium and Mercury in Heilongjiang Province and Its Effect on Body of Beef Cattle
- A Novel Bacillus thuringiensis Cry57 Protein Domain Swap In fluence on Insecticidal Activity
