Molecular Cloning and Expression Analysis of AcFT3 in Allium cepa
2018-10-10ZhangHuihuiSongCeYangCuicuiWeiHeyuanChenDianandWangYong
Zhang Hui-hui, Song Ce, Yang Cui-cui, Wei He-yuan, Chen Dian, and Wang Yong
College of Horticulture, Northeast Agriculture University, Harbin 150030, China
Abstract: Onion (Allium cepa L.) was an economic vegetable and a strictly biennial herb, which was widely distributed in the world. In the past, it was a strictly biennial plant, the studies had shown that FT (FLOWERING LOCUS T) gene was involved in the photoperiod pathway to regulate flowering in the model plant. In this study, transcriptome sequencing method was used to obtain cDNA sequence of FT homologous gene in onion, named AcFT3 (KF864665). AcFT3 had a full-length of 540 bp, encoded 179 amino acids, with 98.31% homology to AfFT (Allium fistulosum), and 63.0%-84.0% homology to other higher plants. Phylogenetic tree analysis indicated that AcFT3 had the closest relationship with AfFT. The results of quantitative RT-PCR showed the expression pattern of AcFT3 both in vegetative growth of onion and in different organs of bolting and flowering, the expression level of AcFT3 reached the highest in the leaves before bolting and in the flower organs after bolting.
Key words: onion (Allium cepa L.), AcFT3, quantitative RT-PCR, flowering time
Introduction
Onion (Allium cepa L.) is a member of Liliaceae Alliaceae family, which is one of the world's largest consumption of vegetables, with rich nutritional value and health function. As a strict biennial plant,onion is sensitive to photoperiod. According to the different reactions of the photoperiod, onion can be divided into long daylight ecological type and short daylight ecological type (George et al.,2014). Under suitable growth conditions, the bulb of onion expands during the vegetative growth period induced by photoperiod in the first year, then it bolts and flowers after vernalization in the next year to complete the plant reproductive developmental period. The genes associated with the photoperiod play a decisive role in this process. Studies find that the plants synthesize signal molecules can be induced by external environmental factors; such as temperature and light, then the expression of transcription factor CO (CONSTANS) is activated by the biological rhythm clock output products GI(GIGANTEA) after a series of signal transduction,subsequently, activate the transcription of FT(Kobayashi et al., 1999; Hayama et al., 2007). For example, in Arabidopsis thaliana, the transcripts of FT (mRNA) move from leaves to SAM by appropriate photoperiod induction. FT protein interactes with a transcription factor protein FD (specific expressed in SAM), which is a basic region leucine zipper domain, and then they promote the expression of downstream flowering gene AP1 together (Abe et al.,2005). Afterward, they regulate other floral organ characteristic genes LFY (Leafy) and AG24 (Agamous-Like 24), ultimately to complete floral organ development (Poehig, 1990; Jaeger et al., 2007). FT and its homologous genes are involved in the effects of plant age and stress induce flowering. Therefore, FT is one of the important integration factors and key genes, during flowering pathway (Zhang, 2012; Liu et al., 2016).
Since Chailakhyan proposed the concept of florigen in 1936, recent studies confirmed that the main component of the florigen was a protein encoded by FT and its homologous genes (Chailakhyan, 1936). The protein is produced in leaves and then moves to stem apex through phloem to induce flowering. At present, FT gene has been isolated from a variety of plants, such as AtFT gene in Arabidopsis thaliana, Hd3a gene in rice, VcFT gene in blueberry, smFT gene in eggplant and DcFT gene in Carrot (Tamaki et al., 2007; Guo et al., 2013; Guo et al., 2016; Zhan et al., 2017). In this study, the coding region of the "florigen" FT gene was coloned from onion, AcFT3, and a bioinformatic analysis and expression profile of AcFT3 in different onion tissues and organs were performed. A theoretical basis was provided for FT gene in regulating flowering time in Onion (Allium cepa L.) breeding process.
Materials and Methods
Plant materials
The plant materials used in this experiment were from the higher-generation inbred line "1007", which was a long day onion with yellow skin. The seed were cultured in March, 2012, and planted in experimental plot in May, got bulb and storaged it in the refrigerator at mid-August, the bulb of the strain was planted in the greenhouse No. 5 of Horticultural Experimental Station of Northeast Agricultural University in November, 2012. Collected leaf sheath and blade before and after bolting, and stem, inflorescence and flower after bolting bulb, materials were quickfrozen by liquid-nitrogen and storaged at -80℃,RNA extraction was used for AcFT3 gene cloning and fluorescence quantitative analysis.
RNA extract and synthesis of the first strand cDNA
The total RNA was extracted using TRIzol reagent(Invitrogen) from inflorescence meristem and leaves during early bolting, flower buds, flower stalks,receptacles and blades during bolting, and flowers,flower stalks, receptacles and leaves during blooming, respectively. The first strand cDNA was synthesized using M-MLV reverse transcriptase (MBI) introduction.
Cloning of AcFT3 gene
cDNA was obtained from the total RNA of onion leaves as templates to obtain AcFT3 gene fragment by transcriptome sequencing. According to the results of transcriptome sequencing, AcFT3 gene clone upstream and downstream primers were designed.The full-length AcFT3 gene was cloned and sent to Huada for sequencing. The reaction system was(20 μL): 10×Taq Buffer 2 μL; dNTP (2.5 mmol · L-1each) 2 μL; upstream primer AcFT3-F (10 μmol · L-1)0.5 μL; downstream primer AcFT3-R (10 μmol · L-1)0.5 μL; Tap DNA polymerase (5 U · μL-1) 0.2 μL;cDNA template 1 μL; ddH2O 13.8 μL. Reaction procedure: pre-denaturation at 94℃ for 1 min; denaturation at 9℃ for 1 min; 52℃ annealing 45 s; 72℃extension 30 s; 72℃ extension 30 s; repeat 30 cycles;72℃ 10 min; storaged at 4℃.

Table 1 Primers and their sequences
Bioinformatics analysis of AcFT3
The obtained AcFT3 gene sequence was analyzed by BLAST for homology gene retrieval, and DNAMAN software was used for sequence comparison and amino acid translation, the open reading frame of Finder ORF was used to predict the sequence. MEGA (5.2.1 version) software was used to build the plant FT protein system evolution tree, and then DNAstar and other software systems on the software program was used for AcFT3 gene function predict and structural analysis.
Analysis of AcFT3 gene expression by qRT-PCR
A TRIzol reagent was used to extract RNA from different organs of vegetative growth and reproductive growth stage. A cDNA template was synthesized by Toyobo reverse transcription kit. Using fluorescence quantitative reference primers (AcActin-F and AcActin-R) and gene specific primers (AcFT3-1 and AcFT3-2), the specificity of the primers was identified by the solubility curve and amplification curve, and the relative expression of the gene was detected,according to Bio-Rad iQ5 fluorescence quantitative PCR according to SYBR ®Premix Ex TaqTMⅡ instructions.
Results
Cloning of AcFT3 gene
cDNA was obtained from the total RNA of onion leaves as template, AcFT3-F and AcFT3-R primers were used for PCR amplification, and the 540 bp of AcFT3 fragment was obtained (Fig. 1).

Fig. 1 RNA agarosegel electrophoresis and PCR amplification of AcFT3
Homology comparison and evolution analysis of AcFT3
cDNA was used as template, and the sequences were obtained from transcriptome sequencing, which suggested that the sequences encoded 179 amino acid residues, and the gene was named AcFT3 GenBank accession number was KF864665, suggesting that the encoding of a protein with a molecular mass of 19.8 ku.Using the protein module of DNAstar to analyze the basic characteristics of AcFT3 protein of onion, AcFT3 contained two conserved elements, which was similar to the mammalian binding protein PEBP (phosphatidyl ethanolamine binding protein). The first one was D-P-D-X-P component, and after that the component was about more than 10 residues, which was a histidine residue; the second was G-I-H-R component.The coding regions of amino acids encoded by AcFT3 gene of the onion and other plants were compared by DNAMAN software tool, sequence homology analysis found that AcFT3 amino acid sequences of onion had the highest homology reached 98.31% with the same families scallions. The homology of amino acid sequence of AcFT3 of onion and FT amino acid of wheat, rice, Narcissus, Populus, longan and Arabidopsis thaliana in 63.0%-84.0% (Fig. 2).
FT amino acid sequence of 18 kinds of plants which had logged on Genbank and AcFT3 amino acid sequence constructed NJ tree by MEGA software(5.2.1 version). Further analysis of the relationship between onion FT with other plants. The analysis of phylogenetic tree results showed that AcFT3 had a nearest relationship with scallions, followed by Iris fulva, indicating that the gene evolution in the same family was more conservative (Fig. 3).
Analysis of spatial and temporal expression of AcFT3
AcActin as a reference gene, used the quantitative realtime PCR to analysis the expression pattern of AcFT3.The results showed that AcFT3 gene was expressed in all the tested tissues of onion, but the expression level in each period was extremely different. The expression of AcFT3 was very low in sheath, bulb before and after bolting, flower bulbs after bolting,but the expression in leaves increased rapidly before bolting, and reached the highest in inflorescence(spathe) at the early stage of bolting, maintained a high level of expression in flower last.
For some annual crops, mRNA of AcFT1 also accumulated in leaves before flowering (Yu et al.,2016), but the highest expression level was in inflorescence tissues that had not yet blooming at the early stage of bolting (Fig. 4).

Fig. 2 Alignment of FT amino acid sequences from different plants
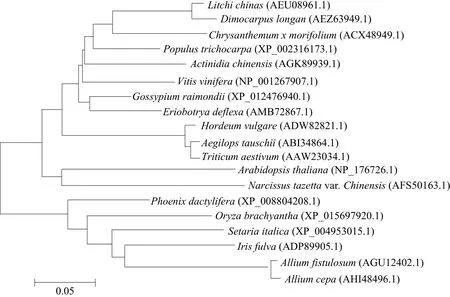
Fig. 3 Phylogenetic analysis of FT amino acid sequences from different plants

Fig. 4 Spatial and temporal expression of AcFT3 gene
Discussion
Eight genes in FT family of PEBP had been identified in onion in previous reports. It was worth mentioning,at the same time when AcFT3 gene was cloned in our laboratory. Robyn also obtained the six FT genes through transcriptome sequencing. They found that the functions of AcFT1 and AcFT4 were opposite to the formation of onion bulbs under the photoperiod.However, the function of AcFT7 might be similar to that of AcFT4, which mainly controlled the formation of onion bulbs. In addition, AcFT4 and AcFT7 might also be involved in the establishment of different mature time between two kinds of onion bulbs. AcFT2 promoted flowering (Robyn et al., 2013; Ranjith et al.,2016). AcFT3 contained two conserved elements.The first one was D-P-D-X-P component and the second was G-I-H-R component, which indicated that AcFT3 belonged to PEBP family members. PEBP family members were a class of regulatory factors which closely related to signal transduction (Duplat-Bermudez et al., 2016; Danilevskaya et al., 2008;Lifschitz et al., 2006). The homology analysis of amino acid sequences showed that FT gene was relatively conservative in the long term evolution process. The phylogenetic tree showed that florigen homologous gene was different with inflorescence meristem specific gene in evolution, the latter form of dicotyledonous plants in evolution clearly was divided into two parallel branches (Yang et al., 2016),and florigen homologous genes were cluster, single dicotyledonous plants and could not be completely separated, therefore, they appeared earlier than disproportionate of the monocotyledons.
AcFT3 was a kind of flowering regulatory gene.It was participate in some approach about flowering in the model plant Arabidopsis thaliana, such as photoperiod, vernalization, GA, autonomous pathway(Zhang, 2012). In a large number of studies on FT,the expression patterns of many plants were different,which had certain express specificity in different periods. Only one of the three FT homologous genes in sunflower was expressed in apical meristem and not expressed in leaves (Blackman et al., 2010). There was a high expression of CsFT gene of cucumber in male and female flowers, but almost wasn't express in cucumber leaves, in plants, such as maize and apple,multiple FT homology genes were expressed in root,leaves, inflorescence and seed, FT homologous genes of premature trifoliate orange were expressed in leaves and the expression in fruits was significantly higher than that in stem and floral organs, it might be that there were multiple FT or FT like genes in the same plant, which had different expression patterns (Zhang et al., 2009). In the research of apple FT homology gene, the content of FT homology gene in the leaves and flowers along with the development of the organ showed a downward trend, it could be speculate that FT homologous gene was not only involved in a flower process, but also involved in flower development. The function was similar with CcLFY of walnut, CcLFY gene regulated flower bud differentiation and floral organ development in transgenic tobacco (Wang et al., 2012). In this experiment, it was found that during the vegetative period of plant growth, the expression of AcFT3 gene in the leaves before bolting was the highest, and during the reproductive growth period,the expression of AcFT3 gene in the ripened leaves decreased sharply. In the early stage of bolting, the expression of the spathaceous was the highest, but the expression in flower was significantly decreased,which was speculated that AcFT3 gene might play a role in regulating plant flowering and affecting plant flowering.
Studies on other plants also indicated that FT's function was involved in the regulation of many developmental processes. A study on the characteristics of the conductible flowering materials and the regulation of reproduction in soybean revealed that the relative expression of the flower promoting gene GmFT and the flower inhibiting gene GmTFL1 coordinated the relationship between source and sink,and was thus involved in the regulation of flowering and pod development (Nan, 2014; Sun et al., 2011).Two different FT-like homologous genes (StSP3D and StSP6A) of potato were controlled by the plant transformation and the formation of tubers under independent environmental factors (Xie and Liu.,2013). Four FT-like genes in tomato genome were highly expressed under the long day condition, which exerted important effects on the promotion of flower bud differentiation (Cao et al., 2015). Plant flowering was a complex process, which was not only related to the photoperiod and vernalization, but also to mutual balance between endogenous genes. In addition, the expression of FT gene and the flowering time were also affected by the environmental factors, such as the length of day and night, and the effect of temperature difference between day and night (Hannah et al.,2016). How AcFT1 gene responded to light and affected vernalization and how it regulated the downstream LEAFY, AP1 and SOCI genes were major directions of the future studies.
Conclusions
The predicted proteins of AcFT3 included D-P-DX-P component and G-I-H-R component, which were characteristic components of PEBP family. Therefore,AcFT3 gene belonged to PEBP protein family, which was involved in the signal transduction of plant flower formation. Onion AcFT3 gene was more homologous to FT gene of onion. The homology analysis of amino acid sequences showed that FT gene was relatively conservative in the long term evolution process. AcFT3 gene was expressed in the plant life cycle combined with the plant's own state as a floral gene, and reached the highest expression level in the spathe. This might be related to the function of AcFT3 gene, which provided a basis for further study on the role of genes in plant flower formation.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Mapping and Candidate Gene Screening of Tomato Yellow Leaf Curl Virus Resistant Gene ty-5
- Mowing Height and Mowing Frequency Interactions on Turf Performance of Kentucky Bluegrass
- Screening of Actinomycetes from Medicinal Plant Rhizosphere Soils for Industrial Enzymes and Antimicrobial Activity
- Comparative Transcriptome Profiling of Glycine soja Roots Under Salinity and Alkalinity Stresses Using RNA-seq
- A Novel Bacillus thuringiensis Cry57 Protein Domain Swap In fluence on Insecticidal Activity
- Distribution of Selenium and Mercury in Heilongjiang Province and Its Effect on Body of Beef Cattle
