Exploring the active ingredients, potential targets and pathways of quassinoids in Simaroubaceae plants by network pharmacology approachs
2018-04-02PengZhoZhiyngYnDnqiLiXioxioHung
Peng Zho, Zhiyng Yn, Dnqi Li, Xioxio Hung*
a Department of Natural Products Chemistry, Shenyang Pharmaceutical University, Shenyang 110016, PR China;
b Institute of Functional Molecules, Shenyang University of Chemical Technology, Shenyang 110142, PR. China
1 Introduction
Traditional Chinese medicine (TCM) has been widely used in China for thousands of years,and they have gradually becoming an option in the treatment of cancer around the world [1,2]. Many clinical studies have shown that TCM is effective in the treatment of breast, gastric, lung and prostate cancers [3,4]. Although the high throughput technologies such as gene expression microarrays,proteomics, and metabolomics were used to study the mechanisms of TCM, the experiments still have limitations which come from complex of TCM [5].With the rapid progress of bioinformatics, network pharmacology was proposed to be a promising way to understand the mechanism of TCM [6,7]. To date,accumulating evidence suggests that the network pharmacology analysis is a powerful way to study the mechanisms of herbal formula [8].
Quassinoids are characteristic compounds of the family Simaroubaceae, and most of them are tetracyclic triterpenic lactone and pentacyclic triterpenic lactone. The representative compounds include: Bruceantin, Brusatol and so on [9]. For example, the brusatol was first separated from the genus brucea in 1968, and related research showed that brusatol is a novel compound that is more effective in the anti-pancreatic cancer in vitro than camptothecin, At present, most of the quassinoids with significant anti-cancer activity were isolated successively from the genus brucea, and their structure-activity relationships had already been systematically studied. Therefore, the quassinoids from these three genera of the family Simaroubaceae were selected as the database of ingredients to study the anti-cancer network pharmacology.
The polytype network pharmacology methods and technologies can facilitate to evaluation the effect of quassinoid, disease treatment, disease prediction and prevention, and practice in the treatment in terms of TCM and promote the further research in drug discovery [10]. With the development of computer science, it is easier and faster to process the mega data than before.With the advent of mega data era, technology and methodology are being transformed [11]. Along with the development and application of the Internet information technology, the information of quassinoids, target proteins, biological pathways,diseases, clinical data of diagnosis and treatments have been accumulated dramatically, which generates mega data in medical field [12]. The research target was shifted from the “causality inference”to the “correlativity analysis” gradually [13].The characteristics of TCM information had strong similarity to these. The advent of the mega data era provided both opportunities and challenges for TCM such as the new computational methods and technologies [14]. Hence, in the current study,86 quassinoids reported from three genera of the family Simaroubaceae were selected, which were docked with 53 different anticancer proteins and 150 pathways in order to study their probable binding models against cancer and predict their therapeutic potential.
2 Material and methods
2.1 Collection and disposal of compounds
Based on the characteristic quassinoids obtained from the separation and retrieved through literature, a database consisting of 86 natural products was established, which was based on the reported quassinoid from the family Simaroubaceae.Then, the structure optimization is optimized by the Structural optimization plug-in included in the Chembiodraw and Sybyl-X (version 2.0, TRIPOS Inc.) software.
2.2 Screening and processing of target protein
First, we searched 53 target proteins of cancer through Therapeutic Target Database, and the target proteins which satisfying the following criteria were selected to construct the drug-target network and the compound-target-pathway network: (a) the source organism is human (b) the protein has a KEGG annotation of pathway in KEGG database [15]. By using blast tools, the structures and binding sites of cancer-related proteins was found through searching the NCBI database involved in CDD database.Then, we used the Uniprot database to search the three-dimensional structure of target proteins, and in consideration of the existence of protein-ligand complexes, crystal structure determination method and resolution, the 53 cancer target proteins were determined ultimately. Finally, the related protein crystal structure was obtained through the RCSB database, and then imported by the Discovery studio 2.5. At the same time, we removed the original ligand and water, hydrogenation, as well as the optimization and repair of the amino acid residues toward target proteins.
2.3 Preparation of data sets and molecular docking
Molecular evaluation was performed by molecular docking software Sybyl-X, which could analyze the binding capacity of each active component with the protein crystallization. Between compound molecular docking and the related protein crystallization, and constantly optimizing the location, configuration, molecular internal rotatable dihedral angle of the receptor compounds and the side chains and skeleton of the amino acid residues of receptors, therefore the optimum conformation of the quassinoids and protein crystallization were found. Finally, the binding ability of each component with protein crystal was evaluated by scoring function analysis.
To keep the accuracy of predicted results and integrity of data, we selected the molecules whose docking scores were higher than that of original ligand in the complex structure, viz. higher than 7.0.Finally, we obtained 86 quassinoids from the family Simaroubaceae and 53 target proteins. The target proteins were associated with 150 human related pathways and 6 related cancer from the KEGG database.
2.4 Construction of compound-target network and target-related cancer network
To find the potential active molecules and target protein, the compound-target network (Fig. 1)was constructed according the docking results in Cytoscape 3.2.1 [16]. We also calculated the node centralities and network properties using the network analyzer tool of Cytoscape 3.2.1. Then, to illuminate the associations between target proteins and the related cancers, we constructed the target-related cancer network based on the data extracted from KEGG database. The target-related relationships were investigated based on the target. One target protein maybe related to many cancers and one cancer could be related to many targets. Several cancers shared the common targets were found through analyzing the parameters of the network.
2.5 Construction of compound-target-pathway network and target-pathway network
Many quassinoids play a crucial role by affecting the related pathways or affecting the metabolisms in the body. Moreover, the state of cancer cell is often the results of a series of variations in our body such as the variations in pathways, instead of the anomaly of compoundtarget interaction [17]. Therefore, clarifying the mechanism of compound-target-pathway associations is a crucial issue of system-based drug discovery. To understanding the action mechanism of the quassinoids, we constructed the compoundtarget-pathway network based on the previous compound-target network and target-pathway network. Several significant pathways were found through analyzing the compound-target-pathway network. Besides, we constructed target-pathway network to find the relation between the pathways and the target proteins. The network properties of the target-pathway network were analyzed and found several important pathways, which could play an important role in the interaction mechanism of the quassinoids of Simaroubaceae plants.
3 Results
3.1 Prediction of potential compounds
The compound-target network (Fig. 1) based on the docking results were constructed, and the network had 139 nodes and 315 edges from Fig. 1,in which colorized circles and blue triangles corresponding to target proteins and quassinoids from the family Simaroubaceae, respectively. Then,86 quassinoids were evaluated and studied for their interaction with anticancer target proteins through network pharmacology. The possible interactions between natural products and cancer proteins were also evaluated by two important topological parameters viz. degree and betweenness centrality.Some quassinoids have many targets, such as Yadanzioside K (15 targets), Yadanzioside P (10 targets), Yadanzioside H (9 targets), Bruceoside D (9 targets), Bruceantinol A (9 targets) and Ailantinol G(9 targets), which meant that these molecules might be the potential active ingredients of the family Simaroubaceae. Through literature research, we found out that the top five quassinoids, Yadanzioside K [18], Yadanzioside P [18], Yadanzioside H [19],Bruceoside D [20], Bruceantinol A [18], were isolated from the plant Brucea javanica, and Ailantinol G [21] was derived from the plant Ailanthus altissima. The specific structures of these quassinoids are shown in the Fig. 2 and Table 1 listed the degree and betweenness of the six potential quassinoids.
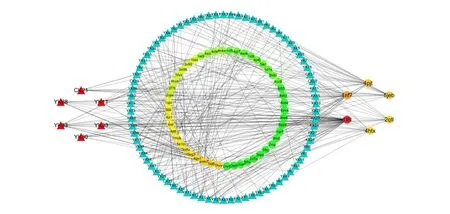
Fig. 1 The compound-target network. The blue and red triangles represent the quassinoids, while the evolving circles stand for protein targets
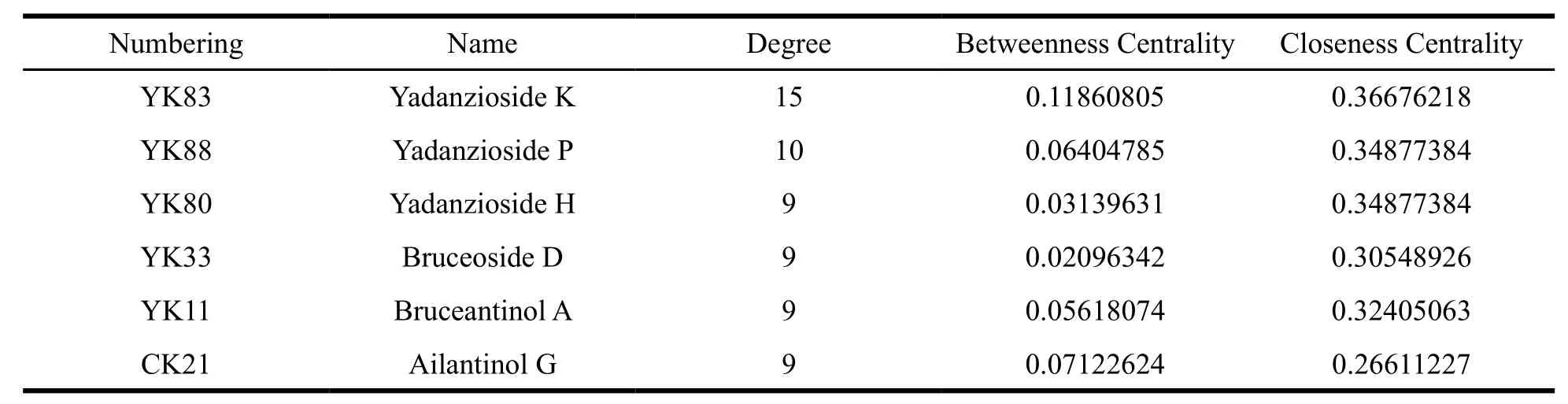
Table 1 Listed the degree and betweenness of the six potential quassinoids. “YK”, “CK” represent the quassinoids derived from Brucea javanica, and Ailanthus altissima, respectively
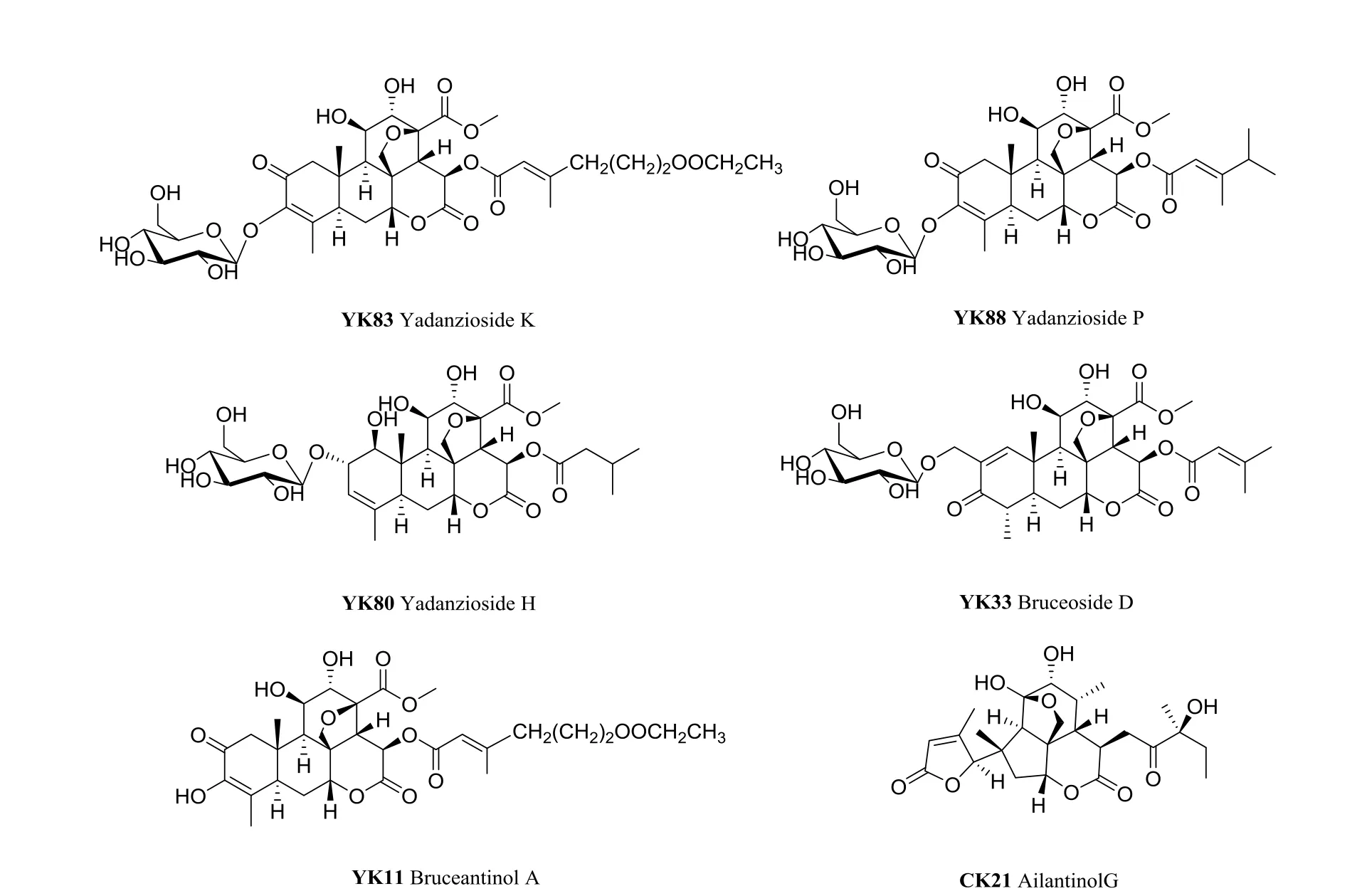
Fig. 2 The specific structure of top six potential quassinoids. “YK”, “CK” represent the quassinoids derived from Brucea javanica, and Ailanthus altissima respectively
Furthermore, the higher degree of nodes meant highly connected molecules which may play important roles in the compound-target interaction network. In other words, the high degree quassinoids of the family Simaroubaceae suggested that they could interact with multiple targets and have better effect on the treatment of cancer.
3.2 Prediction of Target proteins
In order to expound target proteins which are most closely associated with quassinoids and related cancer, the compound-target network and the target-cancer network were established. Among the 53 targets, which linked to 86 quassinoids,many candidate proteins were targeted by more than one quassinoid. According to the compound-target protein network, DNA topoisomerase I (1t8i) was found to have the highest degree values, followed by the Inosine-5'-monophosphate dehydrogenase 2 (1nf7), Poly [ADP-ribose] polymerase 1 (4pjb),Epidermal growth factor receptor (5jeb). The parameters of the ten most promising targets were shown in Table 2.

Table 2 The parameters of the ten most promising targets
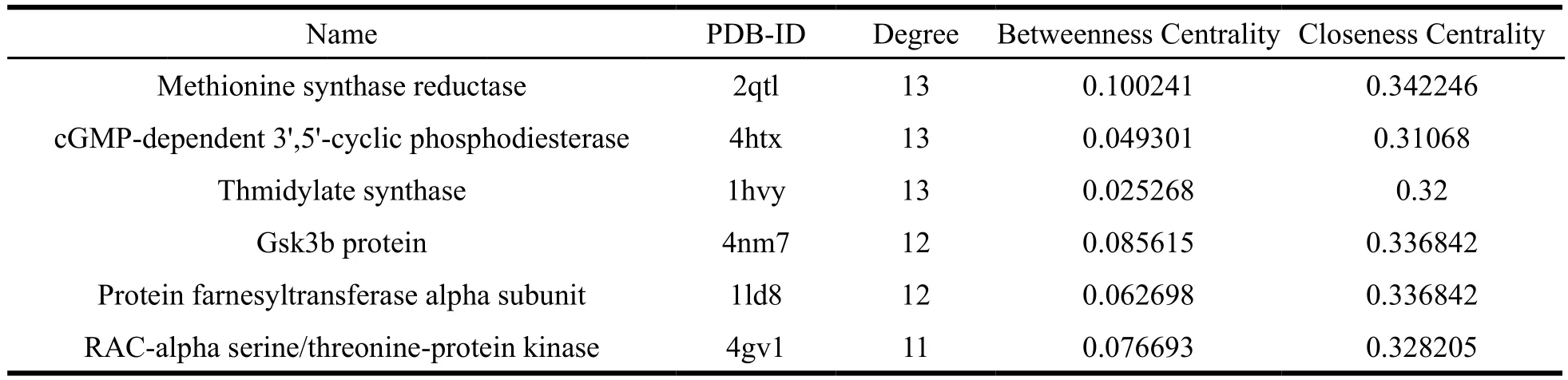
Continued Table 2
As we all know, breast cancer, prostate cancer,colorectal cancer, lung cancer, liver cancer and gastric cancer were the main cancer categories [22].According to the American Cancer Society,12 million cases of cancer were diagnosed in 2013,with 7 million deaths worldwide, and these numbers are expected to more than double by 2030, i.e.27 million cases with 17 million deaths [23]. Then the interaction between target proteins and the related cancers was analyzed, the results were listed in Fig. 3. Large amounts of experimental results reported that the quassinoids of the Simaroubaceae plants possess high anti-cancer effect, and Fig. 3 also indicated that quassinoids might have connection with relevant cancer, which are in accordance with previous researches. In addition, DNA topoisomerase I (1t8i) and epidermal growth factor receptor (5jeb)all have the highest degree values according to the target-cancer network. The higher degree of proteins meant highly connected relevant cancer which might play important roles in the target- relevant cancer interaction network. Through the analysis of the aforesaid network pharmacology, DNA topoisomerase I (1t8i) and epidermal growth factor receptor (5jeb) were viewed as the most promising targets, and might be acted as the key target proteins to achieve the therapeutic effects. Table 3 listed the parameters of the target-cancer network.
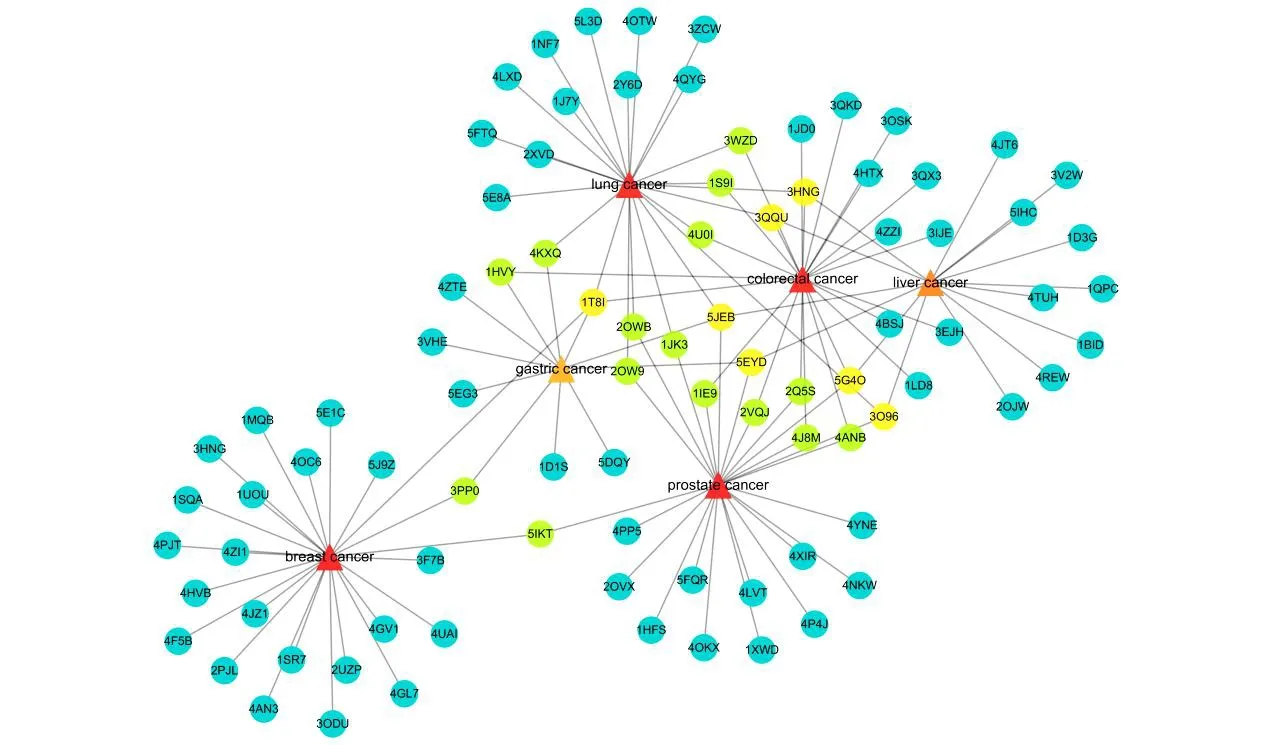
Fig. 3 The target- related cancer network. Red triangles and circles correspond to related cancers and target proteins,respectively, and the evolving color stand for degree of the relationship

Table 3 The parameters of the target-cancer network
3.3 Prediction of important pathways
To further investigate the important relations between target protein and the important pathway,the target-pathway network (Fig. 4) based on the docking results and the data extracted from Therapeutic Target Database (TTD) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were constructed, and the network had 226 nodes and 273 edges. There were several target proteins in one pathway, and one target protein always existed in many pathways. Logically, the role of one pathway which contains many target proteins and interacted with quassinoids is more vital than the role of one target protein that interacted with quassinoids in many pathways. The impact of one target protein on the whole pathway maybe little, and the impact of a pathway which contained many target proteins interacted with the drugs on the body could be huge.Therefore, we tried to find the important pathways through analyzing the target-pathway network.In Fig. 3, the degree of the pathway showed that the number of the target protein in the pathway.The higher degree of the pathway, the more target proteins existed in the pathway. The parameters of target-pathway network were shown in Table 4.
Then the paired relationships information(link between quassinoids and target proteins, link between target proteins and pathways) was imported to Cytoscape software (Cytoscape 3.2.1; http://www.cytoscape.org/), and a compound-target-pathway network was constructed (Fig. 5). The three-level network consisted of 289 nodes (86 quassinoids, 53 target proteins and 150 pathways) and 536 edges.The detailed information of the pathway names and their categories can be found in Table 2.Through the visualization and network analysis of the relationships among quassinoid, target proteins and pathway in the constructed network, PI3K-Akt signaling pathway (hsa04151) and HIF-1 signaling pathway (hsa04066) may play an important role in the mechanism of the potential compounds.
Among the 53 target proteins, DNA topoisomerase I (1t8i) was influenced by 48 quassinoids, and it is associated with Integrated Pancreatic Cancer Pathway. Epidermal growth factor receptor (5jeb) was affected by 15 quassinoids,which maybe related with HIF-1 signaling pathway,ErbB signaling pathway and Calcium signaling pathway. Besides, PI3K-Akt signaling pathway(hsa04151) and HIF-1 signaling pathway (hsa04066)were influenced by 5 quassinoids. It is speculated that the quassinoids in the family Simaroubaceae may be used to control the downstream proteins such as Akt, HIF-1 and mTOR by acting on these target proteins to achieve the anticancer effects.

Fig. 4 The target-pathway network. Blue inverted triangles and circles correspond to signaling pathways and target proteins, respectively, and the evolving color stand for degree of the relationship between them
Fig. 5 The compound-target-pathway network. The blue inverted triangles, red circles and green rhombi represent signaling pathways, quassinoids and target proteins, respectively

Table 4 The parameters of target-pathway network
4 Discussion
Although the lead compounds extracted from traditional Chinese medicine had exhibited preferable anticancer activity over recent years, there were still many unclear underlying mechanisms.Here the interaction mechanism of quassinoids derived from the family Simaroubaceae was explored and the possible target proteins and pathways were predicted by employing multiscale computer modeling. Polytypic networks provided an effective way, which may help us to explore the underlying mechanisms. Therefore, the compoundtarget network, the target-related cancer network, the compound-target-pathway network and the targetpathway network were constructed and several potential active quassinoids, several promising target proteins and pathways in these three genera were found.
As we all know, Chinese medicine treats diseases in a holistic way to make body recovery from imbalanced state by modulating a complex modes of action including multiple pathways. And diverse target proteins included in signaling pathway play a significant role for the application to treat cancer. Then, we analyzed the relations among the quassinoids, targets, pathways and related cancers and explored the possible quassinoid interaction mechanism of quassinoids derived from the family Simaroubaceae.
The results of our work attempt to hope to offer new insights to predict the pharmacological properties of quassinoids and to provide benefit for their further development and research.
5 Conclusion
In conclusion, through constructing the biological network of the interaction between quassinoids, target proteins and pathways from a molecular or protein level, we firstly predicted the active ingredients, potential targets and pathways of the quassinoids derived from plants of Simaroubaceae related to cancer mentioned above.Based on the network analysis, Yadanzioside K (15 targets), Yadanzioside P (10 targets), Yadanzioside H (9 targets), Bruceoside D (9 targets), Bruceantinol A (9 targets) and Ailantinol G (9 targets) have the higher correlation with target proteins, which were considered to be the potential antitumor lead compounds, and the most likely potential target might be DNA topoisomerase I (1t8i) and epidermal growth factor receptor (5jeb). Then the quassinoids in the Simaroubaceae may be used to control the pathways such as PI3K-Akt signaling pathway(hsa04151) and HIF-1 signaling pathway (hsa04066)to achieve the therapeutic effects.
[1] Hyodo I, Amano N, Eguchi K, et al. Nationwide survey on complementary and alternative medicine in cancer patients in Japan. J Clin Oncol, 2005, 23: 2645-2654.
[2] Evans MA, Shaw AR, Sharp DJ, et al. Men with cancer:is their use of complementary and alternative medicine a response to needs unmet by conventional care? Eur J Cancer Care, 2007, 16: 517-525.
[3] Lee YW, Chen TL, Shih YR, et al. Adjunctive traditional Chinese medicine therapy improves survival in patients with advanced breast cancer: A population-based study.Cancer, 2014, 120: 1338-1344.
[4] Xu Y, Zhao AG, Li ZY, et al. Survival Benefit of Traditional Chinese Herbal Medicine (a Herbal Formula for Invigorating Spleen) for Patients With Advanced Gastric Cancer. Integr Cancer Ther, 2013, 12: 414-422.
[5] Liu X, Guo DA. Application of proteomics in the mechanistic study of traditional Chinese medicine.Biochem Soc Trans, 2011, 39: 1348-1352.
[6] Liang X, Li H, Li S. A novel network pharmacology approach to analyse traditional herbal formulae: the Liu-Wei-Di-Huang pill as a case study. Mol BioSyst, 2014,10: 1014-1022.
[7] Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application.Chin. J Nat Med, 2013, 11: 110-120.
[8] Zhao J, Jiang P, Zhang W. Molecular networks for the study of TCM pharmacology. Brief Bioinform, 2010, 11:417-430.
[9] Alves IABS, Miranda HM, Soares LAL, et al.Simaroubaceae family: botany, chemical composition and biological activities. Revista Brasileira de Farmacognosia, 2014, 24: 481-501.
[10] Li G, Zuo X, Liu B. Scientific computation of big data in real-world clinical research. Front Med, 2014, 8:310-315.
[11] Westerhoff HV. Network-based pharmacology through systems biology. Drug Discovery Today: Technologies,2015, 15: 15-16.
[12] Fang Luo, Jiangyong Gu, Xinzhuang Zhang, et al.Multiscale Modeling of Druginduced Effects of ReDuNing Injection on Human Disease: From Drug Molecules to Clinical Symptoms of Disease. Scientific Reports, 2015, 5: 10064.
[13] Vicini P. Multiscale modeling in drug discovery and development: future opportunities and present challenges. Clin Pharmacol Ther, 2010, 88: 126-129.
[14] Cui M, Li H, Hu X. Similarities between “Big Data” and traditional Chinese medicine information. J Tradit Chin Med, 2014, 34: 518-522.
[15] Ogata H, Goto S, Sato K, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res, 1999, 28: 29-34.
[16] Saito R, Smoot ME, Ono K, et al. A travel guide to Cytoscape plugins. Nat Methods, 2012, 9: 1069-1076.
[17] Pujol A, Mosca R, Farres J, Aloy P. Unveiling the role of network and systems biology in drug discovery. Trends in Pharmacological Sciences, 2010, 31: 115-123.
[18] Ye QM, Bai LL, Hu SZ, et al. Isolation, chemotaxonomic significance and cytotoxic effects of quassinoids from Brucea javanica. Fitoterapia, 2015, 105: 66-72.
[19] Sakaki T, Yoshimura S, Ishibashi M, et al. New quassinoid glycosides: yadanziosides A-H, from Brucea javanica. Chemical & Pharmaceutical Bulletin, 1984,32: 4702-4705.
[20] Ohnishi S, Fukamiya N, Okano M, et al. Bruceosides D,E, and F, three new cytotoxic quassinoid glucosides from Brucea javanica. ChemInform, 1996, 58: 1032-1038.
[21] Tamura S, Fukamiya N, Okano M, et al. Three new quassinoids, ailantinol E, F and G, from Ailanthus altissima. Chem Pharm Bull, 2003, 51: 385-389.
[22] Miklos K, Szabolcs O, Istvan K. The current situation of cancer morbidity and mortality in the light of the National Cancer Registry. Orvosi Hetilap, 2017, 153:84-89.
[23] Aggarwal BB, Danda D, Gupta S, et al. Models for prevention and treatment of cancer: Problems vs.promises. Biochem Pharmacol, 2009, 78: 1083-1094.
杂志排行
Asian Journal of Traditional Medicines的其它文章
- Absolute configuration of curdione and its three isomers by NMR, ECD and DFT calculations: an insight into the scope of unsaturated ketone helicity rule based on an ECD study
- Main chemical constituents and pharmacological properties of Harrisonia perforata (Blanco) Merr.
- Chemical constituents of the genus Pithecellobium:a systematic review
- Chemical constituents and pharmacological effects of the fruits of Camptotheca acuminata: a review of its phytochemistry
- Studies on the Chemical Components and Biological Activities of Stellera chamaejasme L.
