MOLECULAR CLONING AND CHARACTERIZATION OF TRYPSINOGEN GENE IN GOLDFISH (CARASSIUS AURATUS) AND ITS EXPRESSION PROFILES IN RESPONSE TO CADMIUM AND OXIDATIVE STRESS
2018-01-24CHENWenBoYANFangFangQINShaoZongandLOUBenJie
CHEN Wen-Bo, YAN Fang-Fang, QIN Shao-Zong and LOU Ben-Jie
(Department of Biology, Institute of Resources and Environment, Henan Polytechnic University, Jiaozuo 454000, China)
As a member of the serine protease family, trypsin (EC3.4.21.4), which is synthesized and secreted by pancreas as trypsinogen, is well known as a pancreatic enzyme that is typically secreted into the intestine to digest proteins[1—3]. In intestine, trypsinogen can be activated by enterokinase or activated trypsin by removal of a short activation peptide in the N-terminal. The activated trypsin not only specifically cleaves the peptide bond on the carboxyl side of arginine or lysine residues, but also can activate other digestive enzymes including chymotrypsin, elastase,collagenase and lipase[4,5]. The specificity for cleaving peptide bonds on the carboxyl side of arginine or lysine residues depends on a catalytic triad composed of a histidine (His-57), an aspartic acid (Asp-102) and a serine (Ser-195)[6].
To date, the complete or partial cDNAs of trypsinogen have been cloned in many marine and freshwater fish[3,7—15]. In these studies, the spatiotemporal expression patterns of trypsinogen mRNA have been extensively examined. There are growing evidences that, as a proteolytic enzyme, trypsin plays im-portant roles in protein digestion, growth and survival rates of fish larvae[12—17]. In teleosts, the digestion and absorption are the fundamental routes of acquiring the nutrients and energy. In view of the greatest importance of protein digestion, trypsin indeed plays vital roles in nutrient intake and growth in teleosts.Thus, the information on mRNA expression patterns and enzymatic activities of trypsinogen under different nutritional status is crucial for optimizing feeding regimes and aquaculture protocols in fish.
Along with the rapid development of industrialization and urbanization, environmental pollution resulting from releasing the anthropogenic chemicals is a growing problem and has attracted worldwide attention. Heavy metals, such as cadmium, occur in the aquatic environment naturally as pollutants and have highly toxic effect to even at relatively lower doses.They can affect multiple biological processes including physiology, biochemistry and development in aquatic organisms[18]. Digestive enzymes, which can indicate the metabolic status of the animals and the degree of adaptation to the environment, can be used as biomarkers in environmental monitoring[19]. For example, the expression of amylase genes increased under cadmium exposure inDaphnia magna[20]; Cadmium,temperature and pH had separated or combined effect on digestive enzymes in teleosts[21]. In addition, cadmium also stimulates the production of the reactive oxygen species (ROS), and excessive ROS further cause oxidative damage at the molecular, cellular and tissue levels[22]. However, no information about the gene expression patterns of trypsinogen responding to cadmium and oxidative stress at the transcript level or their abundance is available in fish.
To better investigate the expression patterns of fish trypsinogen under different nutritional status and environmental stresses, we used the good teleost model, goldfish (Carassius auratus). In this study, the complete cDNA sequence of goldfish trypsinogen(gfTryp) was cloned, and then the tissue distribution ofgfTrypmRNA was analyzed. Furthermore, the hepatopancreatic transcriptional expressions ofgfTrypwere detected under nutritional status, cadmium and H2O2exposure. The results may help to gain further insight into the physiological expression of fish trypsinogen, and thus better promote fish growth and serve healthy aquaculture.
1 Materials and methods
1.1 Fish and experiments
Goldfish (C. auratus), obtained from the local aquatic market, were cultured in indoor tanks with circulating water at room temperature under a 10h dark: 14h light cycle photoperiod and acclimated to a feeding schedule for 2 weeks. For cadmium exposure,goldfish were transferred into 50 L water tanks containing 0.5, 5 or 10 μg/mL cadmium (CdCl2, Sigma)for 24h under static conditions, respectively. In H2O2exposure, goldfish were transferred into 50 L water tank containing 100 μmol/L H2O2. Before these experiments, the fish were fasted for two days and no food was provided during the exposure time. For fasting experiment, goldfish were raised with the feeding schedule (once daily at 10:00 am, with 5% body weight per fish per day) for two weeks, and then divided into two groups with 50 L tanks (n=10/group).In one group, the fish were fed following the normal feeding schedule, and in the other, the fish were fasted for two weeks. The fish were sacrificed and sampled at 10:00 am. The periprandial experiment was followed as our previous report[23]. Goldfish were sampled at 2h before feeding time (8:00 am,t=-2h),2h after feeding time (12:00 am,t=+2h), and 4h after feeding time (14:00 pm,t=+4h), respectively. For sample collection, four or five individuals were randomly selected at each time point, and sacrificed by anesthesia in 0.05% MS222 (Sigma). The hepatopancreas were sampled, frozen immediately in liquid nitrogen, and then stored at -80℃ prior to RNA extraction.
1.2 Molecular cloning and structural analysis of gfTryp cDNA
Total RNA was extracted using TRIzol reagent(Invitrogen) and reversely transcribed into the firststrand cDNA by using ReverTra Ace-α kit (Toyobo).For amplifying the conserved cDNA fragment ofgfTryp, PCR was performed with degenerate primers designed based on the conserved sequences of Cyprinidae fish. The corresponding full-length cDNA sequence ofgfTrypwas obtained by3′-/5′-rapid amplification of cDNA ends (RACE) with 3′-/5′-RACE System (Invitrogen), respectively. All primers for gene cloning were listed in Tab. 1. The cleavage site of the signal peptide was predicated by the SignalP4.1 Server program (http://www.cbs.dtu.dk/services/SignalP/). The hydrophobicity plot, theoretical isoelectric point and molecular weight were calculated using proteomics tools (http://www.expasy.org/proteomics).Alignment of the trypsinogen amino acid sequences among different species was performed with Clustalx1.83. Phylogenetic tree was constructed with Mega 6.05 by Neighbor-Joining method and the reliability was evaluated by bootstrap method with 1000 replicates.
1.3 Tissue distribution of gfTryp gene
Transcriptional tissue distribution ofgfTrypgene was examined by using RT-PCR in selected tissues including the brain, gill, skin, hepatopancreas, kidney,heart, spleen, muscle, anterior intestine, middle intestine, posterior intestine, fat and testis from adult goldfish. Total RNAs were isolated by using TRIzol and their concentrations were determined using a spectrophotometer at 260 and 280 nm. Then the samples with an absorption ratio (260/280 nm) between 1.8 and 2.1 were digested with DNase I (Invitrogen) and reverse-transcribed with ReverTra Ace-α (Toyobo).The PCR cycle and temperature forgfTrypwere 34 and 62℃, respectively, after semi-quantitative validation. In this case, 18S rRNA was used as a reference gene.

Tab. 1 The primer sequences used in the present study
1.4 Measurement of gfTryp mRNA by real-time PCR
ForgfTryp mRNAmeasurement in response to nutritional status, cadmium and H2O2exposure, total RNA from hepatopancreas were isolated by using TRIzol (Invitrogen), digested with DNase I (Invitrogen), and reverse transcribed with ReverTra Ace-α(Toyobo). Transcriptional expression ofgfTrypwas detected using THUNDERBIRD SYBR qPCR Mix(Toyobo) in the Bio-Rad MiniOpticon Real-Time PCR detection system (Bio-Rad) as described previously[23]. The standard curve for each gene was generated through a 5-fold dilution series of cDNA to compare with amplification efficiency of the target gene and reference gene. The procedure of real-time PCR was carried out as follows: initial denaturation 95℃for 1min, 40 cycles of 95℃ for 15s, 58℃ for 15s,72℃ for 30s. Fluorescence data were collected after the third step of every cycle. After reactions, the identity of the PCR products was routinely confirmed by analysis of melting curve. The relative expression levels of gfTryp were calculated according to the 2-ΔΔCtmethod[24].
1.5 Statistical analysis
Quantitative data are presented as means±SD.The significant differences were analyzed by one-way ANOVA followed by the LSDt-test analysis using the SPSS, and indicated with * (P<0.05) and **(P<0.01).
2 Results
2.1 cDNA cloning and sequence analysis of gfTryp cDNA
The full-length ofgfTrypcDNA was successfully cloned from the hepatopancreas using RT-PCR and RACE.gfTrypcDNA was 864 bp long with a 21 bp 5′-untranslated region (UTR), a 114 bp 3′-UTR containing the consensus polyadenylation signal AATAAA, and a 729 bp open reading frame encoding a protein of 242 amino acid residues (GenBank Accession No. KU574644). The predicted pI and charge at pH 7.0 were 5.28 and -6.45, respectively.gfTryp had a signal peptide of fifteen amino acid residues, MRSLVFLVLLGAAFA, an activation peptide of five amino acid residues, LDDDK and a matured trypsin of 222 amino acid residues. Multiple sequence alignment indicated that gfTryp possessed the conserved structural features of vertebrate trypsinogens, including the catalytic triad (His-57, Asp-102,and Ser-195), Asp-189 at the bottom of the substratebinding pocket, Gly-216 and Gly-226 lining the sides of the binding pocket, Tyr-172 involved in trypsin substrate specificity, and 12 cysteine residues responsible for the formation of six disulfide bonds(Fig. 1). Phylogenetic analysis by neighbor-joining method revealed that the trypsinogens reported in vertebrate species could be divided into three separated clades (Fig. 2). According to the classification of vertebrate trypsinogens by Roach (2002)[25], they were named group I, group II and group III, respectively.Our newly cloned gfTryp was clustered into the branch of group I and was closer to its counterparts from Cypriniformes (such as grass carp).
2.2 Tissue distribution of gfTryp gene
Tissue expression profile ofgfTrypgene was de-tected by RT-PCR. As shown in Fig. 3,gfTrypmRNA could be found in all tissues examined. In fat,anterior intestine, middle intestine, posterior intestine and hepatopancreas,gfTrypmRNA expression levels were shown to be relatively higher. However,gfTrypmRNA expression levels were very low in brain, gill and kidney.
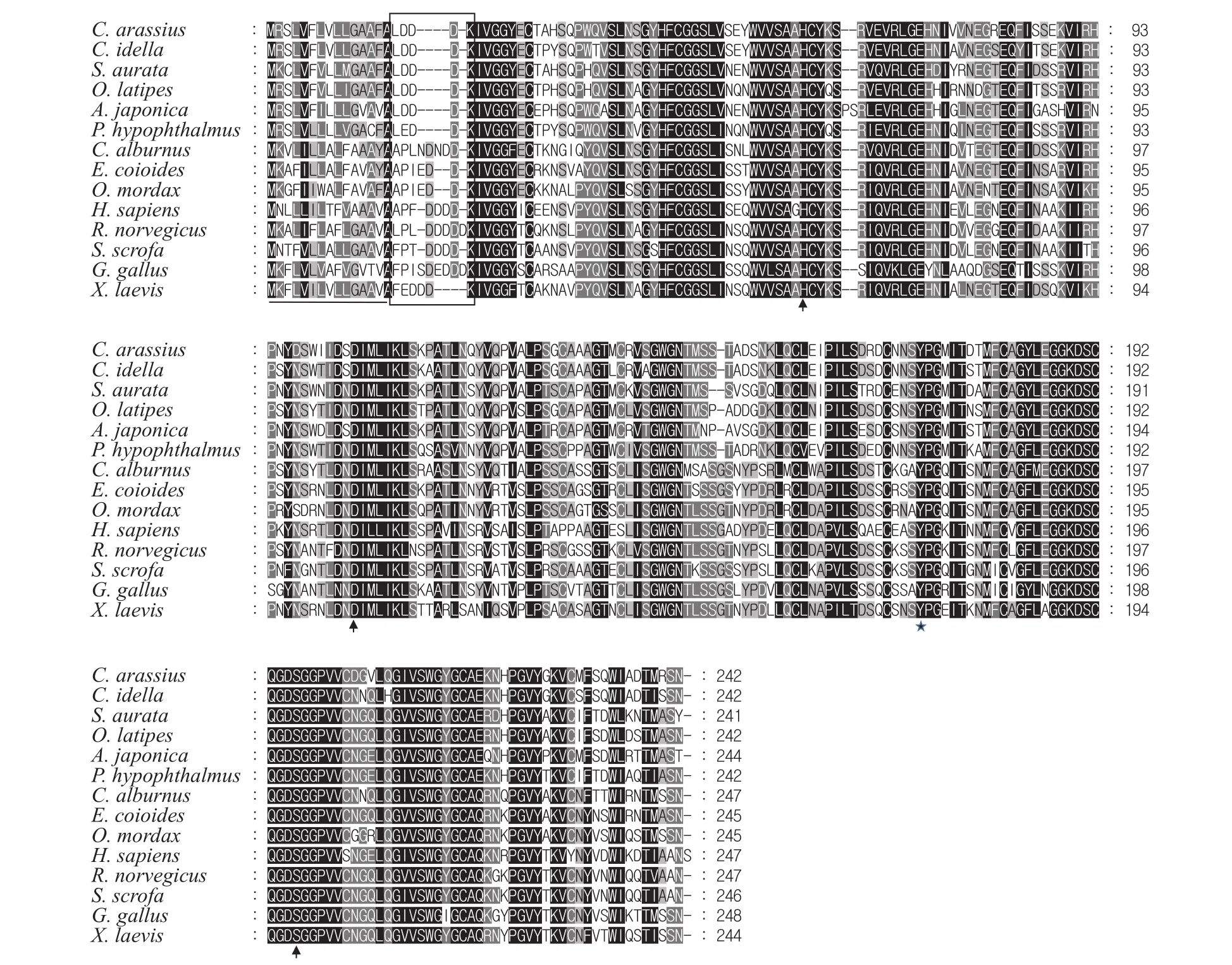
Fig. 1 Amino acid sequence alignment of trypsinogens among different species
2.3 Expression profiles of gfTryp gene in food deprivation and periprandial change
Food deprivation with one week significantly decreased the hepatopancreaticgfTrypmRNA level(Fig. 4A). Periprandial expression analysis showed thatgfTrypmRNA expression level in hepatopancreas was dramatically up-regulated after meal compared to the pre-prandial level (Fig. 4B).
2.4 Expression profiles of gfTryp gene in response to cadmium and H2O2 exposure
To preliminarily understand the expression patterns of trypsinogen gene under heavy metal and oxidative stress,gfTrypexpression levels under cadmium and H2O2exposure were investigated by real-time PCR.gfTrypmRNA expression levels in hepatopancreas increased significantly at 0.5 and 5 μg/mL cadmium (3.2 and 4.7 fold, respectively). However,gfTrypmRNA expression level decreased when the concentration up to 10 μg/mL. As shown in Fig. 5B,gfTrypmRNA expression levels in hepatopancreas decreased significantly at 3h, 6h, 12h and 24h after H2O2exposure (with the maximal response of 0.21 fold at 6h).
3 Discussion
In the present study, thegfTrypcDNA was initially cloned and identified. Alignment analysis showed that gfTryp had the conversed structural characteristics, including catalytic triad residues (His-57,Asp-102, and Ser-195), 12 cysteines forming 6 disulfide bonds, Asp-189 at the bottom of the substratebinding pocket and Gly-216 and Gly-226 lining the sides of the binding pocket. In addition, the con-sensus repeat, GDSGG, around the active site Ser-195, which is usually considered as the diagnostic of a serine protease[26], was also well conserved in goldfish (194GDSGG198). The Tyr residue (Tyr-170), which was shown to be a key residue in determining substrate specificity of trypsins[27], was also present in goldfish. These results reveal that gfTryp may possess the conserved function of protein digestion.Moreover, vertebrate trypsinogens can be classified into three groups: group I or anionic trypsinogens,group II or cationic trypsinogen and group III or psychrophilic trypsinogens[5,25,28]. The gfTryp obtained in this study was shown to group with teleost trypsinogen I based on the phylogenetic tree.

Fig. 2 Phylogenetic tree based upon the alignment of amino acid sequences of trypsinogens from a wide range of vertebrates using the Neighbor-joining bootstrap method by MEGA 6.05 with 1000 bootstrap replicates
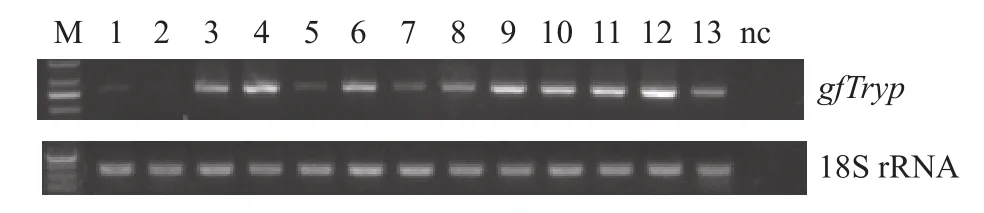
Fig. 3 Tissue distribution of trypsinogen mRNA in different tissues of adult goldfish
gfTrypmRNA was widely expressed in the tested tissues, and the higher levels were observed in the fat, anterior intestine, middle intestine, posterior intestine, hepatopancreas, testis, spleen, heart and skin. The relatively higher expression levels in the hepatopancreas and intestine indicate its conserved function of protein digestion in teleosts. In the following study, we observed that thegfTrypmRNA expression level in hepatopancreas was significantly downregulated when fasting but sharply up-regulated after meal, indicating its nutritional regulation. Meanwhile,the broad tissue expression pattern also suggests that the trypsinogen or its activated trypsin might be involved in other biological actions in teleosts. We are particularly interested in the mild expression ofgfTrypin the testis. In fact, it had previously reported that the trypsinogen mRNA was abundantly expressed in the testis of medaka and eel[29—31]. Their excellent data had confirmed that trypsin was an essential and multifunctional factor in spermatogenesis,and was involved in the male reproductive process.
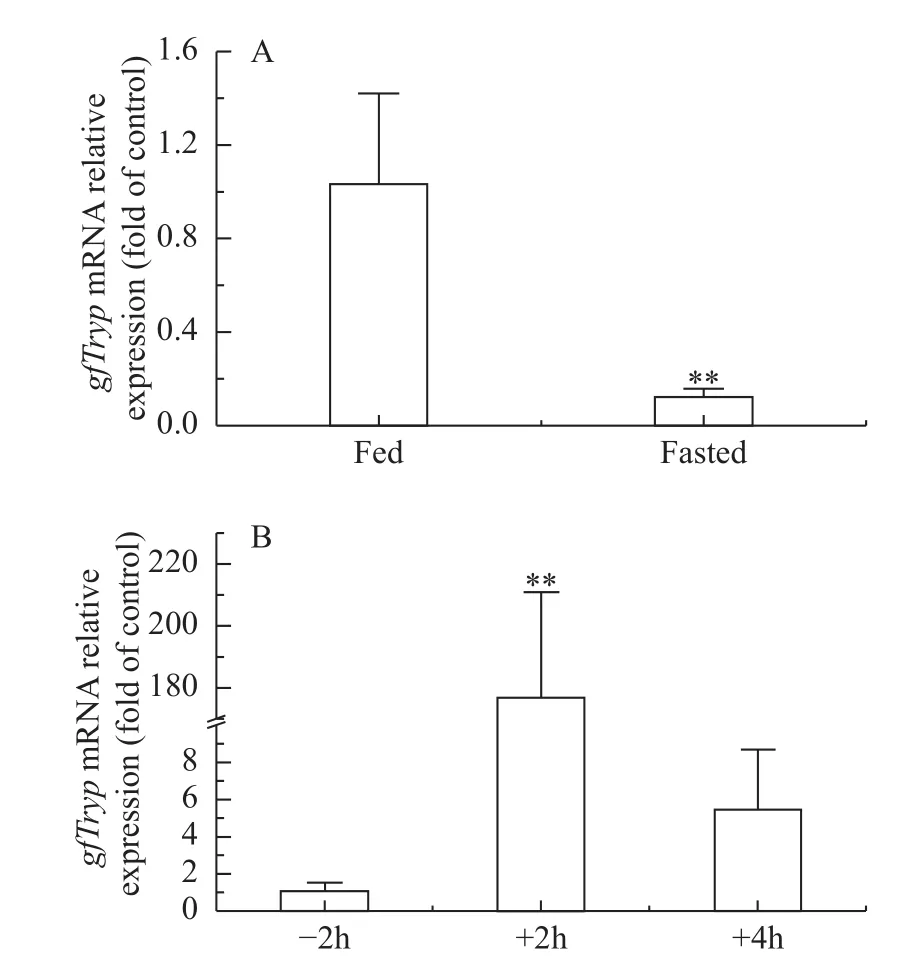
Fig. 4 mRNA expression of gfTryp gene in food deprivation (A)and periprandial change (B) in hepatopancreas
Cadmium is one of the most abundant and ubiquitously distributed toxic elements in aquatic environment[32]. It can trigger potentially highly toxic effect even at relatively lower doses. Using experimental animals and cell model systems, toxic effects of cadmium have been well established in many tissues including liver, kidney, lung, intestine and testes[33,34].However, the relevant information on the heavy mental effecting on digestive enzyme activity and mRNA expression in teleosts has not been well studied. As reported, the trypsin activity was decreased significantly at different cadmium concentrations changing from 0.5 to 5 μmol/L, and furthermore, the secondary structure of trypsin changed greatly with cadmium treatment[35]. Recently, it had been observed that the trypsin activity increased significantly in hepatopancreas, intestine and stomach of the freshwater crab(Sinopotamon henanense) at a low concentration of cadmium (7.25 mg/L), and dropped when increasing cadmium concentration up to 29 mg/L[36]. In mouse pancreas, Shimadaet al.[37]had reported the trypsin activity decreased significantly at one day after cadmium injection. However, the activity of trypsin increased significantly by cadmium treatmentin vitro,and the reason of the increasing trypsin activity may be due to the binding of cadmium at the calcium binding sites[37]. In conclusion, the regulation pattern of cadmium on trypsin activity may be related to its concentration or exposure method. In the present study,we observed that, cadmium at the concentrations of 0.5 and 5 μg/mL could significantly increasegfTrypmRNA expression (~3.2- and ~4.7 fold, respectively).It had been reported that cadmium could regulate gene expression depending on the transcriptional factor of MTF-1[38]. Whether or not the up-regulation ofgfTrypexpression by low cadmium concentration depending on MTF-1 needs to be further explored.
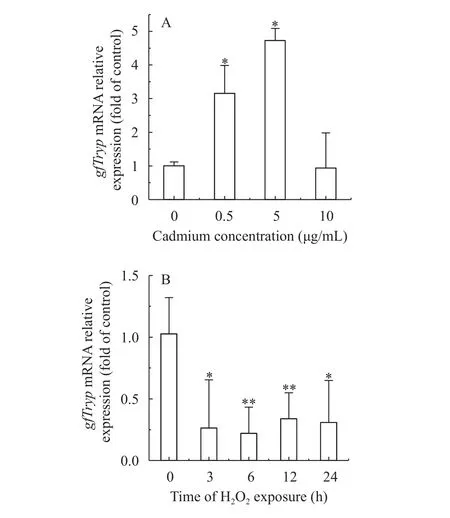
Fig. 5 Effect of cadmium (A) and H2O2 (B) on gfTryp mRNA levels in hepatopancreas of goldfish
In addition, cadmium stimulates the production of the ROS, and excessive ROS can further cause oxidative damage at the molecular, cellular and tissue levels[22]. In this study,gfTrypmRNA expression level decreased when the cadmium concentration up to 10 μg/mL. The observed decreasing expression ofgfTrypmRNA may be caused by the excessive ROS resulting from the high cadmium concentration exposure. As reported, H2O2is the most stable intermediate that generates intracellular non-radical ROS[39],and can induce many gene expression mainly associating with the reduction of peroxides and free radicals, including glutathione, glutathione reductase,glutathione S-transferase, catalase, CuZn SOD, Mn SOD, heat shock proteins[40—43]. Furthermore, it had been reported that oxidative stress could influence the activity of digestive enzymes[44]. Here, we further testedgfTrypmRNA expression level in response to H2O2exposure that generating the oxidative stress.The results showed thatgfTrypmRNA expression levels in hepatopancreas were significantly decreased at 3h, 6h, 12h and 24h under H2O2exposure. This is consistent with the result of high cadmium concentration exposure. Here, although the regulation mechanism needs to be further investigated, we have yet preliminarily validated the expression patterns ofgfTrypmRNA responding to oxidative stress.
Taken together, our studies successfully cloned thegfTrypcDNA from goldfish. The results showed that the nutritional status, cadmium and H2O2exposure could regulate the transcript level ofgfTrypmRNA. Our findings in the present study suggest that the gfTryp, as the mainly digestive enzyme, possesses the conserved digestive function, and further the expression patterns responding to environmental stresses shed new light on the physiological expression of trypsinogen in fish.
[1]Rawlings N D, Barrett A J. Families of serine peptidases [J].Methods in Enzymology, 1994, 244(1): 19—61
[2]Mithöfer K, Fernández-del Castillo C, Rattner D,et al. Subcellular kinetics of early trypsinogen activation in acute rodent pancreatitis [J].American Journal of Physiology, 1998,274(1): G71—G79
[3]Psochiou E, Sarropoulou E, Mamuris Z,et al. Sequence analysis and tissue expression pattern ofSparus auratachymotrypsinogens and trypsinogen [J].Comparative Biochemistry Physiology Part B, 2007, 147(3): 367—377
[4]Rypniewski W R, Perrakis A, Vorgias C E,et al. Evolutionary divergence and conservation of trypsin [J].Protein Engineering, 1994, 7(1): 57—64
[5]Chen J M, Kukor Z, Le Maréchal C,et al. Evolution of trypsinogen activation peptides [J].Molecular Biology and Evolution, 2003, 20(11): 1767—1777
[6]Gráf L, Jancsó A, Szilágyi L,et al. Electrostatic complementarity within the substrate-binding pocket of trypsin [J].Proceedings of the National Academy of Sciences of the United States of America, 1988, 85(14): 4961—4965
[7]Gudmundsdóttir A, Gudmundsdóttir E, Oskarsson S,et al.Isolation and characterization of cDNAs from Atlantic cod encoding two different forms of trypsinogen [J].European Journal of Biochemistry, 1993, 217(3): 1091—1097
[8]Genicot S, Rentier-Delrue F, Edwards D,et al. Trypsin and trypsinogen from an Antarctic fish: molecular basis of cold adaptation [J].Biochimica et Biophysica Acta, 1996,1298(1): 45—57
[9]Douglas S E, Gallant J W. Isolation of cDNAs for trypsinogen from the winter flounder,Pleuronectes americanus[J].Journal of Marine Biotechnology, 1998, 6(6): 214—219
[10]Suzuki T, Srivastava A S, Kurokawa T. cDNA cloning and phylogenetic analysis of pancreatic serine proteases from Japanese flounder,Paralichthys olivaceus[J].Comparative Biochemistry Physiology Part B, 2002, 131(1): 63—70
[11]Kurokawa T, Suzuki T, Ohta H,et al. Expression of pancreatic enzyme genes during the early larval stage of Japanese eelAnguilla japonica[J].Fisheries Science, 2002, 68(4):736—744
[12]Manchado M, Infante C, Asensio E,et al. Molecular characterization and gene expression of six trypsinogens in the flatfish Senegalese sole (Solea senegalensisKaup) during larval development and in tissues [J].Comparative Biochemistry Physiology Part B, 2008, 149(2): 334—344
[13]Ruan G L, Li Y, Gao Z X,et al. Molecular characterization of trypsinogens and development of trypsinogen gene expression and tryptic activities in grass carp (Ctenopharyngodon idellus) and topmouth culter (Culter alburnus) [J].Comparative Biochemistry Physiology Part B, 2010, 155(1):77—85
[14]Liu C H, Chen Y H, Shiu Y L. Molecular characterization of two trypsinogens in the orange-spotted grouper,Epinephelus coioides, and their expression in tissues during early development [J].Fish Physiology and Biochemistry, 2013, 39(2):201—214
[15]Mata-Sotres J A, Martos-Sitcha J A, Astola A,et al. Cloning and molecular ontogeny of digestive enzymes in fed and food-deprived developing gilthead seabream (Sparus aurata)larvae [J].Comparative Biochemistry Physiology Part B,2016, 191: 53—65
[16]Gamboa-Delgado J, Le Vay L, Fernández-Díaz C,et al. Effect of different diets on proteolytic enzyme activity,trypsinogen gene expression and dietary carbon assimilation in Senegalese sole (Solea senegalensis) larvae [J].Comparative Biochemistry Physiology Part B, 2011, 158(4): 251—258
[17]Tillner R, Rønnestad I, Harboe T,et al. Evidence for a regulatory loop between cholecystokinin (CCK) and tryptic enzyme activity in Atlantic cod larvae (Gadus morhua) [J].Comparative Biochemistry Physiology Part A, 2013, 166(3):490—495
[18]Zhu B, Wu Z F, Li J,et al. Single and joint actiontoxicity of heavy metals on early developmental stages of Chinese rare minnow (Gobiocypris rarus) [J].Ecotoxicology and Environmental Safety, 2011, 74(8): 2193—2202
[19]Lai T H, He B Y, Fan H Q,et al. Effects of cadmium stress on the activities of antioxidant enzymes, digestive enzymes and the membrane lipid peroxidation of the mangrove mud clamGeloina coaxans(Gmelin) [J].Acta Ecologica Sinica,2011, 31(11): 3044—3053
[20]De Coen W M, Janssen C R. The use of biomarkers in Daphnia magna toxicity testing II. Digestive enzyme activity inDaphnia magnaexposed to sublethal concentrations of cadmium, chromium and mercury [J].Chemosphere, 1997,35(5): 1053—1067
[21]Kuzmina V V, Golovanova I L, Kovalenko E. Separate and combined effects of cadmium, temperature and pH on digestive enzymes in three freshwater teleosts [J].Bulletin of Environmental Contamination and Toxicology, 2002, 69(2):302—308
[22]Wang Q, Ju X, Chen Y,et al. Effects of L-carnitine against H2O2-induced oxidative stress in grass carp ovary cells(Ctenopharyngodon idellus) [J].Fish Physiology and Biochemistry, 2016, 42(3): 845—857
[23]Chen W, Zhang Z, Dong H,et al. Molecular cloning and sequence analysis of selenoprotein W gene and its mRNA expression patterns in response to metabolic status and cadmium exposure in goldfish,Carassius auratus[J].Comparative Biochemistry Physiology Part B, 2015, 184: 1—9
[24]Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCTmethod [J].Methods, 2001, 25(4): 402—408
[25]Roach J C. A clade of trypsins found in cold-adapted fish [J].Proteins, 2002, 47(1): 31—44
[26]Krem M M, Rose T, Cera E. The C-terminal sequence encodes function in serine proteases [J].Journal of Biological Chemistry, 1999, 274(40): 28063—28066
[27]Hedstrom L, Perona J J, Rutter W J. Converting trypsin to chymotrypsin: residue 172 is a substrate specificity determinant [J].Biochemistry, 1994, 33(29): 8757—8763
[28]Roach J C, Wang K, Gan L,et al. The molecular evolution of the vertebrate trypsinogens [J].Journal of Molecular Evolution, 1997, 45(6): 640—652
[29]Ogiwara K, Takahashi T. Specificity of the medaka enteropeptidase serine protease and its usefulness as a biotechnological tool for fusion-protein cleavage [J].Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(17): 7021—7026
[30]Miura C, Ohta T, Ozaki Y,et al. Trypsin is a multifunctional factor in spermatogenesis [J].Proceedings of the National Academy of Sciences of the United States of America,2009, 106(49): 20972—20977
[31]Rajapakse S, Ogiwara K, Takahashi T. Characterization and expression of trypsinogen and trypsin in medaka testis [J].Zoological Science, 2014, 31(12): 840—848
[32]Almeida J A, Novelli E L, Dal Pai Silva M,et al. Environmental cadium exposure and metabolic responses of the Nile tilapia,Oreochromis niloticus[J].Environmental Pollution,2001, 114(2): 169—175
[33]Zhou T, Zhou G, Song W,et al. Cadmium-induced apoptosis and changes in expression of p53, c-jun and MT-I genes in testes and ventral prostate of rats [J].Toxicology, 1999,142(1): 1—13
[34]Kuriwaki J, Nishijo M, Honda R,et al. Effects of cadmium exposure during pregnancy on trace elements in fetal rat liver and kidney [J].Toxicology Letters, 2005, 156(3): 369—376
[35]Hong F, Wang L, Wang X,et al. Effects of Ce3+, Cd2+, and Hg2+on activities and secondary structure of trypsin [J].Biological Trace Element Research, 2003, 95(3): 233—240
[36]Wu H, Xuan R, Li Y,et al. Effects of cadmium exposure on digestive enzymes, antioxidant enzymes, and lipid peroxidation in the freshwater crabSinopotamon henanense[J].Environmental science and pollution research, 2013, 20(6):4085—4092
[37]Shimada H, Funakoshi T, Waalkes M P. Acute, nontoxic cadmium exposure inhibits pancreatic protease activities in the mouse [J].Toxicological Sciences, 2000, 53(2):474—480
[38]Wimmer U, Wang Y, Georgiev O,et al. Two major branches of anti-cadmium defense in the mouse: MTF-1/metallothioneins and glutathione [J].Nucleic Acids Research, 2005,33(18): 5715—5727
[39]Rosa C E, Bianchini A, Monserrat J M. Antioxidant responses of Laeonereis acuta (Polychaeta) after exposure to hydrogen peroxide [J].Brazilian Journal of Medical and Biological Research, 2008, 41(2): 117—121
[40]Rhee J S, Kim R O, Choi H G,et al. Molecular and biochemical modulation of heat shock protein 20 (Hsp20) gene by temperature stress and hydrogen peroxide (H2O2) in the monogonont rotifer,Brachionus sp[J].Comparative Biochemistry Physiology Part C, 2011, 154(1): 19—27
[41]Zhang Y, Fu D, Yu F,et al. Two catalase homologs are involved in host protection against bacterial infection and oxida-tive stress inCrassostrea hongkongensis[J].Fish and Shellfish Immunology, 2011, 31(6): 894—903
[42]Yang J, Dong S, Jiang Q,et al. Characterization and expression of cytoplasmic copper/zinc superoxide dismutase(CuZnSOD) gene under temperature and hydrogen peroxide(H2O2) in rotiferBrachionus calyciflorus[J].Gene, 2013,518(2): 388—396
[43]Lu X, Wang C, Liu B. The role of Cu/Zn-SOD and Mn-SOD in the immune response to oxidative stress and pathogen challenge in the clamMeretrix meretrix[J].Fish and Shellfish Immunology, 2015, 42(1): 58—65
[44]Lackeyram D, Mine Y, Widowski T,et al. Thein vivoinfusion of hydrogen peroxide induces oxidative stress and differentially affects the activities of small intestinal carbohydrate digestive enzymes in the neonatal pig [J].Journal of Animal Science, 2012, 90(Suppl. 4): 418—420
