两个含二齿席夫碱配体单核镍和铜配合物的合成、结构及抗菌活性
2017-11-01李沙沙于清泉
刘 超 李沙沙 杨 敏 毕 蓉 于清泉 张 莉
刘 超*李沙沙 杨 敏 毕 蓉 于清泉 张 莉
(宿州学院化学化工学院,宿州 234000)
在溶剂热条件下合成了2种含HL(HL=(N-(3-甲基亚水杨基)吲哚乙胺)配体的金属配合物Ni(L)2(1)和Cu(L)2(2),并通过热重分析、红外光谱、元素分析和单晶X射线衍射表征其结构。配合物1和2均属于单斜晶系,空间群为P21/n。配合物1的晶胞参数为 a=1.481 6(7)nm,b=1.310 0(6)nm,c=1.668 7(7)nm,β=111.346(7)°,配合物 2 的晶胞参数为 a=1.479 7(2)nm,b=1.299 41(19)nm,c=1.667 6(2)nm,β=111.537(3)°。配合物1和2具有类似的单核结构,通过分子内C-H…O氢键和分子间D-H…π氢键作用分别将其进一步延伸为三维超分子结构。而且,通过琼脂扩散法对配合物和配体的抗菌活性进行了细致的筛选,实验结果表明配合物1和2比配体具有更优异的抑菌效应。
镍配合物;铜配合物;晶体结构;席夫碱;抗菌活性
During the past decades,transition metal complexes have attracted the attention of more and more researchers because of not only their rich coordination modes,but also their application in many fields of catalyst[1],photochemistry[2],magnetic material[3],medicine[4]and so on[5].Recently,a great deal of effort hasbeen directed toward the synthesis of novel transition metal complexes with excellent biological activities and rich fruits have been obtained[6-7].The design and synthesis of new ligand is the key to search for the metal complexes serving as organic drugs,and some other factors such as central metal ion and hydrogen bonding week interaction also play important roles.A variety of organic ligands are able to form biologically active transition metal complexes,but Schiff base ligands are better candidates for their hard donor-atom frameworks,stable structures,and easy modification[8-9].As two important transition metal complexes,mononuclear Niand Cucomplexes,especially those with Schiff base as the ligands,are inextricably linked to the pharmaceutical science,and have been widely used intheresearch and developmentof novel drugs[10-11].
Up to now,some mononuclear Niand Cucomplexes constructed by the Schiff bases with monodentate,bidentate and polydentate structures have been synthesized,and their applications in the drug field have been widely reported[12-14].In this paper,two novel mononuclear Niand Cucomplexes with a bidentate Schiff base ligand derived from tryptamine have been synthesized and characterized.In addition,their antibacterial activities against Escherichia coli,Bacillus subtilis,and Staphylococcus aureus were measured and discussed.
1 Experimental
1.1 Materials and methods
Tryptamine and 3-methyl-2-hydroxybenzaldehyde were commercially purchased and used as received.All other reagents and solvents were of analytical reagent grade and used without further purification.IR spectra were determined on a Perkin-Elmer 377 FTIR spectrometer by the KBr pressed disc method within the 4 000~400 cm-1region.Thermalgravimetric analyses(TGA)were performed on Perkin-Elmer TGA7 thermogravimetric analyzer under nitrogen atmosphere with a heating rate of 10℃·min-1.1H NMR spectra of HL were measured with a Bruker AV-400 spectrometer using CDCl3solvent.Elemental analyses of C,H and N were carried out with a Vario ELⅢelemental analyzer.
1.2 Synthesis of the HL ligand
The ligand HL was synthesized according to the literature[15].To a methanolic solution(40 mL)of 3-methyl-2-hydroxybenzaldehyde (2 mL,16.66 mmol)wasadded a methanolic solution(10 mL)of tryptamine(2.67 g,16.66 mmol)with stirring.The resulting mixture was refluxed for 36 h and then evaporated to dryness.The yellow crystalline solid was obtained after standing 20 mL of ethanol solution in the refrigerator overnight.Yield:89.0%.m.p.89~90 ℃.1H NMR(400 MHz,CDCl3):δ2.26(s,3H,CH3),3.19(t,J=6.9 Hz,2H,Indol-CH2),3.95 (t,J=3.6 Hz,2H,CH2N),7.02~6.75(m,2H,4,5-Ar-H),7.22~7.10(m,2H,5,6-Indol-H),7.28(m,1H,6-Ar-H),7.34(m,1H,2-Indol-H),7.96~7.71(m,2H,4,7-Indol-H),8.21(s,1H,N=CH),13.98(br,1H,NH).
1.3 Synthesis of complex 1
An ethanol solution(5 mL)of HL(0.55 g,2.00 mmol)was added dropwise to an ethanol solution(50 mL)of Ni(OAc)2·4H2O(0.25 g,1.00 mmol)under room temperature.The resulting mixture was refluxed with stirring for 24 h and the reaction solution was filtered after cooling to room temperature.After removing the solvent,the obtained powder was dissolved in a mixed solvent of CH2Cl2and CH3OH(VCH2Cl2/VCH3OH=2),and dark brown single-shaped crystals of 1 suitable for X-ray analysis were obtained in two days under air evaporation.Yield:85.0%.m.p.112~113 ℃.Anal.Calcd.for C36H34N4NiO2(%):C,70.49;H,5.59;N,9.13.Found(%):C,70.23;H,5.61;N,9.31.IR(KBr,cm-1):3 552(m),3 476(m),3 399(vs),3 028(w),2 940(w),2 901(w),1 618(s),1 596(s),1 571(s),1 450(s),1 427(s),1 312(s),1 236(m),1 019(m),864(w),750(s),710(m),678(w),635(w),587(w).
1.4 Synthesis of complex 2
Complex 2 was prepared and crystallized by the identical procedure as described for complex 1,with Ni(OAc)2·4H2O replaced by Cu(OAc)2·H2O(0.20 g,1.00 mmol),and the mixed solvent of CH2Cl2and CH3OH replaced by 15 mL of DMF.The blue block crystals of 2 suitable for X-ray analysis were obtained in three days under air evaporation.Yield:69.5%.m.p.85~86 ℃.Anal.Calcd.for C36H34N4CuO2(%):C,69.94;H,5.54;N,9.06.Found(%):C,70.17;H,5.38;N,8.94.IR(KBr,cm-1):3 552(m),3 481(s),3 394(vs),3 049(w),2 923(w),1 618(s),1 596(s),1 547(s),1 415(s),1 328(m),1 219(m),1 088(m),1 027(w),863(w),749(m),711(m),678(w),623(m),492(w).
1.5 Crystal structure determination
Suitable single crystals of complexes 1 and 2 with dimensions of 0.19 mm×0.22 mm×0.26 mm and 0.03 mm ×0.16 mm ×0.27 mm were selected under an optical microscope and glued to thin glass fibers,respectively.Diffraction data were collected on a Bruker SMARTAPEX-ⅡCCDdiffractometer at 296(2)K byφ~ω scan mode,equipped with a graphite-monochromatized Mo Kα radiation(λ=0.071 073 nm).The structures of the complexes were solved by direct methods and refined by full-matrix least-squares method on F2with the SHELXS-97 and SHELXL-97 programs[16-17].All non-hydrogen atoms were refined anisotropically,while the hydrogen atoms attached to C,N and O atoms were positioned geometrically and refined isotropically using a riding model.Detailed crystallographic data and structure refinement for complexes 1 and 2 are given in Table 1,the selected bond lengths and bond angles are listed in Table 2,and the relevant hydrogen bonding parameters are summarized in Table 3.
CCDC:1511747,1;1511743,2.

Table 1 Crystallographic data and structure refinement for complexes 1 and 2

Table 2 Selected bond lengths(nm)and bond angles(°)of complexes 1 and 2

Continued Table 2

Table 3 Structural parameters of C-H…O bonds and D-H…πbonds for complexes 1 and 2
1.6 Antibacterial activity
The complexes and HL were tested in vitro to assess their antibacterial activities against three bacterial strains using the agar diffusion assay.Antibacterial activities of the compounds were evaluated by measuring the diameters of inhibition rings.It was found that the bigger the diameter of inhibition ring was,the better the antibacterial activities of the compound were.These compounds were dissolved in DMSO to prepare three different concentrations (10.0,5.0 and 2.5 mg·mL-1)for the evaluation of dose response and stocked after sterilization,respectively.The test strains included Escherichia coli,Bacillus Subtilis and Staphylococcus aureus.The sterilized filter papers with 6 mm diameter were placed on the culture medium after being soaked for 2 h in the solution.After an incubation period of 24 h at 37℃for the strains,the diameters of inhibition rings were determined in the absence of microbial growth.
2 Results and discussion
2.1 Crystal structures of complexes 1 and 2
The crystal structure of complex 1,similar to complex 2,crystallizes in the monoclinic system with space group P21/n and exhibits a slightly distorted square-planar geometry.As shown in Fig.1,both complex 1 and complex 2 consist of one metal ion and two deprotonated ligands in the molecule structures.The two metal ions are tetra-coordinated by two hydroxyl oxygen atoms from two Schiff base ligands and two imino nitrogen atoms from the two ligands in aη2mode,forming two six-membered chelate rings of NiNOC3and CuNOC3,respectively.The metal ion lies approximately in the center of the square-planar geometry constructed by the four donor atoms and the metal atom with a slight deviation.Interesting,two MO bonds and two M-N bonds are in opposite positions of the central Mion,resulting in the trans-planar structure.The six bond angles around the Niion fall in the range of 87.38(11)°~175.92(12)°and the six bond angles around Cuion are from 88.22(13)°to 174.67(13)°,indicating that the metal ion and the four coordination atoms are nearly coplanar,respectively.In complex 1,the average bond distances of Ni-N and Ni-O are 0.191 0(3)and 0.184 0(3)nm,respectively.In complex 2,the average bond distances of Cu-N and Cu-O are 0.198 9(3)and 0.189 6(3)nm,respectively.These data in complex 1 and complex 2 are consistent with those found in similar four-coordinated complexes with square-planar geometries[18-19].In the complexes,the hydroxyl oxygen atoms and the imino nitrogen atoms take part in coordinating with the metal ions of Niand Cu,which makes the CO bond lengths in the complexes become shorter than those in the free ligand(C-O 0.129 7(2)nm)and makes the C=N bond lengths in the complexes longer than those in the free ligand(C=N 0.129 3(2)nm).

Fig.1 Molecular structure of the complex with 30%thermal ellipsoid level(M=Ni or Cu)
In complex 1,the chelate ring plane P1(composed of atoms Ni(1),O(1),C(17),C(12),C(11)and N(3))and the phenyl ring plane P2(composed of atoms C(12)~C(17))are nearly coplanar with a dihedral angle of 13.17(15)°.To avoid the steric hindrance between the aromatic rings of the ligand,the phenyl ring plane P2 and the five-membered ring plane P3 (composed of atoms N(1),C(7),C(8),C(1)and C(6))of the ligand are rotated with respect to each other with a dihedral angle of 64.6(2)°.In complex 2,the dihedral angle between the chelate ring plane P1(composed of atoms Cu(1),O(1),C(17),C(12),C(11)and N(3))and the phenyl ring plane P2(composed of atoms C(12)~C(17))is 14.89(19)°,and the dihedral angle between the phenyl ring plane P2 and the five-membered ring plane P3(composed of atoms N(1),C(7),C(8),C(1)and C(6))of the ligand is 64.8(3)°.
The non-classical hydrogen bonds,such as C-H…O bonds and D-H…πbonds,are very important in the molecular structural stabilization.Similar hydrogen bonding interactions and the results of the structure analyses confirm that complexes 1 and 2 possess the similar supramolecular frameworks.Thus,only the crystal packing diagram of complex 1 is described in detail.As shown in Fig.2,there are three types of CH…O hydrogen bonds between the hydroxyl oxygen atom from one ligand and the methylene carbon atom from another ligand in complex 1,which are C(10)-H(10A)…O(2)(0.274 7(4)nm),C(27)-H(27B)…O(1)(0.310 1(5)nm)and C(28)-H(28A)…O(1)(0.276 0(4)nm),respectively.In addition,three types of C-H…π hydrogen bonds and two types of N-H…πhydrogen bonds,which are C(11)-H(11)…Cg(8)#1(0.383 2(4)nm),C(21)-H(21)…Cg(5)#3(0.373 5(5)nm),C(22)-H(22)…Cg(1)#3(0.364 1(5)nm),N(1)-H(1)…Cg(7)#1(0.322 2(5)nm)and N(2)-H(2A)…Cg(6)#2(0.326 5(5)nm),are also observed in complex 1.As a result,the complex molecules are extended into a threedimensional supramolecular network structure through the above hydrogen bonding interactions.The hydrogen bonds in complex 2,with the C-H…O bond distances in the range of 0.284 2(5)~0.312 9(6)nm and the DH…π bond distances in the range of 0.320 4(5)~0.377 7(5)nm,are consistent with complex 1 in the types of hydrogen bonds.

Fig.2 Crystal packing diagram of complex 1
2.2 Thermal analysis of complexes 1 and 2
To estimate the thermal stability of complexes 1 and 2,thermal gravimetric analyses (TGA)were carried out on crystalline samples in the temperature range of 30~800 ℃.As shown in Fig.3,the TGA curve of complex 1 reveals that it is thermally stable up to 295℃,which indicates that complex 1 can retain structural integrity below 295℃.After that,complex 1 undergoes a one-step weight loss process from 295 to 675℃.The weight lose of 87.78%(Calcd.87.82%)is ascribable to the decomposition of the HL ligand in complex 1,suggesting that the final residue was NiO.Complex 2 also undergoes a onestep weight loss process from 250 to 555℃,which is similar to that of complex 1.As shown in Fig.4,the weight loss of 86.87% (Calcd.87.13%)is also ascribable to the decomposition of the HL ligand in complex 2,suggesting that the final residue was CuO.

Fig.3 TGA curve of complex 1

Fig.4 TGA curve of complex 2
2.3 Antibacterial activity
As shown in Table 4,all of the compounds except DMSO exhibited antibacterial activities against Escherichia coli,Bacillus Subtilis and Staphylococcus aureus.It has been observed from the data that both complex 1 and complex 2 had stronger antibacterial activities than the HL ligand against the same bacteria,which might be related to the biological function of nickel and copper elements.Hence,the complexation with the metal ion increased the antibacterical activities of the HL ligand.The antibacterial activities of the HL ligand were slightly different for the three bacteria,while those of the complexes had notable differences.Meanwhile,with the increase of concentration of these compounds(2.5,5.0 and 10.0 mg·mL-1),the greater diameter of inhibition ring was observed.When the concentration of these compounds was 10.0 mg·mL-1,complex 1 showed the most effective antibacterial activitiesagainst Escherichia coli,followed by Staphylococcus aureus and Bacillus subtilis in order.However,complex 2 showed the most effective antibacterial activities against Escherichia coli,followed by Bacillus subtilis and Staphylococcus aureus in order.In addition,the ligand exhibited better inhibitory activities against Escherichia coli with inhibition ring for 8.7 mm,and complexes 1 and 2 exhibited better inhibitory activities against Escherichia coli with inhibition rings for 17.2 mmand 16.7 mm,respectively.Taking into account their excellent inhibitory activities,the complexes could be considered as antibacterial compounds for further research and development.
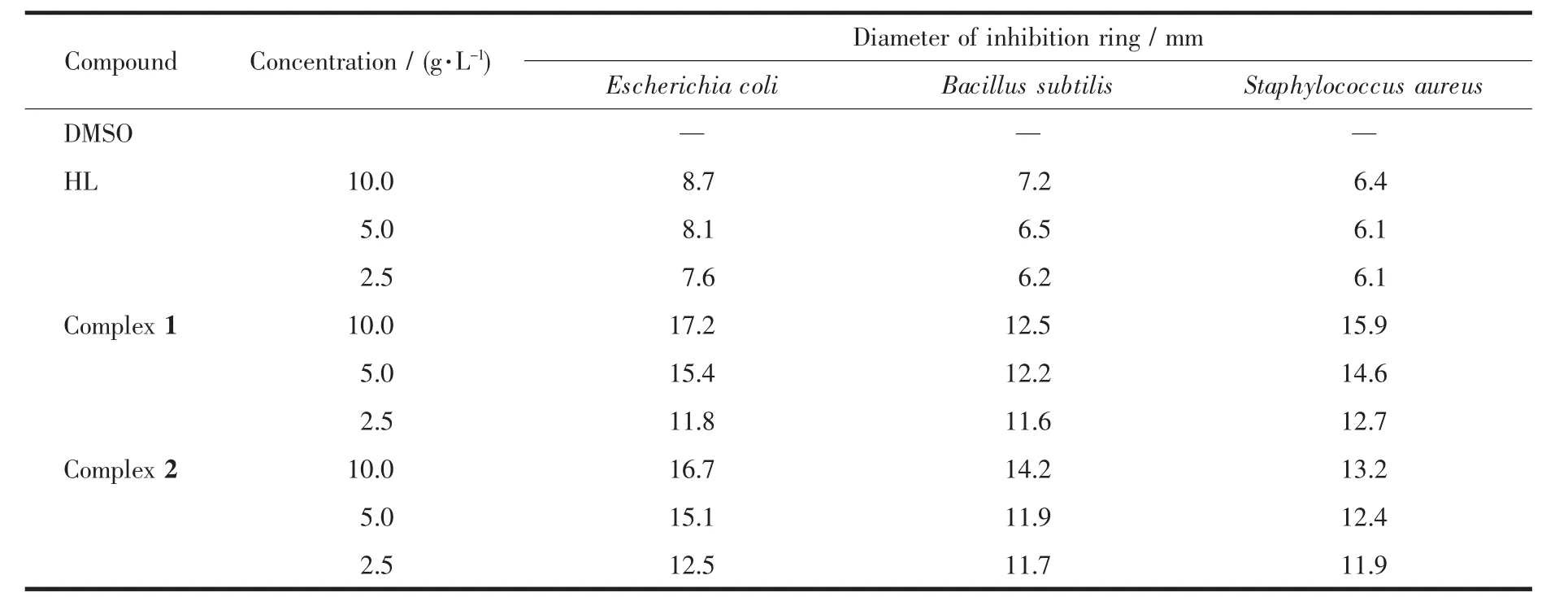
Table 4 Data on antibacterial activity for HL and the complexes
3 Conclusions
In summary,two mononuclear complexes based on N-(3-methylsalicylidene)tryptamine)ligand,Ni(L)2(1)and Cu(L)2(2),have been synthesized and confirmed successfully.Single-crystal X-ray diffraction analyses showed that the metal atoms in complexes 1 and 2 coordinate to two ligands to form a slightly distorted square-planar geometry around the metal atoms,respectively.The complex molecules are stabilized by intramolecular C-H…O hydrogen bonds,and then further extended into an infinite 3D supramolecular frameworks by intermolecular D-H…π hydrogen bonds,respectively.The ligand as well as complexes 1 and 2 exhibited antibacterial activities against the three test strains:Escherichia coli,Bacillus subtilis and Staphylococcus aureus,while complexes 1 and 2 displayed more excellent inhibiting effects than the free ligand.
[1]Kargar H.Transition Met.Chem.,2014,39:811-817
[2]MAO Pan-Dong(毛盼东),YAN Ling-Ling(闫玲玲),WU Wei-Na(吴伟娜),et al.Chinese J.Inorg.Chem.(无机化学学报),2016,32(5):879-883
[3]Wang K,Chen Z L,Zou H H,et al.Transition Met.Chem.,2017,42:17-23
[4]Luo W,Meng X,Xiang J,et al.Inorg.Chim.Acta,2008,361:2667-2676
[5]Hegde D,Naik G N,Vadavi R S,et al.Inorg.Chim.Acta,2017,461:301-315
[6]Alagesan M,Bhuvanesh N S,Dharmaraj N.Dalton Trans.,2013,42:7210-7223
[7]Patel R N,Singh A,Sondhiya V P,et al.J.Coord.Chem.,2012,65:795-812
[8]Sanyal R,Dash SK,Kundu P,et al.Inorg.Chim.Acta,2016,453:394-401
[9]Fekri R,Salehi M,Asadi A,et al.Polyhedron,2017,128:175-187
[10]Budige G,Puchakayala M R,Kongara S R,et al.Chem.Pharm.Bull.,2011,59:166-171
[11]Keypour H,Jamshidi A H,Rezaeivala M,et al.Polyhedron,2013,52:872-878
[12]Creaven B S,Czeglédi E,Devereux M,et al.Dalton Trans.,2010,39:10854-10865
[13]Dong X,Li Y,Li Z,et al.J.Inorg.Biochem.,2012,108:22-29
[14]ZHAO Hai-Yan(赵海燕),YANG Xiao-Dong(杨晓东),LI Na(李娜).Chinese J.Inorg.Chem.(无机化学学报),2017,33(4):685-691
[15]LIU Chao(刘超),ZHUO Xin(卓馨),WU Yi-Kang(吴怡康),et al.J.Suzhou Univ.(宿州学院学报),2011,26(5):60-62
[16]Sheldrick G M.SHELXL-97,Program for Crystal Structure Refinement,University of Göttingen,Germany,1997.
[17]Sheldrick G M.SHELXS-97,Program for Crystal Structure Solution,University of Göttingen,Germany,1997.
[18]Deng J Q,Liang H,Lu X C,et al.Chin.J.Struct.Chem.,2016,35:559-565
[19]Zhao P S,Song J,Sun X J,et al.Chin.J.Struct.Chem.,2011,30:346-353
The title complexes,Ni(L)2(1)and Cu(L)2(2)based on HL(HL=N-(3-methylsalicylidene)tryptamine)have been synthesized under solvothermal conditions and characterized structurally by TGA,infrared spectra,elemental analysis and single-crystal X-ray diffraction.Complexes 1 and 2 both crystallize in monoclinic system,and the space groups are P21/n with the unit cell parameters for complex 1:a=1.481 6(7)nm,b=1.310 0(6)nm,c=1.668 7(7)nm,β=111.346(7)°,and for complex 2:a=1.479 7(2)nm,b=1.299 41(19)nm,c=1.667 6(2)nm,β=111.537(3)°.Complexes 1 and 2 possess the similar mononuclear structures,which are further extended into infinite 3D supramolecular frameworks by intramolecular C-H…O hydrogen bonds and intermolecular D-H…π hydrogen bonds,respectively.Furthermore,the complexes and the ligand have been investigated for their preliminary antibacterial activities in detail by agar diffusion method,and the results demonstrate that complexes 1 and 2 display more excellent inhibiting effects than the ligand.CCDC:1511747,1;1511743,2.
Nicomplex;Cucomplex;crystal structure;Schiff base;antibacterial activity
O614.81+3;O614.121
A
1001-4861(2017)10-1861-08
10.11862/CJIC.2017.220
LIU Chao*LI Sha-Sha YANGMin BI Rong YU Qing-Quan ZHANGLi
(School of Chemistry and Chemical Engineering,Suzhou University,Suzhou,Anhui 234000,China)
2017-05-10。收修改稿日期:2017-07-20。
国家自然科学基金(No.21271136)和安徽省高校自然科学重点研究基金(No.KJ2016A772,KJ2017A436)资助项目。*
。 E-mail:ahliuchao333@163.com
