基于线粒体控制区的中国南海海域卵形鲳鲹遗传多样性
2017-07-12吕金磊杨喜书宫亚运
吕金磊,章 群,杨喜书,宫亚运,曹 艳
(暨南大学水生生物研究中心,教育部热带亚热带水生态工程研究中心,广州 510632)
基于线粒体控制区的中国南海海域卵形鲳鲹遗传多样性
吕金磊,章 群,杨喜书,宫亚运,曹 艳
(暨南大学水生生物研究中心,教育部热带亚热带水生态工程研究中心,广州 510632)
为探讨中国南海重要经济鱼类卵形鲳鲹(Trachinotus ovatus)的遗传多样性,测定了广东闸坡、乌石、安铺和广西东兴以及海南新盈等5个地理群体97 ind样品的线粒体控制区5′端359 bp序列,发现47个变异位点,32个单倍型,总体呈现高单倍型多样性(h=0.951)和高核苷酸(π=0.020 9)多样性的特点。在邻接树和单倍型网络图中出现2个分化显著但不存在明显地理聚群的分支,推测二者的分化时间约为60~18万年前(中更新世),可能是中更新世冰期海平面下降形成边缘海而导致隔离,间冰期海平面上升后出现二次接触。不同地理群体间的遗传分化不显著(Fst=-0.022 4~0.045 3),AMOVA分析也显示97%以上的遗传变异来源于群体内个体间。卵形鲳鲹2个谱系及总体的核苷酸错配图呈现多峰,中性检验均为负值不显著(P>0.05),表明都未经历过大规模的种群扩张,处于相对稳定的状态。
卵形鲳鲹;中国南海;线粒体控制区;遗传多样性
卵形鲳鲹(Trachinotus ovatus)隶属鲈形目(Perciformes)鲹科(Carangidae),俗称金鲳[1];因其体表无鳞,无肌间刺且肉质细嫩、味道鲜美,具有较高的食用和营养价值[2],而成为中国和东南亚等产地广受消费者欢迎的名贵鱼类,也是中国华南地区养殖规模最大、产量最高的主导品种之一[3-4]。目前养殖群体未经选育,就已出现生长慢、成活率低、品质差等种质衰退的现象[4]。同时,随着海洋环境的恶化和过度捕捞,南海渔业资源在不断的衰减,卵形鲳鲹也不例外[5]。因此,亟待开展种质资源的保护和良种选育研究,而了解经济鱼类的遗传背景,是良种选育和资源保护的基础[6]。
目前,国内外卵形鲳鲹的研究报道多集中在人工养殖育种、胚胎发育、疾病诊断和治疗等方面,种群遗传方面的研究较少,仅有孙立元等[7]采用微卫星标记分析海南、广东2个育种群体的遗传多样性;彭敏等[8]利用AFLP技术分析比较了北部湾野生群体和养殖群体的遗传多样性;赵永贞等[9]采用微卫星分子标记技术分析了南海4个群体的遗传多样性;吉磊等[10]分析了海南、广东、福建3个养殖群体的微卫星多态性等。与双亲遗传的核基因组相比,线粒体DNA为母系遗传,结构简单、无基因重组、进化速率快,有效群体大小仅为前者的1/4,是种群遗传学研究的首选分子标记,也是核基因组遗传分析的重要补充[11-12]。其中控制区(control region,CR)是线粒体基因组中进化速率最快的区段,常被用于分析鱼类遗传背景[13]。本研究测定了广东阳江闸坡、廉江安铺镇、雷州乌石镇,海南临高新盈镇,广西东兴等5个地区野生群体的线粒体控制区序列,以分析遗传多样性、种群遗传结构和动态,旨在了解南海卵形鲳鲹的遗传背景,为种质资源的管理保护和合理开发利用提供科学依据。
1 材料与方法
实验用的野生卵形鲳鲹样品分别采集于广东阳江闸坡镇(YZP1~20)、广东廉江安铺镇(YAP1~18)、广东乌石镇(YWS1~19)、海南临高新盈镇(QXY1~20)和广西东兴(GDX1~20)(图1),保存于95%乙醇备用。DNA提取和PCR扩增条件参照乐小亮等[15]的方法,PCR扩增引物为本实验室自行设计的L:5′-tggcttgaaaaaccaccgttg,R:5′-cctgaartaggaaccaaatgccag,将电泳检测为条带清晰明亮的扩增产物送至华大基因切胶纯化,通过ABI-3730 DNA自动测序仪测序。

图1 卵形鲳鲹采样分布图(引自孙湘平[14]。A:中国沿岸流,B:南海暖流)Fig.1 Sam p ling sites for T.Ovatus(cited from SUN Xiang-ping[14].A:the coastal current of China;B:Nanhaiwarm current)
利用MEGA 6.0[16]对测定序列进行校对与对位排列,计算碱基组成、多态位点、简约信息位点、转换与颠换比以及基于Kimura 2-Parameter模型的遗传距离,构建邻接树。利用Network 5.0[17]构建单倍型网络分布图。通过DnaSP 5.1[18]计算单倍型数、单倍型多样性(h)、核苷酸多样性(π),以及遗传分化系数(Fst)和基因流(Nm)值。通过Arlequin 3.5[19]进行AMOVA分析(analysis ofmolecular variance),评估遗传变异在群体内、群体间与组群间的分布;计算Tajima’s D和Fu’s Fs值与核苷酸不配对分析,分析种群历史动态[20]。使用SAMOVA 2.0[21]进行空间分子变异分析(spatial analysis ofmolecular variance,SAMOVA),通过群体的地理位置及序列差异分析群体间的分化。
2 结果与分析
2.1 序列特征及遗传多样性
在5个群体97个样本的线粒体控制区5′端高变区359 bp序列中,没有碱基的插入与缺失;T、C、A、G平均含量分别是30.6%、15.2%、37.4%、16.8%,表现出典型的反G偏倚,吻合脊椎动物线粒体DNA特征[22]。转换与颠换比为14.15,表明序列变异未饱合,可用于种群遗传分析[23]。47个变异位点(13.9%)中39个为简约信息位点,定义32个单倍型,其中16个为多个群体共享。卵形鲳鲹整体呈现出高单倍型多样性(h=0.951)与高核苷酸多样性(π=0.020 9)的特点,其中新盈(h=0.953,π=0.021 8)和安铺群体(h=0.961,π=0.021 9)的遗传多样性最高;乌石群体最低(h=0.895,π=0.018 4),详见表1。

表1 各采样点的采样数量及遗传多样性情况Tab.1 Samp le sizes and genetic diversity indices of T.ovatus in each sampling site
2.2 卵形鲳鲹的遗传分化
邻接树与单倍型网络图拓扑结构大体一致,都出现由不同地理来源个体混杂分布的2个谱系:Lineage A和Lineage B(单倍型网络图见图2)。谱系间分化显著(Fst=0.569,P<0.01),以控制区变异速率为3%~10%/百万年[24]推算的分化时间约为60~18万年前(中更新世)。5个地理群体的群体间和群体内遗传距离相近(0.019~0.023)(表2),遗传分化系数Fst较小(-0.022 4~0.045 3,P>0.05)。将5个地理群体按照琼州海峡东/西两侧,大陆沿海/海南岛,以及根据SAMOVA结果(新盈、乌石、安铺/闸坡、东兴)分组进行分子方差(AMOVA)分析,发现群体内的变异比例均达到97%以上(表3),表明群体间遗传分化不明显,遗传变异主要源于个体之间,地理分布的影响相对较小。

图2 基于线粒体控制区基因序列构建的卵形鲳鲹单倍型网络图Fig.2 Parsimony network of hap lotypes of T.ovatus based on m tDNA control region sequences

表3 不同分组的分子方差分析Tab.3 AMOVA analysis of various groups of different geographic populatons
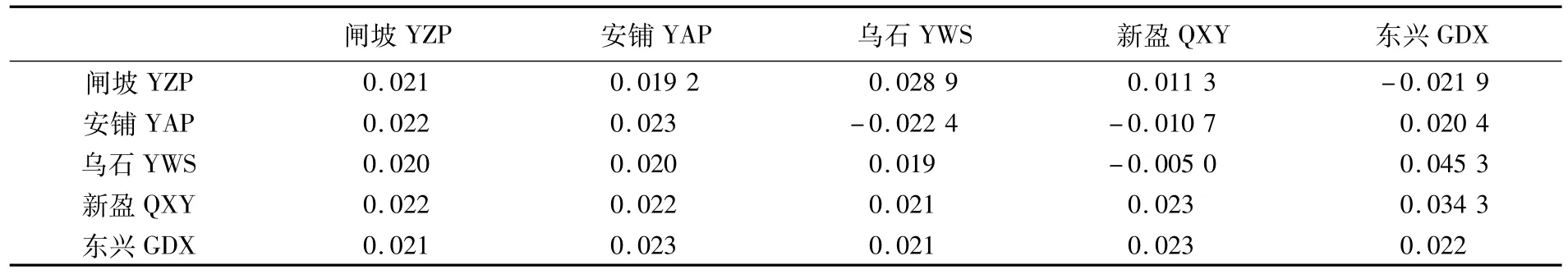
表2 群体内遗传距离(对角线)、群体间遗传距离(对角线下)及群体间遗传分化程度(Fst,对角线上)Tab.2 Genetic distancesw ithin(along diagonal)and among(below diagonal)populations and fixation index(Fst,above diagonal)among populations of T.ovatus
2.3 卵形鲳鲹种群动态分析
中性检验及吻合度检验结果显示,Lineage A的Tajima’s D值和Fu’s Fs的值都为负值(-01.041,-3.824)不显著(P>0.05);Lineage B的Tajima’s D值为正值(0.172)不显著(P>0.05)、Fu’s Fs为负值(-1.739)不显著(P>0.05);两谱系的SSD值(0.014,0.038)和Rg值(0.029,0.072)小且不显著(P>0.05)(表4);核苷酸不配对分析图均为多峰(图3)。卵形鲳鲹总体的Tajima’s D值(-0.616)和Fu’s Fs值(-6.562)都为负值且不显著(P>0.05),SSD值(0.010)和Rg值(0.015)小且不显著(P>0.05)。表明南海卵形鲳鲹总体及2个谱系都没有出现种群扩张的情况。

表4 南海卵形鲳鲹群体的中性检验及吻合度检验Tab.4 Neutrality test and goodness of fit test among T.ovatus populations in the South China Sea

图3 南海卵形鲳鲹不同谱系的核苷酸不配对分布图Fig.3 M ismatch-distribution analysis of different lineage of T.ovatus in the South China Sea
3 讨论
3.1 卵形鲳鲹的遗传多样性
就线粒体序列分析而言,一般常用单倍型多样性(h)和核苷酸多样性(π)来表示遗传多样性的高低程度[25]。南海卵形鲳鲹线粒体控制区总体呈现出高单倍型多样性和高核苷酸多样性的特点(h=0.951,π=0.020 9),单倍型多样性与同海域的真鲷(Pagrusmajor)(h=0.994)、黑鲷(Acanthopagrus schlegeli)(h=0.994)[26]以及多齿蛇鲻(Saurida tumbil)(h=0.957)[27]等相近;核苷酸多样性与同海域的真鲷(π=0.022)相近,高于黑鲷(π=0.007 5)和多齿蛇鲻(π=0.007 4),表明目前南海卵形鲳鲹线粒体序列多样性相对较高。一般认为海洋鱼类出现高单倍型多样性和高核苷酸多样性是因为种群处于持续稳定增长状态或者2个出现相对分化的群体(或亚种)发生二次接触[28]。由于南海海区卵形鲳鲹存在2个谱系,历史上种群数量总体稳定,推测较高的遗传多样性是谱系间二次接触所导致。
就南海海区卵形鲳鲹微卫星分析而言,孙立元等[7]、吉磊等[10]认为育种、养殖群体的遗传多样性较高,而赵永贞等[9]则认为野生群体多样性较低;彭敏等[8]利用AFLP分析,认为北部湾养殖群体和野生群体的多样性都较低。本研究结果与已报道的不完全一致,可能原因有:1)遗传标记的不同:线粒体控制区为母系遗传;微卫星和AFLP是核基因标记,双亲遗传;二者进化速率不同[29],受趋同进化、选择作用等的影响不同[30];同时,由于不同报道分析使用的核基因标记位点也不尽相同,难以直接比较。2)采样群体与采样点不同:已报道的研究样品大多为养殖群体,部分来源或亲本不明确。
3.2 卵形鲳鲹的遗传分化和种群动态分析
南海卵形鲳鲹中存在2个分化时间约在60~18万年前(中更新世)的谱系。历史资料显示,更新世冰期与间冰期旋回,对生物种群的数量和分布有着巨大的影响[31]。冰期海平面下降使得中国近海出现多个封闭的内陆海[32],导致群体间基因交流中断而出现分化。尽管如此,群体间Fst=-0.022 4~0.045 3(P>0.05);97%以上的变异来源于群体内部,表明群体间无明显分化,推测原因如下:1)间冰期海平面上升,不同海域得以连通。2)南海存在复杂洋流(季风漂流、黑潮分支、南海暖流等)及中国沿岸流,琼州海峡的出现也为东西两侧的群体交流提供了通道,卵形鲳鲹的受精卵及刚孵化的仔稚鱼不具备游泳能力,易被海流携带而扩散。3)成鱼游动性强,有聚群索饵和产卵习性[33]。
单倍型网络图呈现不规则图形,没有中心单倍型;中性检验和核苷酸不配对分析表明2个谱系及总体都没有出现扩张的情况,与同一海域分布的鲻(Mugil cephalus)和梭鱼(Chelon haematocheilus)情况相似[34-35],可能原因是南海地处冰期气温变化相对温和的低纬度地区,受冰期的影响相对较小。
3.3 卵形鲳鲹种质资源的保护和利用
物种遗传多样性越高,对环境变化的适应能力就越强,也可为遗传育种提供更丰富的材料[6]。南海卵形鲳鲹虽较同海域的部分鱼类表现出较高的线粒体遗传多样性,但作为南海重要的经济鱼类,受过度捕捞、环境污染等因素影响,捕获量日渐稀少[8];随着卵形鲳鲹人工种苗生产的规模化,相关研究的开展[36-37],养殖业快速兴起,但养殖苗种已出现种质衰退迹象[4],养殖逃逸也可能严重影响野生群体的多样性[38]。因此,在充分利用其资源的同时不应忽视卵形鲳鲹的保护工作。本文仅研究了5个地理群体,没有涉及福建沿海等地理群体,采样点的局限可能没有完全反映出卵形鲳鲹的遗传多样性现状;另外因群体中存在2个谱系,使得随机采集得到的每个谱系的样品数量不足。在今后的工作中,将增加样品采集的地理范围和每个地理群体的样本量,综合运用线粒体序列分析与核基因指纹技术,以期全面了解南海卵形鲳鲹的遗传背景,为种质资源的保护和开发提供更为准确的科学依据。
[1] 朱元鼎,张春霖,张有为,等.南海鱼类志[M].北京:北京科学出版社,1962:392-394.ZHU Y D,ZHANG C L,ZHANG Y W,et al.Fishes in the South China Sea[M].Beijing:Beijing Science Press,1962:392-394.
[2] 杨欣怡.网箱海养卵形鲳鲹肌肉营养品质评价和风味物质研究[D].上海:上海海洋大学,2016:1 -13.YANG X Y.Study on nutritional composition and flavor compounds of sea-cage cultured Trachinotus ovatu muscles[D].Shanghai:Shanghai Ocean University,2016:1-13.
[3] 陈伟洲,徐鼎盛,王德强,等.卵形鲳鲹人工繁殖及育苗技术研究[J].台湾海峡,2007,26(3):435-441.CHENW Z,XU D S,WANG D Q,et al.Study on the spawning and hatching technique for Trachinotus ovatus[J].Journal of Oceanography in Taiwan Strait,2007,26(3):435-441.
[4] 区又君,吉 磊,李加儿,等.卵形鲳不同月龄选育群体主要形态性状与体质量的相关性分析[J].水产学报,2013,37(7):961-969.OU Y J,JIL,LIJE,et al.Correlation analysis of major morphometric traits and body weight of selective group at differentmonth ages of Trachinotus ovatus[J].Journal of Fisheries of China,2013,37(7):961-969.
[5] 鞠海龙.南海渔业资源衰减相关问题研究[J].东南亚研究,2012(6):51-55.JU H L.Studies on the decrementof fishery resource in the South China Sea[J].Southeast Asian Studies,2012(6):51-55.
[6] WARD R D.Genetics in fisheriesmanagement[J].Hydrobiologia,2000,420(1):191-201.
[7] 孙立元,郭华阳,朱彩艳,等.卵形鲳鲹育种群体遗传多样性分析[J].南方水产科学,2014,10(2):67-71.SUN L Y,GUO H Y,ZHU C Y,et al.Genetic polymorphism of breeding populations of golden pompano(Trachinotus ovatus)[J].South China Fisheries Science,2014,10(2):67-71.
[8] 彭 敏,陈晓汉,陈秀荔,等.卵形鲳鲹养殖群体与野生群体遗传多样性的AFLP分析[J].西南农业学报,2011,24(5):1987-1991.PENG M,CHEN X H,CHEN X L,et al.Genetic diversity of wild and cultured Trachinotus ovatus populations by AFLP markers[J].Southwest ChinaJournal of Agricultural Sciences,2011,24(5):1987 -1991.
[9] 赵永贞,陈秀荔,李咏梅,等.南海区卵形鲳鲹遗传多样性的研究[J].西南农业学报,2014,27(4):1786-1790.ZHAO Y Z,CHEN X L,LI Y M,et al.Genetic polymorphism of Trachinotusovatus in Nanhaidistrict[J].Southwest China Journal of Agricultural Sciences,2014,27(4):1786-1790.
[10] 吉 磊,区又君,李加儿.卵形鲳鲹3个养殖群体的微卫星多态性分析[J].热带海洋学报,2011,30(3):62-68.JI L,OU Y J,LI J E.Genetic polymorphism of three cultured populations of golden pompano Trachinotus ovatus as revealed bymicrosatellites[J].Journal of Tropical Oceanography,2011,30(3):62 -68.
[11] AVISE JC.Phylogeography:The history and formation of species[M].London Harvard University Press.2000:9-32.
[12] CANINO M F,SPIES I B,LOWE S A,et al.Highly discordant nuclear and mitochondrial DNA diversity in Atka Mackerel[J].Marine and Coastal Fisheries:Dynamics,Management,and Ecosystem Science,2010(2):375-387.
[13] RAVAOARIMANANA I B,TIEDEMANN R,MONTAGNON D,et al.Molecular and cytogenetic evidence for cryptic speciation within a rare endemic Malagasy lemur,the Northern sportive lemur(Lepilemur septentrionalis)[J].Molecular Phylogenetics&Evolution,2004,31(2):440-448.
[14] 孙湘平.中国近海区域海洋[M].北京:海洋出版社,2006:97-98.SUN X P.China offshore area[M].Beijing:Ocean Press,2006:97-98.
[15] 乐小亮,章 群,赵 爽,等.一种高效快速的鱼类标本基因组DNA提取方法[J].生物技术通报,2010(2):202-204.YUE X L,ZHANG Q,ZHAO S,et al.A fast and efficientmethod for isolation of genomic DNA from fish specimens[J].Biotechnology Bulletin,2010(2):202-204.
[16] TAMURA K,STECHER G,PETERSON D,et al.MEGA6:Molecular evolutionary genetics analysis version 6.0[J].Molecular Biology and Evolution,2013,30(12):2725-2729.
[17] BANDELT H J,FORSTER P,ROHL A.Medianjoining networks for inferring intraspecific phylogenies[J].Molecular Biology and Evolution,1999,16(1):37-48.
[18] LIBRADO P,ROZAS J.DnaSP v5:A software for comprehensive analysis of DNA polymorphism data[J].Bioinformatics,2009,25(11):1451-1452.
[19] EXCOFFIER L,LISCHER H E L.Arlequin v 3.5:A new series of programs to perform population genetics analyses under Linux and Windows[J].Molecular Ecology Resources,2010,10(3):564-567.
[20] NEIM,TAJIMA F.DNA polymorphism detectable by restriction endonucleases[J].Genetics,1981,97(1):145-163.
[21] DUPANLOUP I,SCHNEIDER S,EXCOFFIER L.A simulated annealing approach to define the genetic structure of populations[J].Molecular Ecology,2002,11(12):2571-2581.
[22] KNIGHT A,MINDELL D P.Substations bias,weighting of DNA sequence evolution,and the phylogenetic positions of fea′s viper[J].Systematic Biology,1993,42(1):18-31.
[23] KOICHIRO T,GLEN S,DANIEL P,etal.MEGA6:Molecular evolutionary genetics analysis version 6.0[J].Molecular Biology and Evolution,2013(30):2725-2729.
[24] HOCHACHKA PW,MOMMSEN T.Biochemistry and molecular biology of fishes:Environmental and ecological biochemistry[J].Science,1993(2):1 -38.
[25] BONIN A,NICOLE F,POMPANON F,et al.Population adaptive index:A new method to help measure intraspecific genetic diversity and prioritize populations for conservation[J].Conservation Biology 2007,21(3):697-708.
[26] 曹 艳.基于线粒体控制区序列的中国沿海3种鲷科鱼类遗传多样性分析[D].广州:暨南大学,2016:22-60.CAO Y.Genetic diversity of 3 sparid species in coastal waters of China based on mitochondrial control region sequences[D].Guangzhou:Jinan University,2016:22-60.
[27] 孙冬芳,董丽娜,李永振,等.南海北部湾海域多齿蛇鲻的种群分析[J].水产学报,2010,34(9):1389 -1393.SUN D F,DONG L N,LI Y Z,et al.Population analysis of Saurida tumbil in the Northern South China Sea[J].Journalof Fisheries of China,2010,34(9):1389-1393.
[28] GRANT W S,BOWEN B W.Shallow population histories in deep evolutionary linages of marine fishes:Insights from sardines and anchovies and lessons for conservation[J].Journal of Heredity,1998,89(5):415-426.
[29] UPHOLTW B,DAUID IB.Mapping ofmitochondrial DNA of individual sheep and goat:Rapid evolution in the D-loop region[J].Cell,1997(11):57-583.
[30] LARMUSEAU M H,RAEYMAEKERS J A,HELLEMANSB,et al.Mito-nuclear discordance in the degree of population differentiation in a marine goby[J].Heredity,2010(105):532-542.
[31] DYNESIUSM,JANSSON R.Evolutionary consequences of changes in species'geographical distributions driven by Milankovitch climate oscillations.[J].Proceedings of the National Academy of Sciences,2000,97(16):9115-9120.
[32] WANG P.Response of western pacificmarginal seas to glacial cycles:Paleoceanographic and sedimentological features 1[J].Marine Geology,1999,156(1-4):5-39.
[33] 陈傅晓,唐贤明,谭 图,等.卵形鲳鲹深水网箱养殖风险对策分析[J].中国渔业经济,2011,29(4):145-150.CHEN F X,TANG X M,TAN T,et al.The waveresisted deep-water net-cage breeding of Trachinotus ovatus for risk prevention countermeasures and economic benefit analysis[J].Chinese Fisheries Economics,2011,29(4):145-150.
[34] JAMANDRE B W,DURAND JD,TZENG W N. Phylogeography of the flathead mullet Mugil cephalus in the north-west Pacific as inferred from themtDNA control region.[J].Journal of Fish Biology,2009,75(2):393-407.
[35] LIU J X,GAO T X,WU S F,et al.Pleistocene isolation in the North-Western Pacific marginal seas and limited dispersal in a marine fish,Chelon haematocheilus(Temminck&Schlegel,1845)[J].Molecular Ecology,2007(16):275-288.
[36] 区又君,李加儿,蔡文超.卵形鲳鲹肾脏的显微镜和超显微结构的观察[J].海洋渔业,2015,37(5):434-441.OU Y J,LI J E,CAI W C.Observation on microstructure and ultrastructure of kidney in Trachinotus ovatus[J].Marine Fisheries,2015,37(5):434-441.
[37] 程大川,郭华阳,马振华,等.3月龄卵形鲳鲹形态性状对体质量的影响分析[J].海洋渔业,2016,38(1):26-34.CHENG D C,GUO H Y,MA Z H,et al.Mathematical analysis of morphometric attribute effects on body weight for three-month-old Trachinotus ovatus[J].Marine Fisheries,2016,38(1):26-34.
[38] 张全启,徐晓斐,齐 洁,等.牙鲆野生群体与养殖群体的遗传多样性分析[J].中国海洋大学学报(自然科学报),2004,34(5):816-820.ZHANG Q Q,XU X F,QI J,et al.The genetic diversity of wild and farmed Japanese flounder populations[J].Periodical of Ocean University of China,2004,34(5):816-820.
Genetic diversity of Trachinotus ovatus in the South China Sea inferred from M itochondrial DNA control region sequences
LV Jin-lei,ZHANG Qun,YANG Xi-shu,GONG Ya-yun,CAO Yan
(Institute of Hydrobiology,Jinan University;Engineering Research Center of Tropical and Subtropical Aquatic Engineering,Ministry of Education,Guangzhou 510632,China)
Due to the delicious taste,high nutritional value and increasingmarket demand,Trachinotusovatus has become one of the most economically important maricultured fish in the South China Sea.With the dwindling wild resources,the development of large-scale cultivation and the degraded germplasm of cultivars etc.,the accurate definition of population structure should be assessed for bettermanagement and sustainable exploitation of T.Ovatus resource in the South China Sea.In present study,359 bp mtDNA control region sequences of 97 individuals of T.Ovatus from Zhapo,Anpu and Wushi in Guangdong Province,Xinying in Hainan Province and Dongxing in Guangxi Provincewere analyzed to study the genetic diversity of this species in the South China Sea.A total of 47 polymorphic loci were found,of which 39 sites were parsimonyinformative and 32 haplotypes were defined.The resultant haplotype diversity(h)and nucleotide diversity(π)were 0.951±0.008 and 0.020 9±0.000 8 respectively,indicating the high level of haplotype diversity and nucleotide diversity of T.Ovatus populations in north of the South China Sea.In Neighborjoining tree and TCS network,two lineages were detected,and individuals from various geographical sites scattered in both lineages,thus no geographical cluster was found.The deduced differentiation time was approximately 600~180 ka BP.Despite the presence of two co-existing lineages,the absence of genetic heterogeneity among the 5 sampling siteswas noted,as the genetic fixation was low and not significant(Fst=-0.022 4~0.045 3,P>0.05),and the genetic distances were small(0.020-0.023).The analysis of molecular variance(AMOVA)also revealed that most of genetic variation resided within populations(>97%).This pattern of harboring obvious lineage structure but no geographic structure might have been shaped by glaciation cycles in Pleistocene.During the ice age,the closure ofmarginal seas caused by deeply lowered sea levelmighthave prevented gene flow among seperate seas,resulting in lineages differentiation.In interglacial stage,the rising of sea level might have reconnected marginal seas,promoting the secondary contact of once-isolated lineages,as the warm current of the South China Sea and the China Coastal current might have carried away fish eggs and larvae of T.Ovatus,and the formation of feeding and spawning schools also promoted the gene flow.Both nucleotide mismatch distribution and neutrality tests revealed that the lineage A,lineage B,and all populations as a whole in the South China Sea hadn’t experienced population expansion.In conclusion,although the genetic diversity of T.Ovatus in the South China Sea is relatively high yet,in the contextof over exploitation ofwild resources,degraded the germplastof cultivars,and the potential negative effects of breeding escape on their genetic diversity,propermanagement and sustainable exploitation of this species are urgently deserved.In further study,more populations across the species’range should be analyzed to determine the potential population genetic structure and demographic history with both mitochondrial and nuclear information.
Trachinotus ovatus;South China Sea;mtDNA control region;genetic diversity
Q 349
A
1004-2490(2017)03-0241-08
2017-03-28
国家自然科学基金项目(41071034);中央高校基本科研业务费专项资金项目(21613105)
吕金磊(1990-),男,硕士研究生,研究方向为分子生态学及生物多样性保护。
章 群(1968-),副研究员。E-mail:tqzhang@jnu.edu.cn
