焦化废水活性污泥PAH双加氧酶基因多样性分析
2017-02-22蒙小俊李海波盛宇星曹宏斌安康学院旅游与资源环境学院陕西安康75000中国科学院过程工程研究所绿色过程与工程重点实验室北京100190
蒙小俊,李海波,盛宇星,曹宏斌*(1.安康学院旅游与资源环境学院,陕西 安康 75000;.中国科学院过程工程研究所绿色过程与工程重点实验室,北京 100190)
焦化废水活性污泥PAH双加氧酶基因多样性分析
蒙小俊1,2,李海波2,盛宇星2,曹宏斌2*(1.安康学院旅游与资源环境学院,陕西 安康 725000;2.中国科学院过程工程研究所绿色过程与工程重点实验室,北京 100190)
为分析焦化废水活性污泥中降解PAH双加氧酶的多样性,利用16Sr DNA-PCR-DGGE方法,以实际焦化废水好氧单元活性污泥总DNA为模板,通过引物对污泥中的双加氧酶基因进行了克隆表达和多样性分析.结果表明,以活性污泥总 DNA为模板,利用引物RHD-GN-610F和RHD-GN-916R扩增后有明显的产物,产物大小约为300bp;DGGE指纹图谱显示PCR产物有8条分离条带,丰度分别为2.99%、8.16%、20.75%、28.50%、8.62%、7.26%、10.62%和13.10%;条带经切胶回收PCR扩增后出现明显的扩增产物,TA克隆后成功测试出4条序列,长度分别为305bp,298bp,334bp和294bp,表明焦化废水活性污泥中存在不同降解PAH的RHD酶.这些结果为焦化废水中PAHs的风险评估及其潜在生物降解提供理论基础.
焦化废水;PAHs;活性污泥;双加氧酶基因
PAHs是一类由稠合芳环,不含取代基或杂原子组成的有毒有害化合物,这类物质常以混合的形式存在,广泛分布于不同性质的水环境介质中,如生活废水和工业废水中.在废水的处理过程中PAHs会大量积存于初沉池污泥、活性污泥、消化污泥、脱水污泥和外排污泥中,对微生物活性有负效应,因致突、致畸和致癌使该类物质成为最危险的有机污染物之一.因与固体颗粒有很强的亲和性,在废水处理过程中 PAHs很容易被吸附在固体颗粒表面,吸附作用是 PAHs在废水处理过程中去除的主要途径[1-3].焦化厂是环境中PAHs的主要来源之一,焦化排放的PAHs占我国多环芳烃年排放总量的17.9%,尤其如萘、菲、芘、苯并[a]芘是焦化废水中PAHs的典型代表[4-6].焦化废水生化处理过程中PAHs的迁移转化主要是依赖生物反应器中生物污泥-微生物群落的吸附降解耦合作用来去除,在生物反应器中多环芳烃迁移转化的可能途径主要是生物污泥的吸附,其次是缓慢的生物降解,多环芳烃的水溶解度随着分子量的增大而降低,水相迁移能力也随之变差,多环芳烃会更容易被悬浮物和沉积物吸附[7-10].焦化废水中PAHs的最终去除依靠微生物的降解作用,已从焦化废水活性污泥、土壤或者其他场所鉴定出多株具有降解多环芳烃能力的菌株,如芘降解菌Pseudomonas sp.[11], Burkholderia sp.[11]和Diaphorobacter sp.[12]等,萘降解菌如 Streptomyces sp.[13]和 Geobacillus sp.[14]等,菲降解菌如Pseudomonas sp.[15]和Sphingomonas sp.[16]等.
PAHs的生物降解需要多种酶系的参与,酶系的生成和不同活性都是由相关基因进行编码控制,其中重要的关键启动酶为环羟基双加氧酶(RHD),它决定和控制后续的生物降解步骤和速率.RHD由β和α终端双加氧酶,铁氧化还原蛋白和铁氧化还原蛋白酶组成,在 PAH环裂解过程中另外还需雌二醇双加氧酶的参与,在不同的染色体或质粒位点上,不同的 PAH降解菌具有不同的 RHD编码基因[17-18].已从红杉沉淀物中发现nidA和ndo (nahAc and phnAc)终端双加氧酶基因[19-20],利用实时定量 PCR从土壤和沉积物样品中成功克隆和鉴定出降解 PAHs的环羟化双加氧酶基因RHD[21-22].降解PAHs的双加氧酶基因具有相似性和多样性,如高效降解芘的放线菌 Mycobacterium sp.双加氧酶大亚基和小亚基的基因片段与已知降解芘的分枝杆菌的双加氧酶基因具有高度同源性[23],而降解蒽嗜盐菌AD-3 的双加氧酶基因与 Marinobacter sp.NCE312菌株萘双加氧酶大亚基的部分氨基酸序列同源性相似[24],嗜盐菌群萘双加氧酶(ndo)基因有6种基因型[25].实际焦化废水处理工艺好氧单元多以活性污泥法为主,然而好氧单元活性污泥PAHs双加氧酶基因多样性未见报道.对编码降解PAH启动基因RHD进行克隆表达和多样性分析有利于降解PAHs功能微生物的监测,风险评估及其PAHs的潜在生物降解.本研究利用16Sr DNA- DGGE技术对采自实际焦化废水处理厂好氧单元的活性污泥分析其RHD基因的多样性.
1 材料与方法
1.1 实验试剂
D5625-01E.Z.N.A.Soil DNA Kit(OMEGA)、Q10212Qubit2.0DNA 检测试 剂 盒(Life)、Ep0406Taq DNA Polymerase(Thermo) 、SK8192SanPrep柱式DNA胶回收试剂盒(上海生工).
1.2 实验主要仪器
Pico-2台式离心机(Thermo Fisher)、GL-88B漩涡混合器(海门市其林贝尔仪器制造有限公司)、 TND03-H-H干式恒温器(深圳拓能达科技有限公司)、DYY-6C电泳仪电源(北京市六一仪器厂)、DYCZ-21电泳槽(北京市六一仪器厂)、GelDoc-It310凝胶成像系统(美国 UVP)、Qubit®2.0Q32866荧光计(Invitrogen)、T100TMThermal Cyeler PCR仪(BIO-RAD)、DGGE仪(Bio-Rad).
1.3 采样
活性污泥采自辽宁某焦化厂好氧单元,该焦化废水生物处理系统采用A-A-O工艺,表1为主要水质指标和好氧单元工艺参数,SRT为15~18d,活性污泥中16种EPA优控PAHs单体浓度为18.95~68.50μg/g.

表1 焦化废水主要水质指标和好氧单元工艺参数Table 1 Major indices of coking wastewater and operational parameters of aerobic basin
1.4 样品总DNA提取和PCR扩增
活性污泥基因组 DNA 通过 E.Z.N.A. Soil DNA Kit 提取,Qubit2.0检测DNA浓度,琼脂糖凝胶检测DNA完整性.
所用引物为PAH-RHD,如表2所示[21-22].反应体系50µL:10×Buffer(含2.0mmol/L MgCl2)5μL, dNTP(10mmol/L)1µL,GC(10µmol/L)1µL,NS1(10 µmol/L)1µL,Taq酶(5U/µL)0.25µL,母板 DNA 2µL,ddH2O补至50μL.PCR扩增程序:94℃ 5min预变性;94℃变性 30s,54℃退火 45s,72℃延伸1min,35个循环;延伸7min,4℃保存.取PCR产物各2µL,1.5%琼脂糖,1×TAE缓冲液,120V稳压电泳 15min,利用凝胶成像系统拍摄电泳图谱查看PCR的扩增效果.
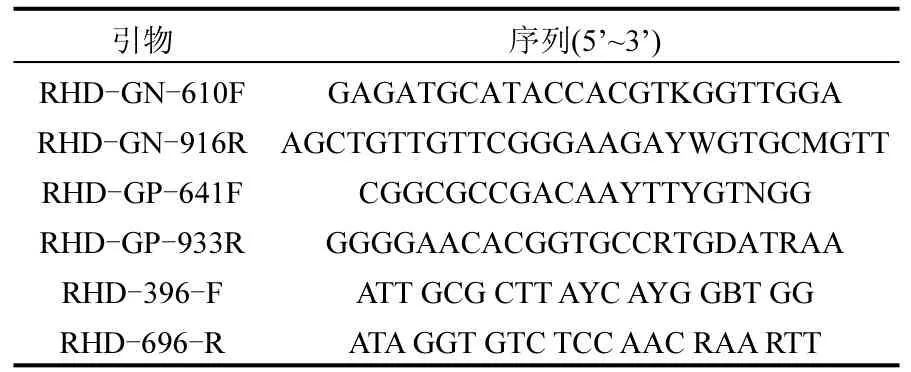
表2 PCR扩增引物Table 2 PCR amplification primers
1.5 变性梯度凝胶电泳DGGE和条带回收
利用D-Code突变检测系统对上述PCR产物(取PCR产物400ng)进行DGGE分析,8%的聚丙烯酰胺凝胶浓度,变性剂浓度梯度 15%~40%,电压条件为70V,60℃恒温,1×TAE中电泳13h.电泳结束后,利用超纯水将胶冲洗干净放入含 TE的染液中染色 30min,然后置于凝胶成像系统上拍摄图谱.待凝胶成像系统拍摄电泳图谱结束后,用洁净的手术刀片将预先挑选的代表性目标DGGE条带完整地切下装入1.5mL离心管中,按上海生工公司试剂盒方法进行回收,备用.
1.6 PCR扩增、产物回收和目标片段测序
PCR反应体系与程序同上.PCR产物按上海生工测序公司 SK8131试剂盒方法切胶回收.依次经过连接、感受态细胞制备、连接产物转化、蓝白斑筛选和质粒提取过程后对目标片段进行测序,测序由上海生工完成.
2 结果与讨论
2.1 PCR扩增产物和DGGE指纹图谱分析
提取活性污泥样品总DNA后,利用表2中的引物以总DNA为模板进行PCR扩增,发现引物RHD-GN-610F和RHD-GN-916R有明显的扩增产物,如图1所示,产物大小为300bp,这与相关研究报道结果一致[21,24],说明 A-A-O处理工艺好氧单元活性污泥中存在降解PAHs的RHD基因酶.
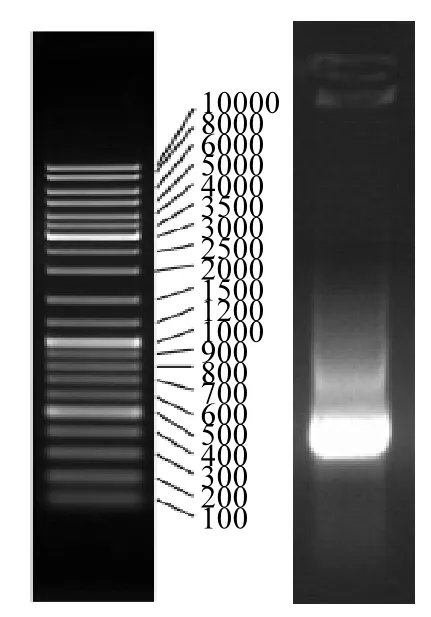
图1 PCR扩增产物Fig.1 PCR amplification product
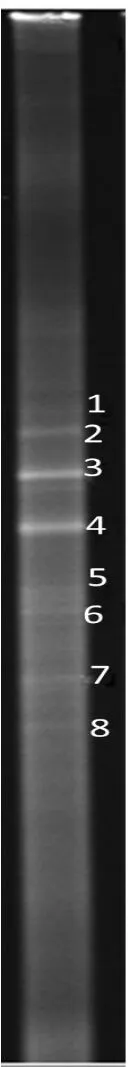
图2 RHD基因DGGE 指纹图谱分析Fig.2 DGGE analysis of RHD genes
对样品PCR产物进行DGGE分析,发现有明显的分离条带(图2),分别编号1、2、3、4、5、6、7、8(图2),说明焦化废水活性污泥中RHD基因存在多样性.RHD基因酶由β和α终端双加氧酶,铁氧化还原蛋白和铁氧化还原蛋白酶组成,在不同的染色体或质粒位点上,不同的PAH降解菌具有不同的 RHD编码基因[17-18].焦化废水活性污泥是一个复杂的微生物群落,存在不同的 PAHs降解菌株,如 Burkholderia sp.和 Pseudomonas sp.、Comamonas sp.和Diaphorobacter sp.等[26-28]. 8条条带的亮度如图2所示,亮度的大小可反映该条带的丰富度,表3所示8条带所占的丰度分别为2.99、8.16、20.75、28.50、8.62、7.26、10.62和13.10%,主要条带为3和4号条带,推测3和4号条带的RHD基因酶是焦化废水中降解PAHs的主要酶.用洁净的手术刀片将1~8号目标DGGE条带完整的切下利用引物 RHD-GN-610F和RHD-GN-916R PCR扩增后进行电泳检测.

表3 RHD基因的多样性和丰度Table 3 Diversities and richness of RHD genes
2.2 DGGE条带PCR产物电泳分析
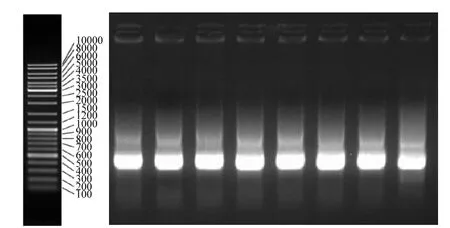
图3 DGGE条带PCR产物电泳图Fig.3 Electrophoresis of DGGE band PCR products
如图3所示,对DGGE指纹图谱中的8条条带进行回收经PCR扩增后发现有明显的扩增产物,进一步证实了活性污泥中存在不同的 RHD基因酶,不同的PAH降解菌具有不同的RHD编码基因[18],说明焦化废水活性污泥中存在不同的PAHs降解菌株,萘、菲、芘的代谢过程分别说明
[18],RHD 加氧酶是 PAHs好氧代谢过程中最重要的关键酶和启动酶,决定和控制后续的生物代谢过程.
2.3 DGGE条带PCR产物序列分析
为了分析 DGGE条带 PCR产物的序列,回收产物依次经过连接、感受态细胞制备、连接产物转化、蓝白斑筛选和质粒提取过程后对目标片段进行测序,成功测试出条带编号分别为4、5、6和7的序列,序列长度分别为305bp、298bp、334bp、294bp,结果如下所示:
条带4:305bp
GAGATGCATACCACGTTGGTTGGACGC ACGCATCGTCTTTGCGCTCGGGGCAGTCGA TATTTACTCCTCTTGCGGGCAACGCGATGCT TCCACCCGAAGGCGCGGGCTTGCAAATGAC CAGCAAGTATGGCAGTGGAATGGGCGTATT GTGGGACGCCTACTCCGGTGTCCACAGCGC TGATCTGGTTCCCGAAATGATGGCATTCGGC GGCGCAAAACAGGAAAAGCTCGCCAAAGA AATCGGCGATGTCCGGGCGCGGATTTACCG CAGCCATCTGAACAGCACTATCTTCCCGAA CAACAGC
条带5:298bp
GAGATGCATACCACGTTGGTTGGACGC ACGCATCGTCTTTGCGCACAGGGCAGTCGA TATTTACCCCTCTTGCGGGCAACGCTATGCC TCCACCCGAAGGCGCGGGCTTACAAGTGA CCAGCAAGTATGGCAGTGGAATGGGCGTAT TGTGGGACGCCTACTCCGGTGTCCACAGCG CAGACCTGGTTCCCGAAATGATGGCATTCG GCGGCGCAAAACAGGAAAAGCTCGCCAAG AAATCGGCGATGTCCGGGCGCGGATTTACC GCAGCCATCGTCGACCTGCAGGCATGCAAG CT
条带6:334bp
GAGATGCATACCACGTTGGTTGGACGC ACGCATCGTCTTTGCGCTCAGGGCAGTCGA TATTTACCCCTCTTGCGGGCAACGCTATGCC TCCACCCGAAGGCGCGGGCTTACAAGTGA CCAGCAAGTATGGCAGTGGAATGGGCGTAT TGTGGGACGCCTACTCCGGTGTCCACAGCG CTGATCTGGTTGCCGAACTGATGGCATTCG GCGCCGCAAGACAGGAAAAACTCGCCAAG GAAATCGGCGTTGTCCGGGCACAGATTTAC CGCAGCCATCTAAACAGCACTATCTTCCCG AACAACAGCTAACCGCACTATCTTCCCGAA CAACAGCT
条带7:294bp
AGATGCATACCACGTTGGTTGGACGCA CGCATCGTCTTTGCGCACAGGGCAGTCGAT ATTTACTCCTCTTGCGGGCAACGCTACGCTT CCACCCGAAGGCGCGGGCTTGCAAATGAC CAGCAAGTATGGCAGTGGAATGGGCGTATT GTGGGACGCATACCACGTAGGTTGGACGCA CGCATCGTCTTTGCGCTCAGGGCAGTCGAT ATTTACTCCTCTTGCGGGCAACGCGATGCTT CCACCCGAAGGCGCGGGCTTGCAAATGAC CAGCAAGTATGGCAGTGGAATGGGCGT
利用BLAST在NCBI数据库(http://blast.stva.ncbi.nlm.nih.gov/Blast.cgi)中进行同源性检索BLAST比对分析显示:条带4,条带5,条带6和条带 7分别与 PAH双加氧酶基因(Accession AM743143.1), (Accession JX297666.1), (Accession AM743159.1),和(Accession AM743143.1)相似,相似度分别为98%,97%,96%和98%,说明克隆表达成功测序的4条的序列基因均为PAH双加氧酶基因.
3 结论
3.1 提取焦化废水活性污泥样品总 DNA,利用引物以总 DNA为模板 PCR扩增,发现引物RHD-GN-610F和RHD-GN-916R扩增后有明显产物,对PCR扩增产物进行DGGE分析,发现有8条条带,丰度分别为2.99%、8.16%、20.75%、28.50%、8.62%、7.26%、10.62%和13.10%.
3.2 8条条带经切胶回收PCR扩增后有明显产物,进一步证实了活性污泥中存在不同的 RHD基因酶.TA克隆后.成功测试出4条序列.长度分别为305bp、298bp、334bp和294bp.
[1] Qiao M, Qi W X, Liu H J, et al. Occurrence, behavior and removal of typical substituted and parent polycyclic aromatic hydrocarbons in a biological wastewater treatment plant [J]. Water Research., 2014,52:11-19.
[2] Yao M, Zhang X W, Lei L C. Polycyclic aromatic hydrocarbons in the centralized wastewater treatment plant of a chemical industry zone: removal, mass balance and source analysis [J]. Science China Chemistry, 2012,55(3):416-425.
[3] Tian W J, Bai J, Liu K K, et al. Occurrence and removal of polycyclic aromatic hydrocarbons in the wastewater treatment process [J]. Ecotoxicology and Environmental Safety, 2012,82: 1-7.
[4] Zhang Y X, Tao S. Global atmospheric emission inventory of polycyclic aromatic hydrocarbons for 2004 [J]. Atmospheric Environment, 2009,43:812-819.
[5] Mu L, Peng L, Cao J J, et al. Emissions of polycyclic aromatic hydrocarbons from coking industries in China [J]. Particuology, 2013,11:86-93.
[6] Ma W, Li Y, Qi H, et al. Seasonal variations of sources of polycyclic aromatic hydrocarbons (PAHs) to a northeastern urban city, China [J]. Chemosphere, 2010,79(4):441-447.
[7] Pavlovich L B, Zhuravleva N V, Bal’tser D V. Polycyclic aromatic hydrocarbons in coke plant wastewater [J].Coke and Chemistry, 2010,53(10):390-395.
[8] Zhang W H, Wei C H, Chai X S, et al. The behaviors and fate of polycyclic aromatic hydrocarbons (PAHs) in a coking wastewater treatment plant [J]. Chemosphere, 2012,88(2):174-182.
[9] Vaessen H A M G, Jekel A A, Wilbers A A M M. Dietary intake of polycyclic aromatic hydrocarbons [J]. Toxicologiacl and Environmental Chemistry, 1988,16:281-287.
[10] 葛晨军,俞花美.多环芳烃在土壤中的环境化学行为 [J]. 中国生态农业学报, 2006,14(1):162-165.
[11] Deng L J, Ren Y, Wei C H. Pyrene degradation by Pseudomonas sp. and Burkholderia sp. enriched from coking wastewater sludge [J]. Journal of Environmental Science and Health, part A, 2012, 47:1984-1991.
[12] Klankeo P, Nopcharoenkul W, Pinyakong O, et al. Two novel pyrene-degrading Diaphorobacter sp. and Pseudoxanthomonas sp. isolated from soil [J]. Journal of Bioscience and Bioengineering, 2009,108(6):488-495.
[13] Balachandran C, Duraipandiyan V, Balakrishna K,et al.Petroleum and polycyclic aromatic hydrocarbons (PAHs) degradation and naphthalene metabolism in Streptomyces sp. (ERI-CPDA-1) isolated from oil contaminated soil [J]. Bioresource Technology, 2012,112:83-89.
[14] Zhang J, Zhang X, Liu J, et al. Isolation of a thermophilic bacterium, Geobacillus sp. SH-1, capable of degrading aliphatic hydrocarbons and naphthalene simultaneously and identification of its naphthalene degrading pathway [J]. Bioresource Technology, 2012,124:83-89.
[15] Lin M, Hu X K, Chen W W, et al. Biodegradation of phenanthrene by Pseudomonas sp. BZ-3, isolated, from crude oil contaminated soil [J]. International Biodeterioration & Biodegradation, 2014,94:176-181.
[16] Tao X Q, Lu G N, Dang Z, et al. A phenanthrene-degrading strain Sphingomonas sp. GY2B isolated from contaminated soils [J]. Process Biochemistry, 2007,42:401-408.
[17] Wongwongsee W, Chareanpat P, Pinyakong O. Abilities and genes for PAH biodegradation of bacteria isolated from mangrove sediments from the central of Thailand [J]. Marine Pollution bulletin, 2013,74:95-104.
[18] Peng R H, Xiong A S, Xue Y, et al. Microbial biodegradation of polyaromatic hydrocarbons [J]. FEMS Microbiology Reviews, 2008,32:927-955.
[19] Zhou H W, Guo C L, Wong, Y S, et al. Genetic diversity of dioxygenase genes in polycyclic aromatic hydrocarbon-degrading bacteria isolated from mangrove sediments [J]. FEMS Microbiology Letters, 2006,262:148-157.
[20] Gomes N C M, Borges L R, Paranhos R, et al. Diversity of nod genes in mangrove sediments exposed to different sources of polycyclic aromatic hydrocarbon pollution [J]. Applied Environmental Microbiology, 2007,73:7392-7399.
[21] Cébron A, Norini M P, Beguiristain T, et al. Real-Time PCR quantification of PAH-ring hydroxylating dioxygenase (PAHRHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples [J]. Journal of Microbiological Methods, 2008,73:148-159.
[22] Ding G C, Heuer H, Zuhlke S, et al. Soil type-dependent responses to phenanthrene as revealed by determining the diversity and abundance of polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenase genes by using a novel PCR detection system [J]. Applied and Environmental Microbiology, 2010,76(14):4765-4771.
[23] 李全霞,范丙全,龚明波,等.降解芘的分枝杆菌M11的分离鉴定和降解特性 [J]. 环境科学, 2008,29(3):763-766.
[24] 崔长征,冯天才,于亚琦,等.降解蒽嗜盐菌AD-3的筛选、降解特性及加氧酶基因的研究 [J]. 环境科学, 2012,33(11):4062-4068.
[25] 何 芬,王立华,宁大亮,等.嗜盐菌群对菲的降解及萘双加氧酶基因的表达规律 [J]. 中国环境科学, 2012,32(9):1662-1669.
[26] Deng L J, Ren Y, Wei C H. Pyrene degradation by Pseudomonas sp. and Burkholderia sp. enriched from coking wastewater sludge [J]. Journal of Environmental Science and Health, part A, 2012,47: 1984-1991.
[27] Ma Q, Qu Y Y, Shen W L, et al. Bacterial community compositions of coking wastewater treatment plants in steel industry revealed by Illumina high-throughput sequencing [J]. Bioresource Technology, 2015,179:436-443.
[28] Klankeo P, Nopcharoenkul W, Pinyakong O. Two novel pyrene-degrading Diaphorobacter sp. and Pseudoxanthomonas sp. isolated from soil [J]. Journal of Bioscience and Bioengineering, 2009,108:488-495.
Diversity of PAH dioxygenase genes of activated sludge from coking wastewater.
MENG Xiao-jun1,2, LI Hai-bo2, SHENG Yu-xing2, CAO Hong-bin2*
(1.School of Tourism and Environment, Ankang University, Ankang 725000, China;2.Key Laboratory of Green Process and Engineering, Institute of Process Engineering, Chinese Academy of Sciences, Beijing 100190, China). China Environmental Science, 2017,37(1):367~372
In order to analyze the biodiversity of PAH dioxygenase in activated sludge from a full-scale coking wastewater, primers were used for dioxygenase gene cloning expression and diversity analysis by 16Sr DNA-PCR-DGGE method using total DNA of aerobic activated sludge as the template. The results showed that significant amplification product was found using primers RHD-GN-610F and RHD-GN-916R, and the product size was 300bp. PCR product was conducted by DGGE analysis and eight separate bands were found, the abundance was 2.99%, 8.16%, 20.75%, 28.50%, 8.62%, 7.26%, 10.62% and 13.10%, respectively. Obvious amplification products further appeared after strips by gel extraction and PCR amplification, and 4sequences were tested successfully by TA cloning, the length of the sequences was 305bp, 298bp, 334bp and 294bp, respectively, indicating the presence of different PAH RHD enzymes in coking activated sludge. These results provide a theoretical basis for the risk assessment and potential biodegradation of PAHs.
coking wastewater;PAHs;activated sludge;dioxygenase gene
X703
A
1000-6923(2017)01-0367-06
蒙小俊(1981-),男,陕西汉中人,博士,主要从事生物法处理工业废水研究.发表论文16篇.
2016-05-10
安康学院高层次人才科研专项经费(2016AYQDZR09);国家自然科学基金项目(31370281);化学工业废水处理污泥污染特征与污染风险控制研究(201509053)
* 责任作者, 研究员, hbcao@home.ipe.ac.cn
