家蚕感染二分浓核病毒(镇江株)的数字基因表达谱分析
2016-12-01尚梦珂钱荷英覃光星郭锡杰
高 坤,尚梦珂,钱荷英,覃光星,郭锡杰
(江苏科技大学生物技术学院/中国农业科学院蚕业研究所,江苏镇江 212018)
家蚕感染二分浓核病毒(镇江株)的数字基因表达谱分析
高 坤,尚梦珂,钱荷英,覃光星,郭锡杰
(江苏科技大学生物技术学院/中国农业科学院蚕业研究所,江苏镇江 212018)
【目的】筛选家蚕与二分浓核病毒(Bombyx mori bidensovirus Zhenjiang strain,BmBDV-ZJ)感染相关的差异表达基因,鉴定家蚕与病毒感染有关的调控基因,为进一步阐明家蚕抗BmBDV-ZJ的分子机制提供理论依据。【方法】采用Illumina高通量测序技术,构建家蚕品种JS口服感染BmBDV-ZJ的数字基因表达谱,为排除个体间差异,以10头蚕作为一个样本用于DGE检测。样本中基因的差异表达检测通过严格的运算法则进行,对差异检验的P值(P value)作多重假设检验校正,通过控制FDR(false discovery rate)来决定P值的域值。本研究中,差异表达基因定义为FDR≤0.001且差异倍数在2倍及以上(|log2ratio|≥1)的基因。采用基因本体论(GO)分类体系确定所有差异表达基因可能的功能。用GO计算P值和bonferoni校正。选用校正P值≤0.05作为基因组显著富集的阈值。WEGO软件用来视化、比较和绘制GO注释结果。利用KEGG数据库进行通路富集分析,进一步确定显著富集代谢途径或信号传导途径,Q值≤0.05的通路指定为DGEs中的显著富集通路。通过qRT-PCR方法对部分差异表达基因进行验证。【结果】感染组和对照组分别得到4 850 663和4 875 307个原始标签,去除低质量标签后,分别得到4 757 934和4 788 406个清洁标签,对应的标签种类数量分别为62 436和63 680种。两个文库间的清洁标签和清洁标签种类的数量在不同拷贝区间分布类似,感染组和对照组样本的测序量分别为3.5 M和3.7 M,测序深度符合试验的要求,两样本的DGE数据是可信的。将这两个DGE数据库的所有清洁标签与家蚕参考基因库进行比对,在对照组与感染组中,分别有36.39%和45.30%的清洁标签可以比对到基因。另有50.02%和43.34%的清洁标签可以比对到家蚕参考基因组,剩余的未知标签分别占清洁标签总数的13.59%和12.35%。共发现了447个差异表达基因,其中306个上调表达,141个下调表达。分别有218、147和179个差异表达基因涉及GO 3个本体中的分子功能、细胞组分和生物过程。利用KEGG公共数据库进行Pathway显著性富集分析,注释到的基因总数为8 473个。447个差异表达基因经鉴定后,其中的330个基因被归类到151个KEGG路径中。差异表达基因显著富集的Pathway(Q值≤0.05)有19个,其中最显著富集的是细胞质中DNA识别通路。挑选了24个差异表达基因进行qRT-PCR验证,其中20个基因的差异表达趋势与DGE的结果一致。其中在DNA识别通路中共检测到9个差异表达基因,BGIBMGA009408-TA、BGIBMGA004913-TA、BGIBMGA011753-TA均为编码RNA聚合酶III的基因,表达量均上调,是对照组的4.3、2.3、3.4倍。【结论】构建了3龄家蚕JS感染BmBDV-ZJ后28 h感染组及对照组幼虫的数字基因表达谱,Pathway显著性富集分析和qRT-PCR验证显示,家蚕感染BmBDV-ZJ后可能通过启动胞质内DNA识别通路来感应入侵病毒的异源DNA成分并迅速启动天然免疫抵御BmBDV-ZJ病毒感染,为研究BmBDV-ZJ侵染家蚕和家蚕抵御BmBDV感染的分子机制打下了基础。
家蚕;数字基因表达谱;二分浓核病毒;qRT-PCR
0 引言
【研究意义】家蚕二分浓核病毒镇江株(Bombyx mori bidensovirus Zhenjiang strain,BmBDV-ZJ)是感染家蚕的一种重要病毒,主要通过食下感染,引起家蚕的浓核病。患病蚕呈现空头,下痢,吐液等症状,是严重影响蚕桑生产的病毒病之一,每年给养蚕业造成较大的经济损失。目前,对于该病毒感染家蚕的致病机理和家蚕抵御该病毒感染的应答机制均不清楚。筛选鉴定家蚕与该病毒感染相关的差异表达基因,进一步研究病毒对家蚕致病的分子机理,对家蚕抗病毒育种和家蚕浓核病的有效防治具有重要意义。【前人研究进展】中国早在1959年就证实了生产上所发生的家蚕空头性软化病是由病毒引起,随后各国科学家分离得到了该病毒的不同株系,分别命名为伊那株(DNV-1)[1]、佐久株(DNV-2)[2-3]、中国(镇江)株(DNV-3)[4]。之前认为该病毒均属于细小病毒科(Parvoriridae)浓核病毒属(Densovirus,DNV)的蚕浓核病毒(B. mori densonucleosis virus);后来发现DNV-2和DNV-3基因组包含两个节段的单链DNA且不采用滚环复制(细小病毒科常用的复制方式)[5],因此将DNV-2和DNV-3重新划分为一个新设定的二分DNA病毒科二分浓核病毒属(Bidensovirus,BDV)[6]。本研究所用的病毒株是1981年由中国农业科学院蚕业研究所(镇江)钱元骏等分离得到的家蚕二分浓核病毒(镇江)株(BmBDV-ZJ),与伊那株(DNV-1)在蚕品种感受性、病毒的物理化学性状、血清学特性等方面明显不同[7]。BDV-ZJ主要在家蚕幼虫中肠柱状上皮细胞的细胞核中复制,引起幼虫的浓核病,不形成多角体[8]。其病毒基因组包括两段不同的DNA单链(6 543和6 022 bp),一个病毒粒子只包含其中一条单链DNA,因此是两种不同病毒粒子的混合物[9]。成熟的病毒粒子都会随感染细胞的破裂而释放,进而感染邻近细胞。大部分家蚕品种对BmBDV-ZJ是易感的,钱元骏等[10]通过对380多个家蚕品种的抗BmBDV-ZJ性能比较研究和不同抗性品种进行杂交试验证实家蚕对BmBDV-ZJ抗性为隐性遗传,同时也受微效多基因的影响。有的品种即使接种高浓度的病毒也完全不发病。日本科学家[11]研究发现了一个与DNV-2易感性有关的家蚕基因nsd-2,抗性品种中该基因开放阅读框中大约6 000个碱基缺失,导致其编码的具有12个跨膜域的氨基酸转运膜蛋白的缺失,且该蛋白仅在家蚕的中肠中表达;而通过转基因技术修复该基因的缺失后可以使抗性品种对BmBDV-2病毒易感,该研究结果表明病毒识别的位点可能位于nsd-2基因缺失的膜蛋白部分,然而病毒是如何与这种膜蛋白相互作用仍不清楚。裘智勇等[12-13]研究表明,家蚕对BmBDV-ZJ的抵抗性可能与家蚕中肠细胞表面的某些特殊蛋白因子有关。BAO等[14]通过抑制消减杂交技术,研究了两个不同抗性家蚕品种JS(对BmBDV-ZJ感受性)和NIL(对BmBDV-ZJ非感受性)感染BmBDV-ZJ后基因的相对表达变化,发现了在抗性品种中有11个基因明显上调,推测这些基因与NIL的抗BmBDV-ZJ感染复制有关。【本研究切入点】病毒生活史的每一步都受病毒和宿主间分子相互作用的调控,通过家蚕与BmBDV-ZJ病毒感染相关的基因差异表达分析,鉴定可能与病毒复制有关的宿主细胞内调控因子,有助于阐明家蚕抗BmBDV-ZJ的抗病毒机制。【拟解决的关键问题】通过对易感家蚕品种JS的差异基因表达谱和差异基因可能参与的信号通路分析,筛选和寻找更多可能与病毒感染相关的差异表达基因。
1 材料与方法
试验于2013年9月至2015年9月在中国农业科学院蚕业研究所病理研究室完成。
1.1 供试材料与试剂
1.1.1 家蚕及病毒株 家蚕品种JS由中国农业科学院国家蚕种质资源保存中心提供;家蚕二分浓核病毒(BmBDV-ZJ)由中国农业科学院蚕业研究所家蚕病理研究室繁殖保存;BmBDV-ZJ抗血清由中国农业科学院蚕业研究所家蚕病理研究室制备保存。
1.1.2 主要试剂 PrimeScriptTMRT Reagent Kit、
SYBR Premix Ex TaqTMKit购自TaKaRa公司;总RNA提取试剂盒、Trizol、M-MLV反转录酶购自Invitrogen公司;DNA marker DL 2000购自TaKaRa公司;其他均为国产或进口分析纯试剂。
1.2 试验方法
1.2.1 家蚕幼虫添毒感染 取二分浓核病毒感染蚕的中肠干粉0.4 g,加入8 mL 2% Na2CO3溶液研磨匀浆,静止4—5 min,加5 mL蒸馏水至50 mL,3 000 r/min离心20 min,取上清加3倍体积0.2 mol·L-1醋酸,处理30 min,用Na2CO3调pH值至6—7,用蒸馏水稀释至1/1 000备用,过滤除菌备用。
试验用家蚕幼虫JS在标准温度和光照条件下饲养至3龄起蚕。计数40头蚕为感染组(添食BmBDV-ZJ),另40头为对照组(添食灭菌水)。将制备病毒悬浮液均匀涂到桑叶上喂食感染组家蚕,对照组喂食同样体积的灭菌水涂过的桑叶,12 h后全部喂饲正常桑叶。考虑到个体间差异可能带来的影响,每10条蚕收集一管提取RNA,共设置3组生物学重复。最后剩余10头蚕用来检测病毒感染发病情况。
1.2.2 病毒感染的确认 在一块7 cm×10 cm的琼脂板(1%琼脂糖)上打7个孔,中间孔加入抗血清,周围6个孔加待测样品,将琼脂板放入补湿饭盒中,在20—30℃条件下孵育1 d观察有无沉淀带出现。
1.2.3 数字基因表达谱(digital gene expression, DGE)测序及分析 由华大基因公司提供技术支持,对28 h时间点的感染组及对照组JS家蚕幼虫各取10 μg总RNA,经过纯化、反转录、酶切和PCR线性扩增后,使用Illumina HiSeqTM2000进行测序。差异表达基因的筛选及其Gene Ontology(GO)、Pathway显著性富集分析参照文献[15-16]进行。
1.2.4 qRT-PCR分析 通过实时荧光定量PCR(qRTPCR)对DGE筛选得到的部分差异表达基因进行验证。感染组和对照组各3管样品,以每管样品提取的总RNA作为模板,用PrimeScriptTMRT Reagent Kit(TaKaRa)进行反转录合成第一链cDNA。以家蚕Actin 3作为内参基因,使用SYBR® Premix Ex TaqTM(TaKaRa)试剂盒在ABI PRISM® 7300 检测系统上进行实时定量PCR,每个模板做3次重复。反应条件:94℃ 10 min变性,94℃ 15 s,60℃ 31 s,40个循环。数据分析参考文献[17]进行,引物序列见表1。
2 结果
2.1 病毒感染的确定
通过双向免疫扩散法诊断病毒的感染。在BmBDV-ZJ感染28 h后,所有接种感染的家蚕幼虫其中肠研磨液与血清孔之间均出现了白色沉淀带,而与健康蚕(对照组)中肠研磨液及BmCPV、BmNPV感染蚕研磨液均不起反应,表明感染组家蚕幼虫全部被BmBDV-ZJ感染。
2.2 DGE数据库分析
采用Illumina高通量测序技术,感染组和对照组分别得到4 850 663和4 875 307个原始标签,去除低质量标签后,分别得到4 757 934和4 788 406个清洁标签,对应的标签种类数量分别为62 436和63 680种(表2)。两个文库间的清洁标签和清洁标签种类的数量在不同拷贝区间分布类似(图1),感染组和对照组样本的测序量分别为3.5 M和3.7 M(图2),测序深度符合试验的要求,两样本的DGE数据是可信的。
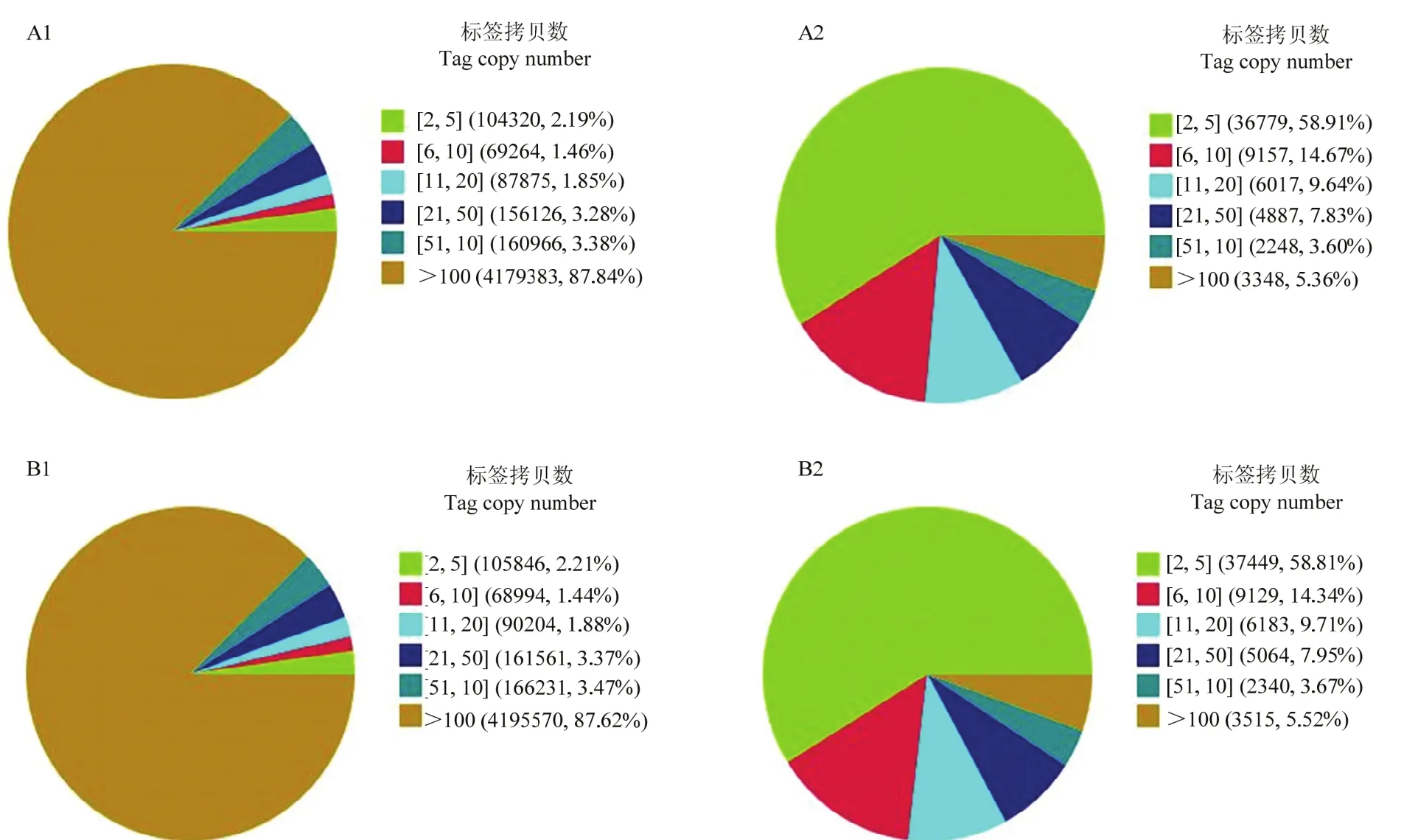
图1 对照组和感染组的清洁标签和清洁标签种类在不同拷贝区的分布Fig. 1 Distribution of total clean tags and distinct clean tags in each library
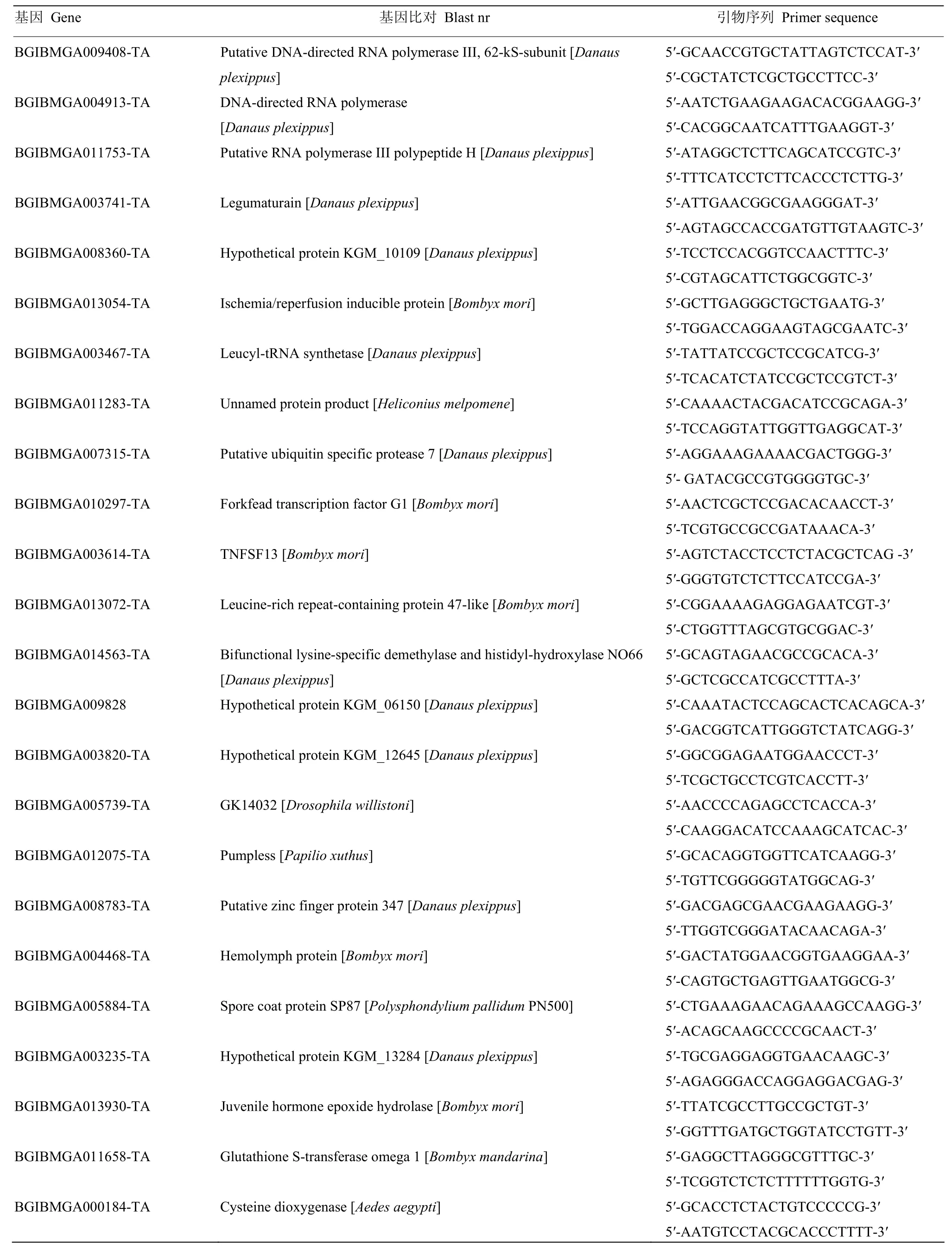
表1 定量PCR引物Table 1 Primers for qRT-PCR
2.3 标签比对及标准化处理
将所有清洁标签与家蚕参考基因库进行比对,在对照组与感染组中,分别有36.39%和45.30%的清洁标签可以比对到基因,占总清洁标签种类数的30.56%和37.41%。另有50.02%和43.34%的清洁标签可以比对到家蚕参考基因组,剩余的未知标签分别占清洁标签总数的13.59%和12.35%(表2)。
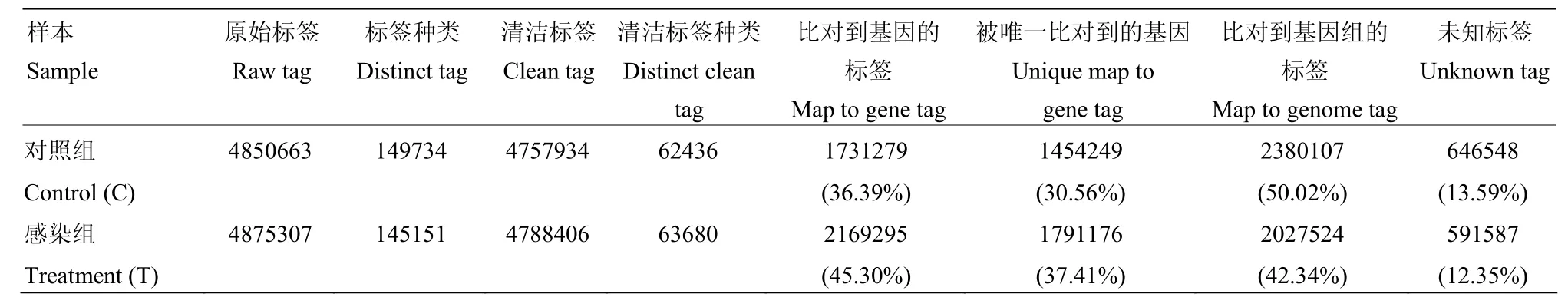
表2 两个样本中标签分布表Table 2 Distributions of tags between the two libraries
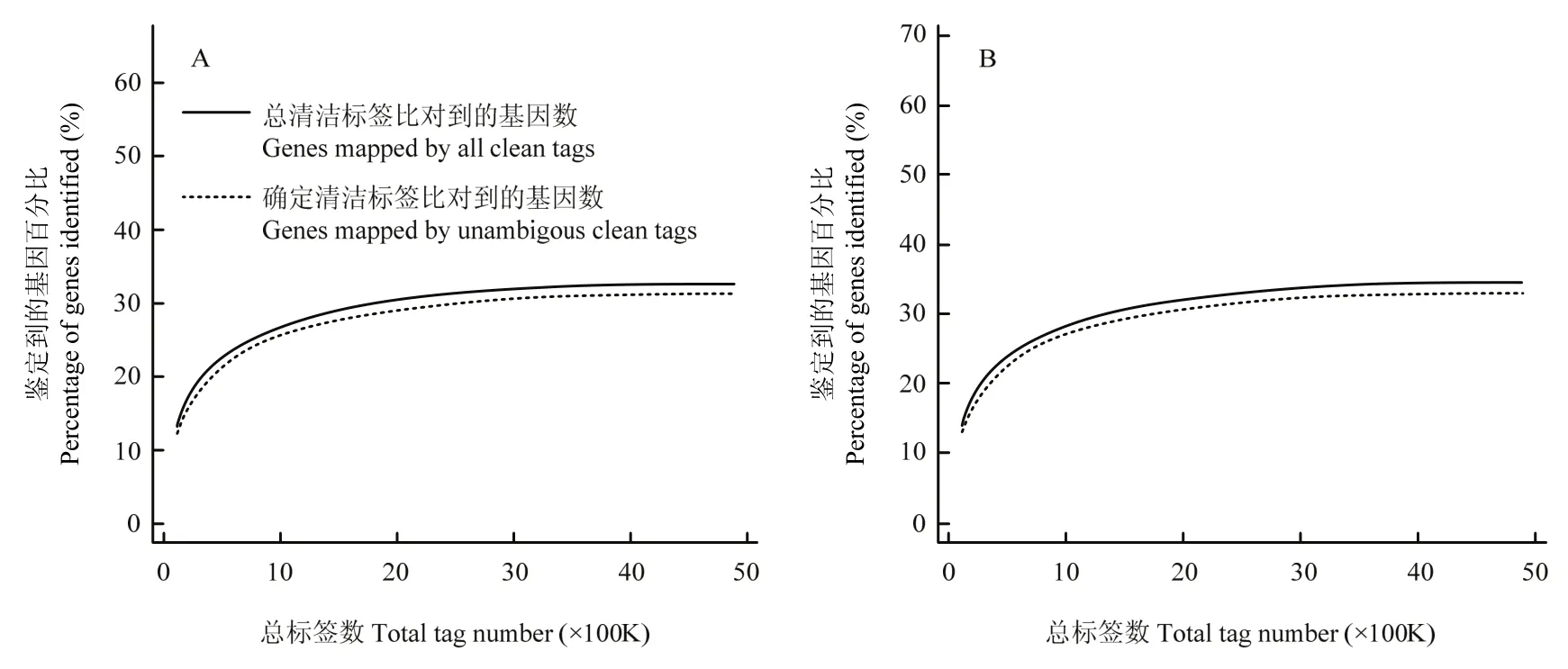
图2 数字基因表达谱的测序饱和趋势图Fig. 2 Trends of saturation of DGEs
2.4 差异基因分析及其验证
筛选了发现错误率(false discovery rate)FDR≤0.001且差异倍数在2倍及以上(|log2ratio|≥1)的基因作为差异表达基因,共发现差异表达基因447个。在感染组中,上调表达基因306个,下调表达基因141个。为了验证DGE数据的准确性,挑选了24个差异表达基因进行RT-PCR验证,其中20个基因表达情况与DGE的结果一致(图3),说明DGE数据基本可以准确反映样本的基因表达情况。
2.5 GO和KEGG分析
GO共有3个本体,分别描述基因的分子功能、所处的细胞组分、参与的生物过程。GO结果显示分别有147、179、218个差异基因分配在这3个本体中,其中在核糖核蛋白复合体、核糖体、核糖体亚基、大分子复合物、细胞内组分、蛋白酶体调控组分、代谢过程、基因表达和结构分子活性中基因富集程度较高,校正后的P值≤0.05(表3)。
为了进一步了解基因的生物学功能,利用KEGG公共数据库进行Pathway显著性富集分析,注释到的基因总数为8 473个,447个差异表达基因经鉴定后,其中的330个基因被归类到151个KEGG路径中。定义为在差异表达基因中显著富集的Pathway(Q值≤0.05)有19个(表4),其中最显著富集的是细胞质中DNA识别通路,其次是RNA聚合酶、核糖体、嘧啶代谢、蛋白酶体、抗原加工与呈递、代谢途径等。
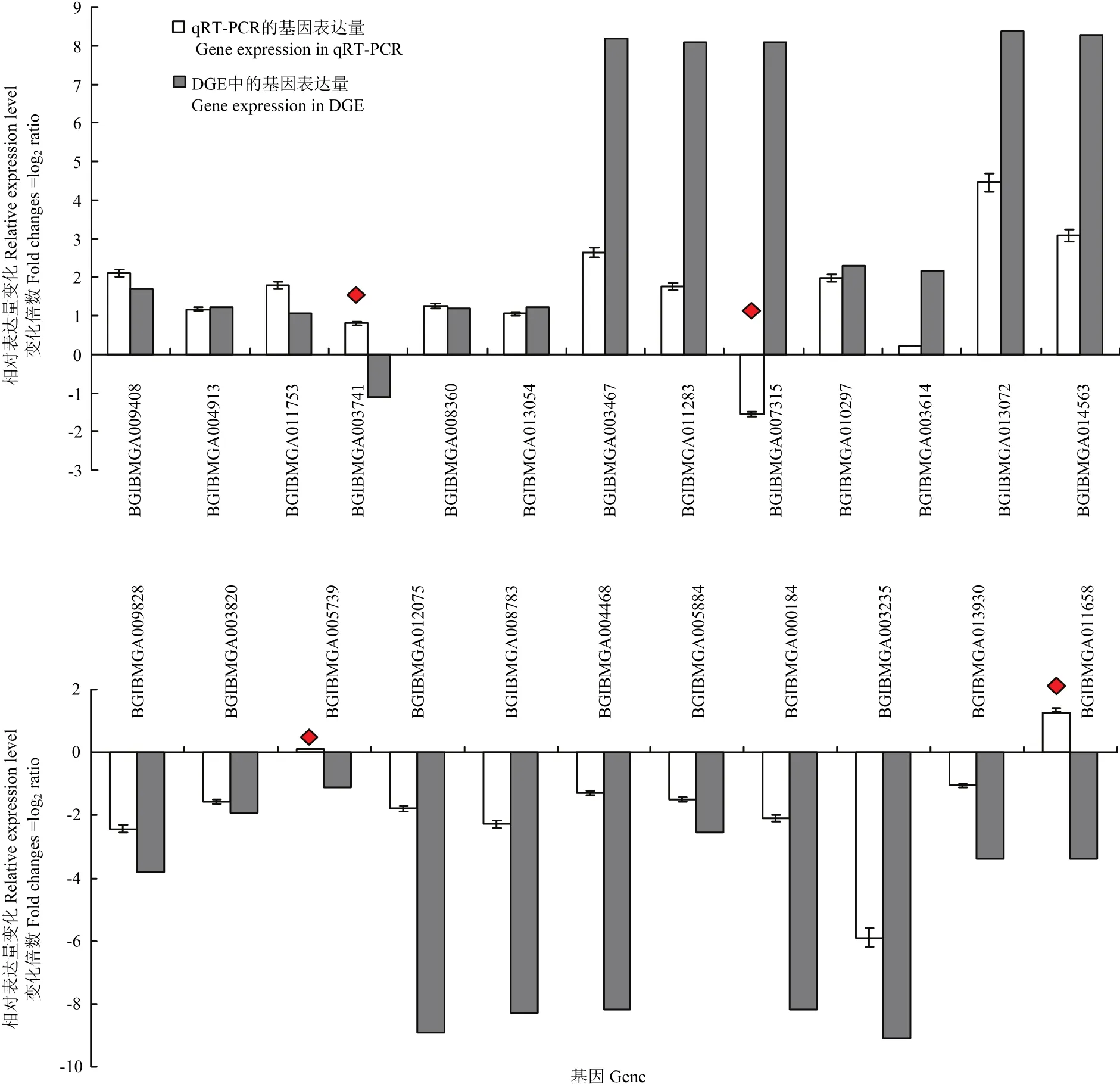
图3 候选基因在对照和感染BmBDV-ZJ的家蚕幼虫中的差异表达Fig. 3 Differential expression levels of candidate genes in BmBDV-ZJ-infected and control B. mori larvae
3 讨论
为了研究BmBDV-ZJ感染家蚕的可能分子机制和宿主的应答反应,基于本研究构建JS家蚕感染组和对照组的两个DGE文库分析,筛选得到447个差异表达基因,其中感染组有306个上调表达基因和141个下调表达基因,该差异基因的上调和下调对比结果与之前家蚕感染质型多角体病毒的DGE结果类似,都是上调表达的基因数目明显高于下调表达的基因数目[15-16,18]。相关研究表明,家蚕在感染大部分病原微生物时的差异表达基因都有上调基因明显多于下调基因的趋势,LIU等[19]通过基因芯片研究发现家蚕感染革兰氏阳性菌(Serratia marcescens)后有172个基因上调表达,61个基因下调表达;感染革兰氏阴性菌(Staphylococcus aureus)后125个基因上调表达,104个基因下调表达;感染真菌(Beauveria bassiana)后有133个基因上调表达,24个基因下调表达;而被PBS处理后有201个基因上调表达和40个基因下调表达。可见无论是病毒感染、细菌感染还是真菌感染都会引起家蚕更多基因的上调表达来应对病原的入侵。
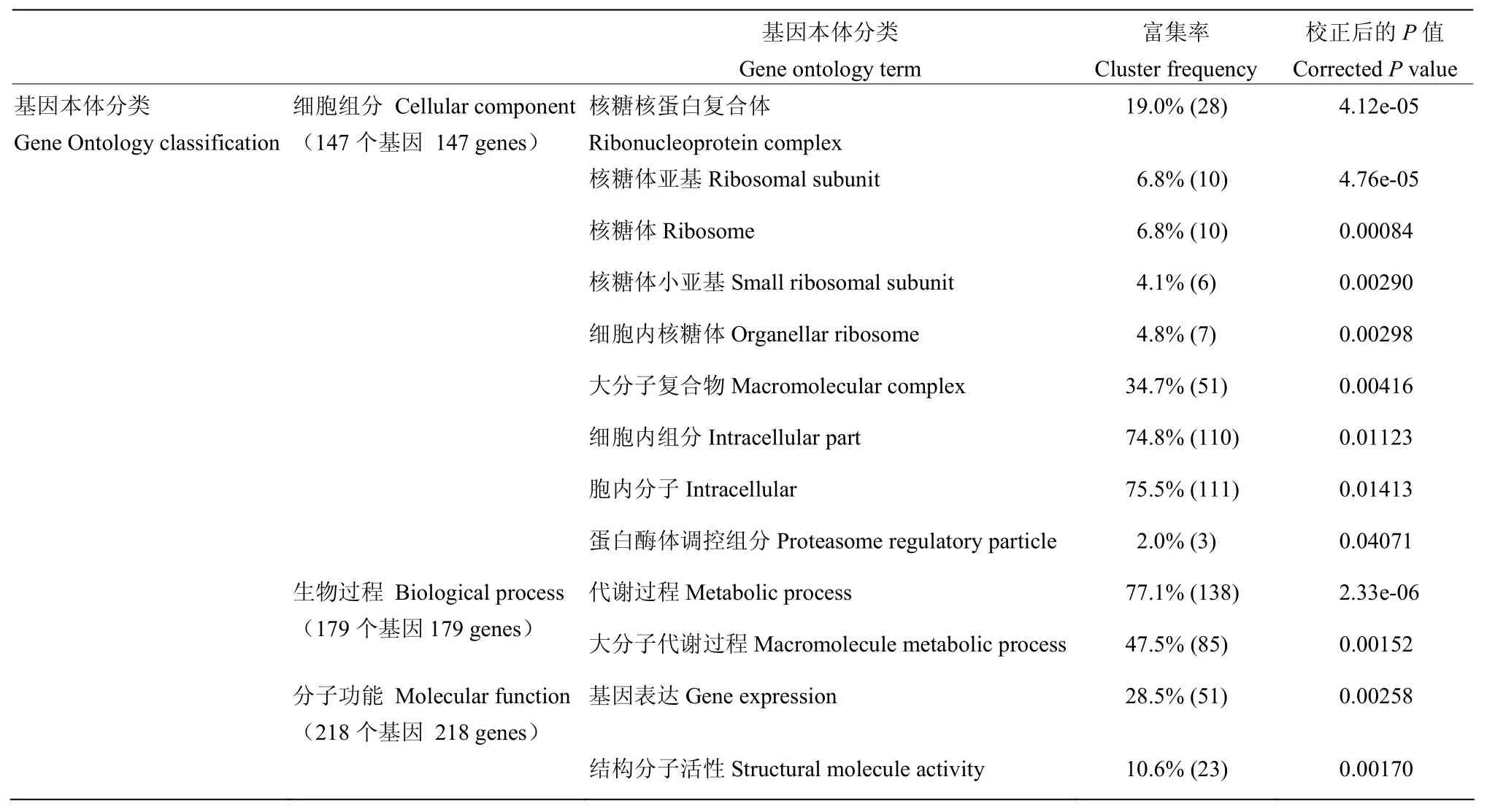
表3 基因本体分类Table 3 Gene Ontology classification
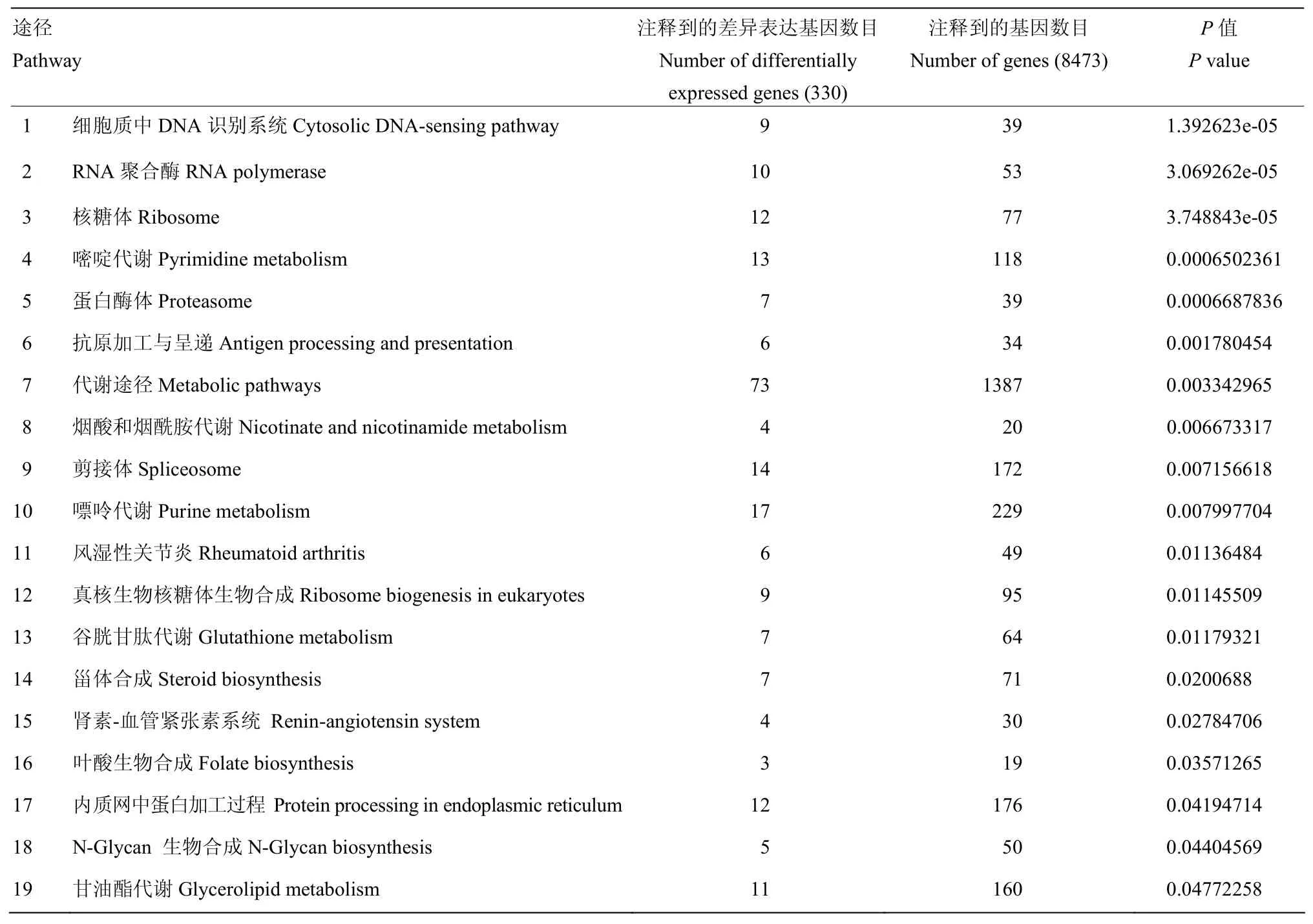
表4 显著富集的PathwayTable 4 Significantly enriched pathways
利用KEGG公共数据库进行Pathway显著性富集分析,其中最显著富集的是细胞质中DNA识别通路(cytosolic DNA-sensing pathway),该通路可以有效识别入侵病毒的异源DNA成分并迅速启动天然免疫,以及随后的特异性免疫应答来对病毒进行清除,是机体抵抗病毒感染的重要机制[20-21]。DNA感受器(DNA sensor)是宿主感受外源入侵DNA和免疫防御的桥梁,可以特异地识别病毒等外源DNA进而激活下游的免疫信号途径,并通过诱导表达抗病毒蛋白来抑制病毒的复制并向周围的细胞示警[22-23]。目前已经有超过10种DNA感受器被发现。例如,Toll样受体9(TLR9)家族[24]、视黄酸诱导基因蛋白RIG-I相关受体(RLRs)家族[25]、DNA依赖的RNA聚合酶III[26]、干扰素诱导蛋白16(IFI16)[27];DExD-H框解旋酶超家族[28-30]等。RNA聚合酶III可以识别侵入细胞内的DNA病毒,然后以病毒DNA为模板,将病毒信息转录合成一种5′三磷酸化的特殊的双链RNA,进而被RIG-I分子所识别。RIG-I可以激活I型干扰素产生,也可以激活NF-κB等信号通路并诱导抗病毒基因的表达,从而抑制病毒等病原体复制[31-32]。家蚕感染BmBDV-ZJ后在DNA识别通路中共检测到9个差异表达基因,其中BGIBMGA009408-TA、BGIBMGA004913-TA、BGIBMGA011753-TA均为编码RNA聚合酶III的基因,表达量均上调,是对照组的4.3、2.3、3.4倍(图2)。其功能可能作为家蚕细胞质中的DNA感受器,通过识别病毒DNA激活家蚕抗BmBDV-ZJ感染的免疫应答反应。
干扰素-γ诱导的溶酶体巯基还原酶(IFN-γ inducible lysosomal thiol reductase,GILT)在脊椎动物适应性免疫中的MHCII类抗原加工和呈递过程中起着关键作用,可以催化未折叠的天然抗原蛋白二硫键的断裂,进而对其进行酶解加工[33]。虽然适应性免疫和干扰素的产生已明确只在脊椎动物中存在,但是GILT基因家族在脊椎动物和无脊椎动物中普遍存在,如在虾[34]、蟹[35]、鲍[36]、果蝇[37]和线虫[38]中均发现了该基因。无脊椎动物中的GILT基因在感染细菌和病毒后也都出现上调表达趋势,推测其功能可能不同于脊椎动物中的抗原加工和呈递,而是参与了无脊椎动物中的某种先天性免疫信号途径。家蚕感染BmBDV-ZJ病毒后基因BGIBMGA003741-TA和BGIBMGA008360-TA均上调表达,且这2个基因都含有GILT保守结构域,推测为家蚕中的GILT基因。其功能是否类似于接头蛋白干扰素刺激基因,其上调表达是否可以进一步诱导相关抗病毒蛋白的产生还需要进一步试验验证。
BGIBMGA013054-TA作为缺血再灌注诱导蛋白的同源基因,其主要功能是调节细胞内物质装配和相关转运蛋白的活性[39]。病毒感染家蚕后,会利用宿主细胞的蛋白合成系统来进行病毒蛋白的复制,这些异源蛋白的产生,加速了细胞的物质运输过程,以增加宿主细胞对病毒蛋白的多重耐受性。家蚕感染BmBDV-ZJ后该基因的上调表达可能与细胞内一些蛋白的定位有关。
BmBDV-ZJ通常引起的是慢性病,感染的中肠上皮细胞不像感染BmBDV-1的中肠上皮细胞那样容易脱落,而是通过增加细胞数目,引起上皮组织折叠,在10—20 d间死亡,少数幼虫可以化蛹。因此本研究分析发现家蚕感染BmBDV-ZJ后,很多与细胞增殖相关的转录因子和蛋白都发生了明显上调,如叉头框转录因子G1(BGIBMGA010297-TA,forkfead transcription factor G1)、富含亮氨酸重复序列蛋白47(BGIBMGA013072-TA,leucine-rich repeat-containing protein 47-like protein)、赖氨酸去甲基化和组氨酸脱氢的双功能酶(BGIBMGA014563-TA,bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66)等基因。初步推测家蚕可以通过对这些基因的上调表达促进细胞增殖来增加细胞的数目,进而替代因感染BmBDV-ZJ而失去正常生理功能或凋亡的中肠柱状细胞。
BGIBMGA007315-TA是家蚕的泛素特异蛋白酶7(Ubiquitin specific protease 7,USP7),定量结果显示为下调,与DGE结果相反。该基因编码的一种泛素化酶,可以水解Mdm2(p53的E3泛素连接酶)对p53蛋白进行去泛素化,保护p53不被S26蛋白酶体降解,进而调节p53与Mdm2的稳定性[40]。相关研究表明,EB病毒的核抗原1蛋白可以通过与USP7结合,破坏p53的稳定性,有利于病毒的潜伏感染[41]。BmBDV-ZJ感染家蚕后病毒是否会与家蚕的USP7相互作用,USP7下调表达是否也会影响到家蚕p53蛋白的稳定性还需要进一步验证。
综上所述,差异表达基因最显著富集的是细胞质DNA识别通路,该通路涉及的基因BGIBMGA009408-TA、BGIBMGA004913-TA、BGIBMGA011753-TA、BGIBMGA003741-TA、BGIBMGA008360-TA和BGIBMGA013054-TA将进一步进行功能验证,明确DNA识别通路在家蚕抗BmBDV-ZJ病毒感染中的先天免疫机制。其他大部分差异基因功能未知,因此后期主要工作是对这些未知功能的差异表达基因做进一步鉴定及其参与的抗病毒机制研究。
4 结论
构建了3龄家蚕JS感染BmBDV-ZJ后28 h感染组及对照组幼虫的数字基因表达谱,Pathway显著性富集分析和qRT-PCR验证显示,家蚕感染BmBDV-ZJ后可能通过启动胞质内DNA识别通路来感应入侵病毒的异源DNA成分并迅速启动天然免疫抵御BmBDV-ZJ病毒感染。
[1] BANDO H, CHOI H, ITO Y, KAWASE S. Terminal structure of a densovirus implies a hairpin transfer replication which is similar to the model for AAV. Virology, 1990, 179(1): 57-63.
[2] BANDO H, CHOI H, ITO Y, NAKAGAKI M, KAWASE S. Structural analysis on the single-stranded genomic DNAs of the virus newly isolated from silkworm: the DNA molecules share a common terminal sequence. Archives of Virology, 1992, 124(1/2): 187-193.
[3] BANDO H, HAYAKAWA T, ASANO S, SAHARA K, NAKAGAKI M, IIZUKA T. Analysis of the genetic information of a DNA segment of a new virus from silkworm. Archives of Virology, 1995, 140(6): 1147-1155.
[4] IWASHITA Y, CHUN C Y. The development of a densonucleosis virus isolated from silkworm larvae, Bombyx mori, of China//AKAI H, KING R C, MOROHOSHI S. The Ultrastructure and Functioning of Insect Cell. Tokyo: Society for Insect Cells Japan, 1982: 161-164.
[5] HAYAKAWA T, ASANO S, SAHARA K, IIZUKA T, BANDO H. Detection of replicative intermediate with closed terminus of Bombyx densonucleosis virus. Archives of Virology,1997, 142: 393-399.
[6] ADAMS M J, CARSTENS E B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Archives of Virology, 2012, 157: 1411-1422.
[7] 钱元骏, 郭锡杰, 胡雪芳, 黄可威, 渡部仁. 我国和日本家蚕DNV的血清学关系. 蚕业科学, 1985, 11(4): 241-242. QIAN Y J, GUO X J, HU X F, HUANG K W, WATANABE H. The serological relationship between China isolate densonucleosis virus and Japan isolate densonucleosis virus. Acta Sericologica Sinica, 1985, 11(4): 241-242. (in Chinese)
[8] 郭锡杰, 钱元骏, 胡雪芳, 王红林. 我国家蚕浓核病毒(DNV)寄生部位研究. 蚕业科学, 1985, 11(2): 93-98. GUO X J, QIAN Y J, HU X F, WANG H L. Studies on locations of Bombyx mori densonucleosis virus (China isolate) invasion. Acta Sericologica Sinica, 1985, 11(2): 93-98. (in Chinese)
[9] WANG Y J, YAO Q, CHEN K P, WANG Y, LU J, HAN X. Characterization of the genome structure of Bombyx mori densovirus (China isolate). Virus Genes, 2007, 35: 103-108.
[10] 钱元骏, 胡雪芳, 孙玉昆, 戴仁鸣. 家蚕浓核病毒的研究. 蚕业科学, 1986, 12(2): 89-94. QIAN Y J, HU X F, SUN Y K, DAI R M. Studies on Bombyx mori densonucleosis virus. Acta Sericologica Sinica, 1986, 12(2): 89-94. (in Chinese)
[11] ITO K, KIDOKORO K, SEZUTSU H, NOHATA J, YAMAMOTO K, UCHINO K, KALYEBI A, EGUCHI R, HARA W, TAMURA T, KATSUMA S, MITA K, KADONO-OKUDA K. Deletion of a gene encoding an amino acid transporter in the midgut membrane causes resistance to a Bombyx parvo-like virus. Proceedings of the National Academy of Sciences of the United Dtates of America, 2008, 105(21): 7523-7527.
[12] 裘智勇, 李木旺, 沈兴家, 郭锡杰. 家蚕对浓核病毒(镇江株)抵抗性和感受性品种的中肠组织蛋白比较分析. 蚕业科学, 2008, 34(2): 244-249. QIU Z Y, LI M W, SHEN X J, GUO X J. Comparative analysis of proteins extracted from midgut of silkworm strains susceptible and non-susceptible to Bomby mori densovirus (Zhenjiang strain). Acta Sericologica Sinica, 2008, 34(2): 244-249. (in Chinese)
[13] 裘智勇, 李木旺, 覃光星, 刘挺, 沈兴家, 郭锡杰. 家蚕对浓核病毒中国镇江株抵抗性机制的初步研究. 蚕业科学, 2007, 33(4): 596-601. QIU Z Y, LI M W, QIN G X, LIU T, SHEN X J, GUO X J. Primary studies on mechanism of silkworm (Bombyx mori) resistance to densovirus China (Zhenjiang) strain. Acta Sericologica Sinica, 2007, 33(4): 596-601. (in Chinese)
[14] BAO Y Y, LI M W, ZHAO Y P, GE J Q, WANG C S, HUANG Y P, ZHANG C X. Differentially expressed genes in resistant and susceptible Bombyx mori strains infected with a densonucleosis virus. Insect Biochemistry and Molecular Biology, 2008, 38(9): 853-861.
[15] GAO K, DENG X Y, QIAN H Y, QIN G X, HOU C X, GUO X J. Cytoplasmic polyhedrosis virus-induced differential gene expression in two silkworm strains of different susceptibility. Gene, 2014, 539: 230-237.
[16] GAO K, DENG X Y, QIAN H Y, QIN G X, GUO X J. Digital gene expression analysis in the midgut of 4008 silkworm strain infected with cytoplasmic polyhedrosis virus. Journal of Invertebrate Pathology, 2014, 115(1): 8-13.
[17] 高坤, 邓祥元, 裘智勇, 覃光星, 郭锡杰. 家蚕感染质型多角体病毒 (BmCPV)后中肠组织差异蛋白质分析. 中国农业科学, 2013, 46(13): 2796-2807. GAO K, DENG X Y, QIU Z Y, QIN G X, GUO X J. Comparative analysis of differential proteins from midgut of silkworm induced by cytoplasmic polyhedrosis virus infection. Scientia Agricultura Sinica, 2013, 46(13): 2796-2807. (in Chinese)
[18] GUO R, WANG S M, XUE R Y, CAO G L, HU X L, HUANG M L, ZHANG Y Q, LU Y H, ZHU L Y, CHEN F, LIANG Z, KUANG S L, GONG C L. The gene expression profile of resistant and susceptible Bombyx mori strains reveals cypovirus-associated variations in host gene transcript levels. Applied Microbiology and Biotechnology, 2015, 99: 5175-5187.
[19] LIU F, LING E, WU S. Gene expression profiling during early response to injury and microbial challenges in the silkworm, Bombyx mori. Archives of Insect Biochemistry and Physiology, 2009, 72(1): 16-33.
[20] YANAI H, SAVITSKY D, TAMURA T, TANIGUCHI T. Regulation of the cytosolic DNA-sensing system in innate immunity: a current view. Current Opinin in Immunology, 2009, 21(1): 17-22.
[21] MANSUR D S, SMITH G L, FERGUSON B J. Intracellular sensing of viral DNA by the innate immune system. Microbes and Infection, 2014, 16(12): 1002-1012.
[22] RATHINAM V A, FITZGERALD K A. Innate immune sensing of DNA viruses. Virology, 2011, 411(2): 153-162.
[23] 邢雅玲, 郑洋, 王凯, 陈晓娟, 陈忠斌. 病原DNA识别及其诱导天然免疫调节机制研究进展. 生物化学与生物物理进展, 2011, 38(12): 1099-1105. XING Y L, ZHENG Y, WANG K, CHEN X J, CHEN Z B. The cellular recognition of pathogenic DNA and the related regulation of innate immunity. Progress in Biochemistry and Biophysics, 2011, 38(12): 1099-1105. (in Chinese)
[24] EBIHARA N, CHEN L, TOKURA T, USHIO H, IWATSU M, MURAKAMI A. Distinct functions between toll-like receptors 3 and 9 in retinal pigment epithelial cells. Ophthalmic Research, 2007, 39(3): 155-163.
[25] CHOI M K, WANG Z C, BAN T, YANAI H, LU Y, KOSHIBA R, NAKAIMA Y, HANGAI S, SAVITSKY D, NAKASATO M, NEGISHI H, TAKEUCHI O, HONDA K, AKIRA S, TAMURA T, TANIGUCHI T. A selective contribution of the RIG-I-like receptor pathway to type I interferon responses activated by cytosolic DNA. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(42): 17870-17875.
[26] CHIU Y H, MACMILLAN J B, CHEN Z J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell, 2009, 138(3): 576-591.
[27] VEERANKI S, CHOUBEY D. Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization. Molecular Immunology, 2012, 49(4): 567-571.
[28] ZHANG Z Q, YUAN B, BAO M S, LU N, KIM T, LIU Y J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature Immunology, 2011, 12(10): 959-965.
[29] KIM T, PAZHOOR S, BAO M S, ZHANG Z Q, HANABUCHI S, FACCHINETTI V, BOVER L, PLUMAS J, CHAPEROT L, QIN J, LIU Y J. Aspartate-glutamate-alanine-histidine box motif (DEAH)/ RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(34): 15181-15186.
[30] ZHANG X, BRANN T W, ZHOU M, YANG J, OGUARIRI R M, LIDIE K B, IMAMICHI H, HUANG D W, LEMPICKI R A, BASELER M W, VEENSTRA T D, YOUNG H A, LANE H C, IMAMICHI T. Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. Journal of Immunology, 2011, 186(8): 4541-4545.
[31] ABLASSER A, BAUERNFEIND F, HARTMANN G, LATZ E, FITZGERALD K A, HORNUNG V. RIG-I dependent sensing of poly (dA-dT) through the induction of an RNA polymerase III transcribed RNA intermediate. Nature Immunology, 2009, 10(10): 1065-1072.
[32] MELCHJORSEN J, RINTAHAKA J, SØBY S, HORAN K A, POLTAJAINEN A, ØSTERGAARD L, PALUDAN S R, MATIKAINEN S. Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS- dependent and MDA5/MAVS/RNA polymerase III-independent pathways. Journal of Virology, 2010, 84(21): 11350-11358.
[33] ARUNACHALAM B, PHAN U T, GEUZE H J, CRESSWELL P. Enzymatic reduction of disulfide bonds in lysosomes: Characterization of a gamma interferon inducible lysosomal thiol reductase (GILT). Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(2): 745-750.
[34] DE ZOYSA M, LEE J. Molecular cloning and expression analysis of interferon-γ inducible lysosomal thiol reductase (GILT)-like cDNA from disk abalone (Haliotis discus discus). Journal of Invertebrate Pathology, 2007, 96(3): 221-229.
[35] HUANG W S, DUAN L P, HUANG B, ZHOU L H, LIANG Y, TU CL, ZHANG F F, NIE P, WANG T. Identification of three IFN-gamma inducible lysosomal thiol reductase (GILT)-like genes in mud crab Scylla paramamosain with distinct gene organizations and patterns of expression. Gene, 2015, 570: 78-88.
[36] KONGTON K, PHONGDARA A, SRITHAWORN M T, WANNA W. Molecular cloning and expression analysis of the interferon-γinducible lysosomal thiol reductase gene from the shrimp Penaeus monodon. Molecular Biology Reports, 2011, 38: 3463-3470.
[37] KONGTON K, MCCALL K, PHONGDARA A. Identification of gamma-interferon-inducible lysosomal thiol reductase (GILT) homologues in the fruit fly Drosophila melanogaster. Developmental and Comparative Immunology, 2014, 44: 389-396.
[38] HASTINGS K T, CRESSWELL P. Disulfide reduction in the endocytic pathway: immunological functions of gamma-interferoninducible lysosomal thiol reductase. Antioxidants and Redox Signaling, 2011, 15(3): 657-668.
[39] PROKOPENKO O, MIROCHNITCHENKO O. Ischemia-reperfusioninducible protein modulates cell sensitivity to anticancer drugs by regulating activity of efflux transporter. American Journal of Physiology-Cell Physiology, 2009, 296: C1086-C1097.
[40] SHENG Y, SARIDAKIS V, SARKARI F, DUAN S, WU T, ARROWSMITH C H, FRAPPIER L. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nature Structral and Molecular Biology, 2006, 13(3): 285-291.
[41] HOLOWATY M N, FRAPPIER L. HAUSP/USP7 as an Epstein-Barr virus target. Biochemical Society Transactions, 2004, 32(5): 731-732.
(责任编辑 岳梅)
Digital Gene Expression Analysis of Silkworm Infected by Bombyx mori Bidensovirus Zhenjiang Strain
GAO Kun, SHANG Meng-ke, QIAN He-ying, QIN Guang-xing, GUO Xi-jie
(College of Biotechnology, Jiangsu University of Science and Technology/Sericultural Research Institute, Chinese Academy of Agricultural Sciences, Zhenjiang 212018, Jiangsu)
【Objective】 The objective of this study is to screen differentially expressed genes in the Bombyx mori larvae infected with BmBDV-ZJ (B. mori bidensovirus Zhenjiang strain) and identify regulatory genes related to the virus infection and the host response so as to provide important clues for better understanding of the mechanism of B. mori resistance against BmBDV-ZJ infection. 【Method】 The differential gene expression profiles in JS B. mori larvae after oral infection with BmBDV-ZJ were constructed using Illumina Genome Analyzer platform. In order to exclude the effects of individual differences, 10 larvae were dissected and pooled as one sample for digital gene expression (DGE) analysis, respectively. The differential expression detection of genes across samples was performed using a rigorous algorithm method. False discovery rate (FDR) was used to determine the P value threshold in multiple tests and analyses. The significance of the gene expression difference was obtained through a FDR≤0.001 and the absolute value of log2ratio≥1. The gene ontology (GO) classification system was used to determine the possible functions of all differentially expressed genes. P value was calculated by GO (http://www.geneontology. org/) and corrected by Bonferoni. A corrected P value≤ 0.05 was selected as a threshold for significant enrichment of the gene sets. WEGO (web gene ontology annotation plot) software was used for visualizing, comparing and plotting GO annotation results. Pathway enrichment analysis was conducted to further identify the significantly enriched metabolic pathways or signal transduction pathways by using the KEGG database. Pathways with a Q value≤0.05 were designated as significantly enriched pathways in DGEs. Then some of the differentially expressed genes were verified by quantitative real-time PCR (qRT-PCR). 【Result】 Totally, 4 850 663 and 4 875 307 raw tags were generated in the control and BmBDV-ZJ infected DGE (digital gene expression) libraries, respectively. There were 4 757 934 and 4 788 406 clean tags corresponding to 62 436 and 63 680 distinct clean tags were filtered from the raw tags. The distribution of the total and distinct tags over the different tag abundance categories showed highly similar patterns in each DGE library. The sequencing depths reached approximately 3.5 and 3.7 million in the two DGE libraries, respectively, which satisfied the requirement for the experiment. So the two DGE libraries were reliable. The tag sequences of the two DGE libraries were mapped to the reference database of B. mori. In the control and BmBDV-ZJ-infected DGE library, 36.39% and 45.30% of the clean tags were mapped to a gene in the reference database, 50.02% and 43.34% of the clean tags could be mapped to genome of B. mori, while 13.59% and 12.35% of the clean tags were unknown tags. A total of 447 differentially expressed genes were detected, of which 306 were upregulated and 141 were downregulated. There were 218, 147, 179 differentially expressed genes have GO categories according to molecular function, cellular component and biological process, respectively. KEGG (http://www.genome.jp/kegg) ontology assignments were used to classify the functional annotations of the identified genes. Among the differentially expressed genes, 330 were mapped to 151 pathways in the KEGG database. Nineteen terms was significantly enriched (Q value≤0.05) and the cytosolic DNA-sensing pathway was significantly enriched. Moreover, 24 differentially expressed genes were verified using qRT-PCR, showing that 20 genes were concordant in the expression with DGE. Among the 9 differentially expressed genes related to cytosolic DNA-sensing pathway, BGIBMGA009408-TA, BGIBMGA004913-TA, BGIBMGA011753-TA, which were the DNA-directed RNA polymerase III genes in B. mori, were all up-regulated in the BmBDV-ZJ infected B. mori with 4.3, 2.3, 3.4-fold change, respectively. 【Conclusion】 The results of this study may serve as a basis for future research not only on the molecular mechanism of BmBDV-ZJ invasion but also on the mechanism of B. mori resistance against BmBDV-ZJ infection.
Bombyx mori; digital gene expression; Bombyx mori bidensovirus; quantitative real-time PCR
2016-04-08;接受日期:2016-06-02
国家自然科学基金(31402141)、江苏省自然科学基金(BK20140508)
联系方式:高坤,E-mail:gkunjn2002@126.com。通信作者郭锡杰,Tel:0511-84401328;E-mail:guoxijie@126.com
