Identification of Sphaeroma terebrans via morphology and the mitochondrial cytochrome c oxidase subunit I(COI) gene
2016-11-15XiuFengLIChongHANCaiRongZHONGJunQiuXUJianRongHUANG
Xiu-Feng LI, Chong HAN, Cai-Rong ZHONG, Jun-Qiu XU, Jian-Rong HUANG,*
1School of Life Sciences, Sun Yat-Sen University, Guangzhou 510275, China
2Management Bureau of Dongzhaigang Mangrove Natural Reserve, Haikou 571129, China
Identification of Sphaeroma terebrans via morphology and the mitochondrial cytochrome c oxidase subunit I(COI) gene
Xiu-Feng LI1, Chong HAN1, Cai-Rong ZHONG2, Jun-Qiu XU1, Jian-Rong HUANG1,*
1School of Life Sciences, Sun Yat-Sen University, Guangzhou 510275, China
2Management Bureau of Dongzhaigang Mangrove Natural Reserve, Haikou 571129, China
Sphaeroma terebrans, a wood-boring isopoda, is distributed worldwide in tropical and subtropical mangroves. The taxonomy of S. terebrans is usually based on morphological characteristics, with its molecular identification still poorly understood. The number of teeth on the uropodal exopod and the length of the propodus of the seventh pereopod are considered as the major morphological characteristics in S. terebrans, which can cause difficulty in regards to accurate identification. In this study, we identified S. terebrans via molecular and morphological data. Furthermore, the validity of the mitochondrial cytochrome c oxidase subunit I (COI) gene as a DNA barcode for the identification of genus Sphaeroma, including species S. terebrans, S. retrolaeve, and S. serratum, was examined. The mitochondrial COI gene sequences of all specimens were sequenced and analysed. The interspecific Kimura 2-parameter distances were higher than intraspecific distances and no intraspecificinterspecific distance overlaps were observed. In addition, genetic distance and nucleotide diversity (π)exhibited no differences within S. terebrans. Our results revealed that the mitochondrial COI gene can serve as a valid DNA barcode for the identification of S. terebrans. Furthermore, the number of teeth on the uropodal exopod and the length of the propodus of the seventh pereopod were found to be unreliable taxonomic characteristics for S. terebrans.
Sphaeroma terebrans; DNA barcode;COI gene; Molecular identification
lNTRODUCTlON
Mangroves are biologically and globally important ecosystems(Giri et al., 2011). Their aerial roots provide an important substrate in which many species of animals live and reproduce(Nagelkerken et al., 2008). Sphaeroma terebrans, a woodboring isopoda, is found worldwide in tropical and subtropical mangroves (Estevez, 1978), where it preferentially burrows into the aerial roots for shelter and reproductive habitat (Harrison & Holdich, 1984; John, 1970). In recent years, substantial S. terebrans outbreaks have seriously affected mangrove stands in China, especially in Hainan island (Fan et al., 2014).1
The effects of S. terebrans on mangroves have been studied by many researchers (Estevez & Simon, 1975; Estevez, 1978;Jones & Icely 1981; Kensley & Schotte, 1999; Perry, 1988;Rehm & Humm, 1973); however, the taxonomic standards of S. terebrans remain poorly understood. Due to some minor morphological differences, including the number and arrangement of the tubercles on the pereonite, the structure of the pereopod, and the presence of tubercles furnished with bristle-like hairs on the abdomen, S. terebrans was previously named as S. vastator (Bate, 1866) and S. destructor(Richardson, 1897). Based on morphological identification,Estevez & Simon (1975) concluded that S. vastator and S. destructor were synonyms of S. terebrans.
The classic use of morphological characteristics for species delimitation can result in under- or over-estimation of biodiversity due to factors such as phenotypic plasticity(Knowlton, 1993). DNA barcode, which can supplement taxonomic datasets in the process of species delimitation(Schindel & Miller, 2005), is a practical tool that can be used for the identification of various species within a known taxonomic framework and for linking different biological life stages of the same species (Feng et al., 2011; Puillandre et al., 2009;Schindel & Miller, 2005). The mitochondrial COI gene has been proposed as a universal barcode, and has been successfully applied in the identification of Portunidae, fish, bivalve molluscs,and hoverflies (Blair et al., 2006; Hebert et al., 2003; Ma et al.,2012; Persis et al., 2009; Ståhls et al., 2009). The COI gene sequences of S. terebrans have been analysed in America and Africa (Baratti et al., 2005, 2011), with results suggesting that cosmopolitan S. terebrans is comprised of more than one species. Therefore, its taxonomic status needs to be revaluated.
The aim of the present study was to provide a reliable and valid way to delimit S. terebrans. In this study, the validity of the mitochondrial COI gene as a DNA barcode marker for the identification of three species of Sphaeroma, namely, S. terebrans, S. retrolaeve, and S. serratum, was examined. To detect if there was any cryptic species in S. terebrans, the COI gene sequences of individuals with morphological differences were analysed. Our study should be useful in the identification of the genus Sphaeroma and for further research on S. terebrans.
MATERlALS AND METHODS
Sampling and scoring of morphological characteristics
The S. terebrans specimens were collected from three localities in China (Figure 1). Prior to DNA extraction, all specimens were examined under an anatomical lens and assigned to groups according to comparison with previous morphological descriptions (Harrison & Holdich, 1984). The morphological differences of S. terebrans were then photographed by a TM3030Plus tabletop microscope. The S. terebrans individuals were sorted according to the following morphological characteristics: the number and arrangement of tubercles on the pereonite, number of teeth on the uropodal exopod, shape of the pleotelson, setae distribution, and length of the second and seventh pereopods. These are considered to be major characteristics for the diagnosis of S. terebrans within Sphaeroma (Harrison & Holdich, 1984). The S. retrolaeve specimens were collected from Hainan and Beihai mangroves. All samples were preserved in 95% alcohol.

Figure 1 Sample collection sites of S. terebransHK: Haikou, Hainan, WC: Wenchang, Hainan, BH: Beihai, Guangxi,ZJ: Zhanjiang, Guangdong
DNA extraction, PCR amplification, and sequencing
The genomic DNA of S. terebrans and S. retrolaeve were obtained from the pereopods. DNA extractions were performed using a TaKaRa MiniBEST Universal Genomic DNA Extraction Kit Ver.5.0 following the manufacturer's protocols. The primers mtd10 5'-TTGATTTTTTGGTCATCCAGAAGT-3' (Roehrdan. 1993) and Florence 5'-CCTAAAAAATGTTGAGGGAA-3' were used for amplification of the mitochondrial COI gene (Baratti et al., 2005). We followed PCR protocols as per Baratti et al.(2005). The PCR products were electrophoresed using 1% agarose gel and sequenced by Shanghai Majorbio Bio-Pharm Technology Co., Ltd.
Data analysis
All sequences were aligned using ClustalW (Thompson et al.,1997). Interspecific and intraspecific sequence divergences were calculated using the Kimura 2-parameter (K2P) model with the pairwise deletion option in MEGA 5.0 (Kimura, 1980). Haplotypes were identified and analysed using DNA SP version 4.1 (Librado & Rozas, 2009). Nucleotide diversity (π) and haplotype diversity(h) were calculated according to Nei (1987)using DNA SP version 4.1 (Rozas & Rozas, 1999). Based on the K2P model, neighbor joining (NJ) and maximum likelihood(ML) trees were constructed using MEGA 5.0 (Kimura, 1980;Tamura et al., 2011), with the Cymodoce fuscina voucher from the NCBI (GenBank Accession No. KJ410468) used as an outgroup. Node supports for the two approaches (NJ and ML)were inferred with bootstrap analysis (1 000 replicates).
RESULTS
Prior to DNA extraction, we assigned S. terebrans specimens into A1-A5 and B morphotypes (Figure 2). The number of teeth on the uropodal exopod and the length of the propodus of the seventh pereopod varied within S. terebrans, which were assigned into seven groups (Table 1).
Partially aligned COI sequences 498 bp in length were obtained from 70 S. terebrans individuals and 10 S. retrolaeve individuals. The COI sequences of S. serratum and C. fuscina voucher were downloaded from the NCBI. Details of these sequences are shown in Table 2. There were fifteen haplotypes for S. terebrans and two for S. retrolaeve. Haplotype sequences were deposited in the NCBI under accession numbers KU558703-KU558719.
All haplotype sequences were aligned and edited, and no insertion or deletion sites were found in any of the sequences. The intraspecific distances in S. terebrans ranged from 0.001 to 0.013 (Table 3). The maximum interspecific distance (1.394)was between S. serratum and C. fuscina voucher, while the minimum interspecific distance (0.24) was between S. serratum and S. retrolaeve. No overlaps between interspecific and intraspecific distances were found, suggesting the existence of a distinct barcoding gap. The NJ phylogenetic tree is shown inFigure 3. Distinct clusters corresponding to species were found with high bootstrap support.
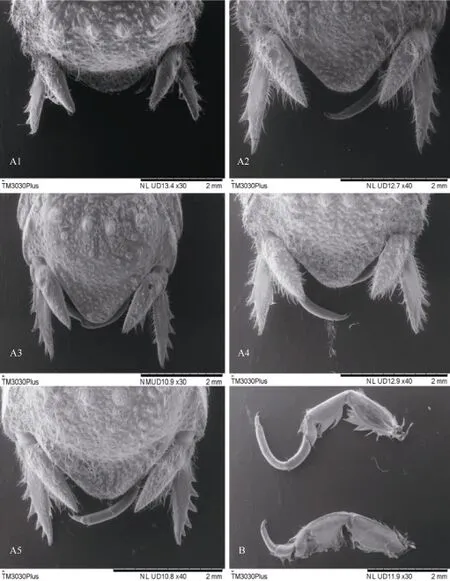
Figure 2 Diagnostic morphological characteristics of S. terebransA1-A5 are uropodal exopods with different numbers of teeth. B is the seventh pereopod with different propodus length.
The S. terebrans individuals were sorted into seven groups according to their morphological traits, and partial COI sequences of S. terebrans were aligned and compiled. The intraspecific distances ranged from 0.001 to 0.003 within the SS,SW, WW, WL, and LL groups. The intraspecific distance between PL and PS was 0.001 (Table 4). The mean haplotype diversity (h) was 0.555%, and ranged from 0.200% (PL group)to 0.866% (WW group) (Table 5). The highest nucleotide diversity (π) was found in the WW group (0.004), while the lowest was found in the PL group (0.000) (Table 5). Results suggested that there were no mitochondrial genetic variations within S. terebrans.
Phylogenetic analysis of genus Sphaeroma was performed using NJ and ML methods, which yielded similar results. The NJ tree revealed that the three species of Sphaeroma and one species of Cymodoce formed monophyletic clusters (Figure 3). The nearest relationship was observed between S. retrolaeve and S. serratum, while the most distant relationship was found between S. terebrans and C. fuscina voucher.
DlSCUSSlON
The rapid and effective identification of closely related woodborer Sphaeroma species is important for the research andrestoration of eroded mangroves. Identification of S. terebrans based on morphological characteristics alone is weak and, to some extent, ambiguous. Based on morphological characteristics,Some individuals of S. terebrans were previously named S. vastator (Bate, 1866) and S. destructor (Richardson, 1897). In this study, clear evidence was provided for the identification of S. terebrans individuals, which exhibited differences in morphological characteristics. The validity of using the mitochondrial COI gene sequence as a DNA barcode for the identification of genus Sphaeroma was examined, and included three Sphaeroma species, namely, S. terebrans, S. retrolaeve and S. serratum,with C. fuscina voucher (Sphaeromatidae) used as an outgroup. A distinct barcoding gap was found between the intraspecific and interspecific distances in each species. The NJ phylogenetic tree consisted of four distinct clusters, each containing individuals from one species only. These results indicate that the partial mitochondrial COI gene is an effective DNA barcode for the identification of the genus Sphaeroma.
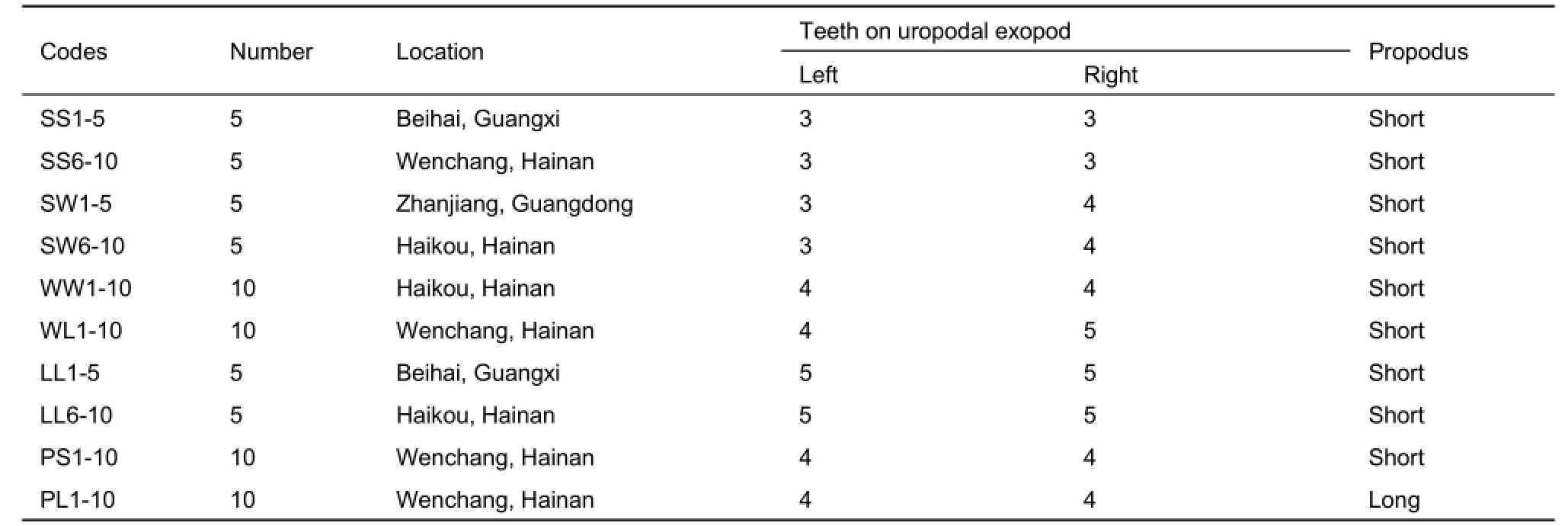
Table 1 List of sampling localities and morphological differences of S. terebrans
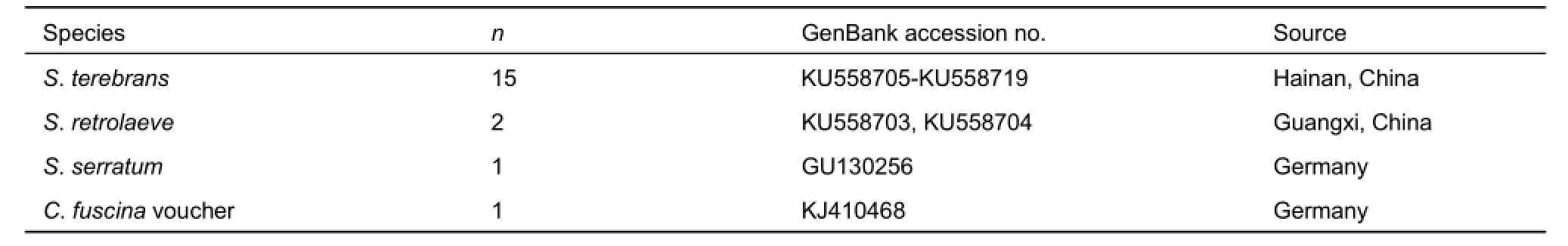
Table 2 List of COl sequences, GenBank accession numbers, and geographic sources of samples

Table 3 Pairwise genetic distances (Kimura 2-parameter) of three Sphaeroma species and one Cymodoce species based on COl sequences

Figure 3 Neighbor-joining phylogenetic tree of individual haplotypes of three species of Sphaeroma and one species of Cymodoce

Table 4 lntraspecific genetic distances (Kimura 2-parameter) of S. terebrans with morphological differences based on COl sequences

Table 5 Number of haplotypes, haplotype diversity(h), and nucleotide diversity(π) of different groups
Individuals of S. terebrans had different numbers of teeth on the uropodal exopod and different lengths of the propodus of the seventh pereopod. These individuals were sorted into seven groups, with each group containing 10 individuals. The genetic distance and nucleotide divergence showed no variation among the different groups. Therefore, these results revealed that the COI gene sequences of individuals with morphological differences were almost no difference. Although Harrison & Holdich (1984) determined that the propodus of the seventh pereopod of subadult males is relatively short, Our investigations showed that the length of the pereopodal propodus in S. terebrans was not necessarily linked with gender. Previous research concluded that cosmopolitan S. terebrans was comprised of more than one species (Baratti et al., 2011,2005), but morphological taxonomic details of S. terebrans were not mentioned. In our research, specimens in China were carefully checked according to morphological characteristics and were assigned into different groups, with molecular methods used for further identification. This combination of morphological taxonomy and molecular divergence should provide results of greater reliable.
CONCLUSlONS
In this study, the mitochondrial COI gene was found to be an effective DNA barcode for the identification of Sphaeroma species, whereas the number of teeth on the uropodal exopod and the length of the propodus of the seventh pereopod were found to be invalid taxonomic characteristics. The phylogenetic relationships determined in this study will be of use for studying the species composition of Sphaeroma in eroded mangroves in China and for establishing a good foundation for the restoration of mangrove ecosystems.
Baratti M, Goti E, Messana G. 2005. High level of genetic differentiation in the marine isopod Sphaeroma terebrans (Crustacea Isopoda Sphaeromatidae) as inferred by mitochondrial DNA analysis. Journal of Experimental Marine Biology and Ecology, 315(2): 225-234.
Baratti M, Filippelli M, Messana G. 2011. Complex genetic patterns in the mangrove wood-borer Sphaeroma terebrans Bate, 1866 (Isopoda,Crustacea, Sphaeromatidae) generated by shoreline topography and rafting dispersal. Journal of Experimental Marine Biology and Ecology, 398(1-2):73-82.
Bate CS. 1866. II.-Carcinological Gleanings.-No. II. Annals and Magazineof Natural History, 17(97): 24-31.
Blair D, Waycott M, Byrne L, Dunshea G, Smith-Keune C, Neil KM. 2006. Molecular discrimination of Perna (Mollusca: Bivalvia) species using the polymerase chain reaction and species-specific mitochondrial primers. Marine Biotechnology, 8(4): 380-385.
Estevez ED. 1978. Ecology of Sphaeroma terebrans Bate, a wood boring isopod, in a Florida mangrove forest. Ph. D. thesis, University of South Florida, Tampa, 1-154.
Estevez ED, Simon JL. 1975. Systematics and ecology of Sphaeroma(Crustacea: Isopoda) in the mangrove habitats of Florida. In: Proceedings of the International Symposium on Biology and Management of Mangroves. Gainesville: Institute of Food and Agricultural Sciences, University of Florida.
Fan HQ, Liu WA, Zhong CR, Ni X. 2014. Analytic study on the damages of wood-boring isopod, Sphaeroma, to China mangroves. Guangxi Sciences,21(2): 140-146, 152. (in Chinese)
Feng YW, Li Q, Kong LF, Zheng XD. 2011. DNA barcoding and phylogenetic analysis of Pectinidae (Mollusca: Bivalvia) based on mitochondrial COI and 16S rRNA genes. Molecular Biology Reports, 38(1):291-299.
Giri C, Ochieng E, Tieszen LL, Zhu Z, Singh A, Loveland T, Masek J, Duke N. 2011. Status and distribution of mangrove forests of the world using earth observation satellite data. Global Ecology and Biogeography, 20(1):154-159.
Harrison K, Holdich DM. 1984. Hemibranchiate sphaeromatids (Crustacea:Isopoda) from Queensland, Australia, with a world-wide review of the genera discussed. Zoological Journal of the Linnean Society, 81(4): 275-387.
Hebert PDN, Ratnasingham S, de Waard JR. 2003. Barcoding animal life:cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society B: Biological Sciences, 270(S1): S96-S99. John PA. 1970. Observations on the boring activity of Sphaeroma terebrans Spence Bate, a wood boring isopod. Zoologischer Anzeiger, 185(5-6): 379-387.
Jones DA, Icely JD. 1981. Excirolana bowmani, a new mangrove-boring isopod from Kenya (Isopoda, Cirolanidae). Crustaceana, 40(3): 266-271.
Kensley B, Schotte M. 1999. New records of isopods from the Indian River Lagoon, Florida (Crustacea: Peracarida). Proceedings of the Biological Society of Washington, 112(4): 695-713.
Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16(2): 111-120.
Knowlton N. 1993. Sibling species in the sea. Annual Review of Ecology and Systematics, 24(1): 189-216.
Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25(11): 1451-1452.
Ma HY, Ma CY, Ma LB. 2012. Molecular identification of genus Scylla(Decapoda: Portunidae) based on DNA barcoding and polymerase chain reaction. Biochemical Systematics and Ecology, 41: 41-47.
Nagelkerken I, Blaber SJM, Bouillon S, Green P, Haywood M, Kirton LG,Meynecke JO, Pawlik J, Penrose HM, Sasekumar A, Somerfield PJ. 2008. The habitat function of mangroves for terrestrial and marine fauna: a review. Aquatic Botany, 89(2): 155-185.
Nei M. 1987. Molecular Evolutionary Genetics. Columbia: Columbia University Press.
Perry DM. 1988. Effects of associated fauna on growth and productivity in the red mangrove. Ecology, 69(4): 1064-1075.
Persis M, Reddy ACS, Rao LM, Khedkar GD, Ravinder K, Nasruddin K. 2009. COI (cytochrome oxidase-I) sequence based studies of Carangid fishes from Kakinada coast, India. Molecular Biology Reports, 36(7): 1733-1740.
Puillandre N, Strong EE, Bouchet P, Boisselier MC, Couloux A, Samadi S. 2009. Identifying gastropod spawn from DNA barcodes: possible but not yet practicable. Molecular Ecology Resources, 9(5): 1311-1321.
Rehm A, Humm HJ. 1973. Sphaeroma terebrans: a threat to the mangroves of southwestern Florida. Science, 182(4108): 173-174.
Richardson H. 1897. Description of a new species of Sphaeroma. Proceedings of the Biological Society of Washington, 11: 105-107.
Roehrdanz RL. 1993. An improved primer for PCR amplification of mitochondrial DNA in a variety of insect species. Insect Molecular Biology,2(2): 89-91.
Rozas J, Rozas R. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics, 15(2): 174-175.
Schindel DE, Miller SE. 2005. DNA barcoding a useful tool for taxonomists. Nature, 435(7038): 17.
Ståhls G, Vujic A, Pérez-Bañon C, Radenkovic S, Rojo S, Petanidou T. 2009. COI barcodes for identification of Merodon hoverflies (Diptera,Syrphidae) of Lesvos Island, Greece. Molecular Ecology Resources, 9(6):1431-1438.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10): 2731-2739.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25(24): 4876-4882.
15 July 2016; Accepted: 08 September 2016
s: This project was funded by the GEF China Wetlands System Project, Science and Technology Foundation of Macao (045/2010/A) and Special Fund for Marine-Scientific Research in the Public Interest (201305021)
, E-mail: lsshjr@mail.sysu.edu.cn
10.13918/j.issn.2095-8137.2016.5.307
杂志排行
Zoological Research的其它文章
- Lamprey: a model for vertebrate evolutionary research
- What is the destiny of a threatened fish, Ptychobarbus chungtienensis, now that non-native weatherfishes have been introduced into Bita Lake, Shangri-La?
- Evolution and phylogenetic application of the MC1R gene in the Cobitoidea (Teleostei: Cypriniformes)
- A new species of the genus Triplophysa (Cypriniformes:Nemacheilidae), Triplophysa daochengensis, from Sichuan Province, China
- A new cave species of the Genus Triplophysa from Yunnan, China
- Identification of candidate piRNAs in the gonads of Paralichthys olivaceus (Japanese flounder)
